Abstract
Animals use their gustatory systems to evaluate the nutritious value, toxicity, sodium content, and acidity of food. Although characterization of molecular identities that receive taste chemicals is essential, molecular receptors underlying sour taste sensation remain unclear. Here, we show that two transient receptor potential (TRP) channel members, PKD1L3 and PKD2L1, are coexpressed in a subset of taste receptor cells in specific taste areas. Cells expressing these molecules are distinct from taste cells having receptors for bitter, sweet, or umami tastants. The PKD2L1 proteins are accumulated at the taste pore region, where taste chemicals are detected. PKD1L3 and PKD2L1 proteins can interact with each other, and coexpression of the PKD1L3 and PKD2L1 is necessary for their functional cell surface expression. Finally, PKD1L3 and PKD2L1 are activated by various acids when coexpressed in heterologous cells but not by other classes of tastants. These results suggest that PKD1L3 and PKD2L1 heteromers may function as sour taste receptors.
Keywords: chemical senses, polycystic kidney disease, gustation, ion channel, acid
Taste reception occurs at the apical tip of taste cells that form taste buds. Each taste bud has an onion-like shape and is composed of 50–100 taste cells that possess microvilli (1). There are four major taste areas in the oral region in which taste buds are concentrated: three taste areas on the tongue (circumvallate papilla, foliate papilla, and fungiform papilla) and a fourth taste area on the palate on the top surface of the mouth. In mammals, taste is generally classified into five distinct taste modalities: bitter, sweet, umami (the taste of some l-amino acids), salty, and sour (1). Much progress has been made in unraveling the molecular mechanisms of bitter, sweet, and umami taste in recent years (2–5). Bitter chemicals are detected by ≈30 T2R receptor family members. Sugars and sweeteners are detected by T1R2 and T1R3 heteromers, whereas umami tasting l-amino acids are detected by T1R1 and T1R3.
In contrast, the molecular mechanisms involved in sensing salty and sour taste are poorly understood and even confusing (6). Regarding sour taste transduction, several candidate receptors have been proposed. For example, acid-sensing ion channel (ASIC)2 is proposed to function as a sour receptor in the rat (7). However, it is not expressed in mouse taste cells and not required for acid sensation (8). HCN1 and HCN4, members of hyperpolarization-activated cyclic nucleotide-gated (HCN) channels, also are putative sour receptor channels (9). However, calcium imaging experiments using taste bud slices did not support this possibility, because Cs+, an inhibitor of HCN channels, did not block Ca2+ response of taste cells to acid stimuli (10). In addition, the proteins are localized on basolateral membranes of taste cells. Members of two pore domain K+ channels also are proposed to have some roles in acid transduction (11, 12). However, their expression levels seem to be low, and the proteins are mainly distributed on basolateral membranes of taste cells. None of these putative sour receptor proteins are shown to be localized at the taste pore region, where tastants are likely to be detected.
Transient receptor potential (TRP) ion channels are implicated as necessary signaling components in various sensory systems of diverse animal species ranging from mammals and fish to fruit flies and nematodes, including vision, smell, pheromone, hearing, touch, osmolarity, thermosensation, and sweet, bitter, and umami taste (13, 14). Some TRP channels, such as TRPV1, function directly as receptors for stimuli (high temperature and capsaicin) by themselves, whereas other TRP channels, such as TRP-melastatin 5 (TRPM5), are downstream effectors of G protein-coupled sensory receptors (13–15).
Among TRP channel families, members of the polycystic kidney disease (PKD) family, also called TRPP or polycystins, have unique properties (16, 17). Their founding members, PKD1 and PKD2, were identified as autosomal dominant polycystic kidney disease genes. PKD1 and its related proteins, PKD1L1–PKD1L3 and PKDREJ, are large proteins with a very long N-terminal extracellular domain, followed by 11 transmembrane domains that include a six-transmembrane TRP-like channel domain at the C terminus. Because of their unique properties, PKD1 and its related proteins are not explicitly included in the TRP channel family (18). In contrast, PKD2 and its related proteins, PKD2L1 and PKD2L2, have six transmembrane domains, similar to other TRP members, and can function as nonselective cation channels (19). PKD1 and PKD2 heteromer association is shown to be required for formation of a functional receptor/channel (20). PKD1 and PKD2 are thought to sense mechanical flow, osmolarity, and unknown extracellular ligand(s), although the biological functions of the PKD-related molecules are poorly understood. In Caenorhabditis elegans, a PKD1 homolog, Lov-1, and a PKD2 homolog are coexpressed by male-specific sensory neurons that are localized at the chemosensory cilia and are required for male mating behavior, suggesting they also form complexes and function as sensory receptors (21).
Identification and characterization of taste receptors is fundamentally important to understand our taste sensation. We hypothesized that additional TRP family members other than TRPM5 might be specifically expressed in taste cells and are involved in taste transduction. We show that two PKD-like TRP ion channel members, PKD1L3 and PKD2L1, may function as sour taste receptors.
Results
Expression of PKD1L3 and PKD2L1 mRNA in Taste Cells.
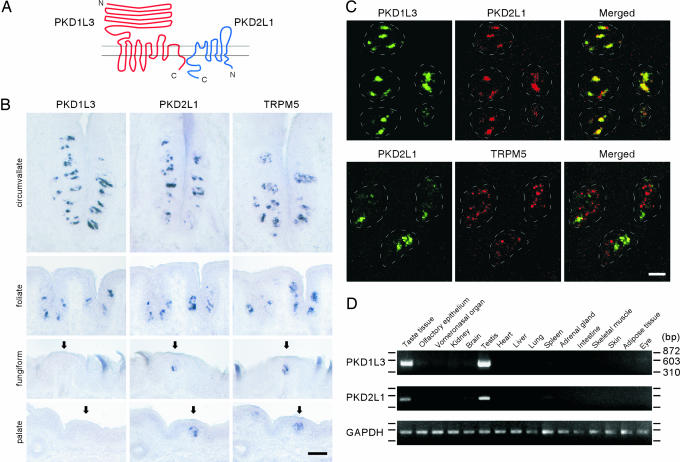
The mouse genome contains at least 33 genes encoding TRP channel-like proteins. To identify TRP ion channel members functioning in taste transduction, we performed in situ hybridization using probes for all 33 genes (22) on sections of mouse circumvallate papillae from the back of the tongue in mouse taste tissue. Consistent with previous reports, probes for TRPM5, a TRP channel member required for normal bitter, sweet, and umami sensation, labeled ≈40% of taste cells (15, 23, 24). In addition, we found that the probes for PKD1L3 and PKD2L1 hybridized to ≈20% of circumvallate taste cells (Fig. 1). A similar expression pattern was observed with rat circumvallate papillae (data not shown). Other TRP members did not show specific and robust expression in circumvallate taste cells in our analysis.
Fig. 1.
Expression of PKD1L3 and PKD2L1 mRNAs in mouse taste tissues. (A) Schematic diagrams of predicted membrane topology of PKD1L3 and PKD2L1. (B) Expression of PKD1L3, PKD2L1, and TRPM5 mRNA in different taste areas. Arrows indicate taste bud regions in the fungiform papillae and the palate. (Scale bar, 50 μm.) (C) PKD1L3 and PKD2L1 are coexpressed in taste cells but not colocalized with TRPM5. Double-label fluorescent in situ hybridization was used to examine PKD1L3, PKD2L1, and TRPM5 expression. The dotted white lines indicate the approximate area of taste buds. (Scale bar, 20 μm.) (D) PKD1L3 and PKD2L1 are specifically expressed in taste tissues. RT-PCR was performed with total RNA from 16 mouse tissues. PCR products were run on agarose gels with markers.
To examine the expression of PKD1L3 and PKD2L1 in different taste areas, we performed in situ hybridization with sections from circumvallate, foliate, and fungiform papillae as well as the palate. PKD2L1 expression was observed in a subset of taste cells in all four different taste areas, as well as TRPM5 (Fig. 1B). In contrast, PKD1L3 expression was observed in circumvallate and foliate papillae but not in fungiform papillae or the palate (Fig. 1B).
To investigate the correlation of TRPM5-, PKD1L3-, and PKD2L1-expressing cells in taste buds, we next performed double-labeled fluorescent in situ hybridization. In circumvallate and foliate papillae, almost all of the PKD1L3-positive cells were also PKD2L1-positive, indicating that these two molecules are coexpressed in the same cells. In contrast, TRPM5 signals did not appear to colocalize with either PKD2L1 or PKD1L3 signals (Fig. 1C and data not shown), demonstrating that PKD1L3- and PKD2L1-expressing cells are likely to be segregated from TRPM5-expressing, bitter-, sweet-, and umami-receptor-expressing taste cells. In fungiform papillae and the palate, PKD2L1-positive cells were PKD1L3-negative, confirming the absence of PKD1L3 expression in these areas (data not shown).
To examine the mRNA expression of PKD1L3 and PKD2L1 in different tissues, we performed RT-PCR with mRNA from 16 different mouse tissues. We found that both PKD1L3 and PKD2L1 are abundantly expressed in taste tissues and testis, whereas they are absent or only faintly expressed in all other tissues examined (Fig. 1D). In contrast, control RT-PCR for GAPDH showed a comparable amount of products from all tissues (Fig. 1D). When we increased the number of the PCR cycles up to 40 cycles, we observed PCR products in many tissues (data not shown), consistent with the previous reports showing low expression levels in various tissues (25, 26). It is worth noting that mRNAs encoding other taste receptors, T1Rs and T2Rs, also are found to be expressed in the testis (27–29). The significance of expression of the taste receptors in the testis is currently unknown.
Protein Localization of PKD2L1 in Taste Cells.
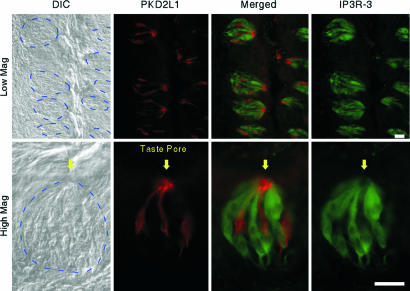
Taste reception occurs in the taste pore that contains an accumulation of apical tips of cell dendrites topped with microvilli. If PKD1L3 and PKD2L1 function as bona fide taste receptors, these proteins are likely to localize at the apical tip of the taste cell dendrite. To address this possibility, we generated antibodies against PKD2L1 and analyzed its cellular localization within taste cells. Immunostaining with rat and mouse circumvallate and foliate taste tissues demonstrated that PKD2L1 specifically localized to the taste pore area at the apical end of a subset of taste cells, with weaker labeling throughout the positive cells (Fig. 2 and data not shown). Similar localization was observed in mouse fungiform papillae and the palate (data not shown). Optical sectioning using confocal microscopy confirmed this observation (Movie 1, which is published as supporting information on the PNAS web site). Preincubation of the antibody with peptide antigen (10 ng/ml) abolished the taste cell staining, confirming the specificity of the antibody (data not shown). An inositol 1,4,5-trisphosphate receptor 3 (IP3R-3) antibody marks phospholipase Cβ2- and TRPM5-expressing bitter-, sweet-, and umami-sensing cells (15, 30, 31). Double staining using antibodies against PKD2L1 and IP3R-3 revealed that distinct sets of taste cells express PKD2L1 or IP3R-3 (Fig. 2 and Movie 1), consistent with mRNA expression patterns. These results, together with interactions between PKD1L3 and PKD2L1 (see Interaction of PKD1L3 and PKD2L1 and Their Cell Surface Expression), are consistent with a role of PKD1L3 and PKD2L1 in taste reception.
Fig. 2.
Localization of PKD2L1 protein at the taste pore. Sections of rat circumvallate taste cells were incubated with anti-PKD2L1 and anti-IP3R-3 antibodies. The dotted blue lines on the differential interference contrast (DIC) images indicate the approximate area of taste buds. Mag, magnification. (Scale bar, 20 μm.)
Interaction of PKD1L3 and PKD2L1 and Their Cell Surface Expression.
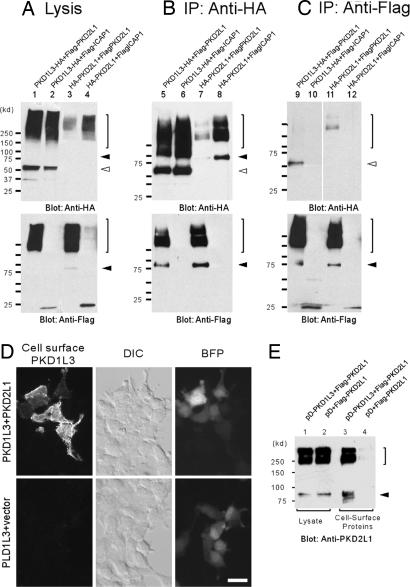
Because PKD1L3 and PKD2L1 are expressed in the same cells, we hypothesized that PKD1L3 and PKD2L1 might also interact with each other to form functional receptors. This hypothesis was assessed by coimmunoprecipitation assays. Mammalian expression vectors encoding the HA-tagged C-terminal half of PKD1L3 protein, including TM6–TM11 and Flag-tagged PKD2L1, were transfected in 293T cells. After the cell extracts were precipitated with anti-HA antibodies, proteins were eluted and Western blotting analysis was performed. Flag-PKD2L1 proteins were detected as ≈90 kDa or higher molecular-mass bands after precipitation of HA-PKD1L3 (Fig. 3B, lane 5). Many multitransmembrane proteins, including ion channels and G protein-coupled receptors, are known to show such high molecular-mass bands, probably caused by oligomer formation in the sample buffer. When the entire mature protein of PKD1L3 with a HA tag was coexpressed with Flag-tagged PKD2L1 and was precipitated, PKD2L1 also was copurified, indicating an association between them (Fig. 6, which is published as supporting information on the PNAS web site). In contrast, we could not detect any coprecipitation when Flag-tagged integrin cytoplasmic domain-associated protein 1, a negative control (32), was used (Fig. 3B, lane 6). Likewise, when PKD2L1 was precipitated, PKD1L3 was specifically copurified (Fig. 3C, lane 9). Homomeric interaction of PKD2L1 proteins was also suggested in our coimmunoprecipitation assays (Figs. 3 B and C, lanes 7 and 11).
Fig. 3.
Association of PKD1L3 and PKD2L1 proteins is required for their cell surface expression. (A) Control Western blot analysis indicating expression of PKD1L3 and PKD2L1. (B) When PKD1L3-HA or HA-PKD2L1 was precipitated, Flag-PKD2L1 proteins were coprecipitated (lanes 5 and 7). Negative control integrin cytoplasmic domain-associated protein 1 (ICAP1) proteins were not precipitated. The filled arrowhead indicates the PKD2L1 monomer. The open arrowhead indicates the PKD1L3 monomer. The bracket indicates high-molecular-mass oligomers. (C) When Flag-PKD2L1 was precipitated, PKD1L3-HA or HA-PKD2L1 proteins were coprecipitated (lanes 9 and 11). (D) HEK 293T cells expressing HA-tagged PKD1L3 in the presence or absence of PKD2L1 were stained with anti-HA antibodies under nonpermeabilized conditions. (Scale bar, 20 μm.) (E) Cell surface expression of PKD2L1 in the presence or absence of PKD1L3 was measured by a cell-surface biotinylation assay.
To test whether interaction between PKD1L3 and PKD2L1 is required for proper trafficking, we first analyzed the cell surface expression of PKD1L3 with or without the presence of PKD2L1. PKD1L3 was tagged with HA at the N-terminal extracellular domain. When PKD1L3 alone was expressed in HEK 293T cells, little cell surface expression was observed, whereas control blue fluorescent protein signals confirmed effective transfection (Fig. 3D). In addition, PKD1L3 signals were observed when the cells were permeabilized and stained (data not shown). In contrast, when PKD1L3 was expressed with PKD2L1, robust cell surface expression of PKD1L3 was observed (Fig. 3D).
We next assessed PKD2L1 cell surface expression with or without PKD1L3 by cell surface biotinylation assay. Intact HEK 293T cells expressing N-terminal Flag-tagged PKD2L1 were labeled with a membrane-impermeable biotinylation reagent. After solubilization, the biotinylated proteins were purified by immobilized streptavidin, and the PKD2L1 proteins were revealed on a Western blot with either anti-PKD2L1 or anti-Flag antibodies. When PKD2L1 alone was expressed in the cells, only faint signals were observed (Fig. 3E, lane 4 and data not shown), consistent with a previous report (26). In contrast, when PKD2L1 was coexpressed with PKD1L3, strong signals were detected, indicating that cell-surface PKD2L1 proteins were dramatically increased (Fig. 3E, lane 3 and data not shown). These observations suggested that interaction between PKD1L3 and PKD2L1 was necessary for their cell surface expression.
PKD1L3 and PKD2L1 Function as Sour Taste Receptors.
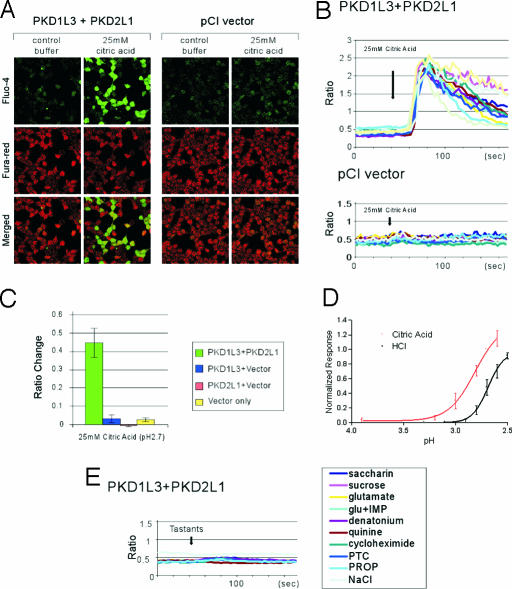
To examine whether PKD1L3 and PKD2L1 function as taste receptors, we first performed calcium imaging experiments using HEK 293T cells transiently expressing PKD1L3 and/or PKD2L1. Cells were loaded with calcium-sensitive dyes Fluo-4 and Fura red and then stimulated with various taste chemicals, including acids and NaCl. When the calcium concentration inside the cells increases upon stimulation with ligands, Fluo-4 signals increase and Fura red signals decrease, allowing us to perform ratiometric measurement of changes in intracellular calcium concentration (33). We found that cells expressing both PKD1L3 and PKD2L1 responded specifically to solutions containing acids, including citric acid, HCl, and malic acid (Fig. 4 and data not shown). In cells expressing PKD1L3 or PKD2L1 alone or not expressing either, acid solutions had little effect in calcium responses (Figs. 4 A–C). Although ASICs are endogenously expressed in the HEK 293 cells (34), the sour tastants we used did not cause significant activation in control cells. However, we occasionally observed a fraction of cells exhibiting nonspecific calcium response with a short duration (<12 s), probably responding to mechanical flow (data not shown). A pH–response curve revealed an EC50 of pH 2.8 for calcium response by citric acid (Fig. 4D). A dose–response curve using HCl showed that HCl was less potent than that of citric acid at the same pH (Fig. 4D), consistent with the notion that weak acids taste more sour than strong acids (35). PKD1L3 and PKD2L1 did not respond to NaCl, bitter chemicals, sucrose, a sweetener (saccharin), or umami compounds l-glutamate and inosine monophosphate (Fig. 4E). These experiments suggest that PKD1L3 and PKD2L1 form a functional receptor specifically activated by sour tastants. PKD1L3 and PKD2L1 functioning was not inhibited by the ASIC and Na+/H+ exchanger inhibitor amiloride or by the HCN and two-pore K+ channel inhibitor Cs+ (data not shown).
Fig. 4.
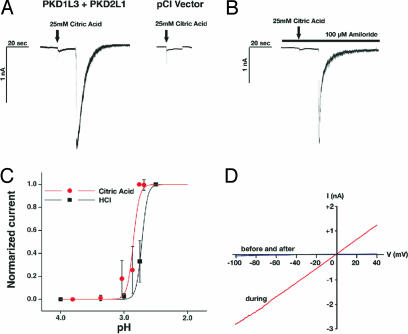
PKD1L3 and PKD2L1 are stimulated by sour tastants. (A) HEK 293T cells expressing PKD1L3 and PKD2L1 or control pCI vector were stimulated with control buffer or buffer containing 25 mM citric acid. (B) Kinetics of the Fluo-4/Fura red ratio changes for 10 representative cells. (C) PKD1L3 and PKD2L1 respond to citric acid. In contrast, cells expressing PKD1L3 or PKD2L1 alone, or neither, do not respond. Fluorescent ratio of the entire field (≈200 cells) before and after stimulation (±SEM) are shown (n = 3–5). (D) Citric acid and HCl dose responses for PKD1L3 and PKD2L1. Bath solutions containing 5, 10, 15, 20, 25, or 30 mM citric acid (corresponding to pH 3.9, 3.2, 3.0, 2.8, 2.7, or 2.7, respectively) were applied. In addition, HCl at 9, 10, 11, 12, 13, 14, or 15 mM (corresponding to pH 3.3, 3.1, 2.9, 2.8, 2.7, 2.6, or 2.5, respectively) was applied. Normalized fractions of responders (±SEM) are shown (n = 4). (E) PKD1L3 and PKD2L1 do not respond to sweet, bitter, umami, and salty chemicals. Averages of the kinetics of 12–31 representative cells for each ligand are shown. These cells responded to 25 mM citric acid (data not shown). glu+IMP, l-glutamate plus inosine monophosphate; PTC, phenylthiocarbamide; PROP, 6-n-propylthiouracil.
Finally, we performed patch-clamp recordings on transfected HEK 293T cells to characterize the electrophysiological properties of PKD1L3- and PKD2L1-mediated currents. In HEK 293T cells expressing PKD1L3 and PKD2L1, application of 25 mM citric acid caused small transient and sustained currents, possibly derived from ASICs endogenously expressed in the cells (34) followed by a robust current (115.2 ± 35.4 pA/pF, n = 14) with a rapid inactivation, whereas the transient and sustained currents were observed in cells transfected with the pCI vector as well (Fig. 5A). The robust current with rapid inactivation also was observed in the Ca2+-free condition (data not shown). The robust currents were not inhibited by 100 μM amiloride, which abolished the transient component of the small inward currents, suggesting that the transient currents are ASIC-mediated (Fig. 5B). We observed some delay in the activation of PKD1L3- and PKD2L1-mediated currents compared with amiloride-sensitive nonspecific currents (Fig. 5A). The mechanism of this delayed activation is currently unknown. A pH–response curve revealed an EC50 of pH 2.9 for activation by citric acid (Fig. 5C). A response curve using HCl showed that HCl appeared to be less potent than that of citric acid at the same pH (Fig. 5C), consistent with the calcium imaging analysis described above (Fig. 4D). The current–voltage relationship is approximately linear like one reported for PKD1- and PKD2-mediated currents (20) with a reversal potential of 1.6 ± 0.5 mV (n = 3), most likely involving a nonselective cation channel (Fig. 5D).
Fig. 5.
Electrophysiological properties of the PKD1L3 and PKD2L1 channel expressed in HEK 293T cells. (A) Representative whole-cell inward currents evoked by 25 mM citric acid (recorded at −60 mV) in cells transfected with PKD1L3 and PKD2L1 cDNAs or vector alone (pCI Vector). Arrows indicate a time point of acid application. (B) A representative whole-cell inward current evoked by 25 mM citric acid in the presence of 100 μM amiloride. (C) Extracellular pH-dependent curves for current activation. Bath solutions containing 5, 10, 15, 20, 25 or 30 mM citric acid (corresponding to pH 3.8, 3.4, 3.0, 2.9, 2.8, or 2.8, respectively) were applied. Currents at each pH were normalized to the currents evoked by application of 25 mM citric acid or HCl at pH 2.5 and averaged across cells (n = 14 for citric acid, n = 9 for HCl). Error bars indicate standard deviation. (D) Current–voltage relationship using a voltage ramp–pulse protocol (−100 to + 40 mV in 100 ms).
Discussion
How is the taste information coded in the taste buds? It is well demonstrated that different sets of taste cells are responsible for bitter, sweet, and umami sensation (15, 36). Our study showed that PKD1L3- and PKD2L1-expressing cells are segregated from bitter, sweet, and umami receptor-expressing taste cells, raising the possibility that a subset of cells may be “labeled” as sour-sensing cells. A specific fraction of taste cells (23–25%) are activated by citric acid with the calcium imaging method using taste bud slices (10). This finding corresponds well to the number of taste cells expressing PKD1L3 and PKD2L1 (≈20%). Our expression analysis of PKD1L3 showed that its expression may be absent in fungiform papillae and the palate, although expression of PKD2L1 was observed at all of the taste areas examined (Fig. 1B). This observation suggests that mechanisms of sour reception may be different in fungiform papillae and the palate.
How are PKD1L3 and PKD2L1 heteromers activated by sour stimuli? PKD1L3 has a large extracellular domain followed by 11 transmembrane domains, similar to PKD1, although their extracellular domains show little similarities. In contrast, PKD2L1 has six transmembrane domains, like most other TRP channel members (Fig. 1A). PKD1 does not seem to function as an ion-conducting channel but rather plays a critical role in sensing mechanical flow and other stimuli, whereas PKD2 is known to form the functional ion-conducting channel (19, 37). PKD2L1 shows 47% identity to PKD2 and has been shown to form functional calcium-permeable channels (38), whereas it is not known whether PKD1L3 alone can form a functional channel. By analogy to PKD1 and PKD2, PKD1L3 might function as a sour-sensing receptor and PKD2L1 might function as an ion-conducting channel. Although we showed that HEK 293T cells reacted to acid solutions only when both PKD1L3 and PKD2L1 were expressed, we cannot rule out the possibility that the PKD1L3 plus PKD2L1 reaction to acid application may be caused by an indirect downstream effect.
Sour sensation does not seem to be a simple measurement of pH in the solution. For example, at the same pH, weak acids such as citric acid or acetic acid taste more sour than HCl (35). Similarly, calcium imaging experiments using mouse taste tissue slices have shown that citric acid is a more potent sour ligand than HCl at the same pH (10). Accordingly, it was proposed that intracellular acidification in the taste bud is the proximate stimulus for sour taste (39). These reports suggest that sour receptors themselves and/or sour signal transduction components are affected by low intracellular pH. Our analysis showed that citric acid may be more potent than HCl at the same pH to activate PKD1L3 and PKD2L1, consistent with the psychophysical data. We observed a delay between application of the acid and channel openings (Fig. 5A). This could potentially be explained by a delayed effect of addition of external acid on the internal pH. Future inside-out and outside-out patch-clamp recordings will determine whether extracellular or intracellular protons are functioning as ligands for PKD1L3 and PKD2L1. Other possibilities include that intracellular acidification in taste cells could affect the function of PKD1L3 and PKD2L1 by modification, such as phosphorylation. Alternatively, heterologous cells may lack important components affecting sour transductions, such as carbonic anhydrase, H+/Na+ exchangers, and voltage-gated calcium channels (39).
What are the roles of PKD1L3 and PKD2L1 in taste sensation in vivo? Although our studies suggest roles for PKD1L3 and PKD2L1 in sour taste transduction, they do not exclude the possibility that other putative sour taste receptors may also play some roles in sour transduction. Previous behavioral studies have shown that mice exhibit moderate avoidance to 15 mM citric acid or 10 mM HCl and exhibit strong avoidance to 30 or 150 mM citric acid or 100 mM HCl (36, 40). These results are in good agreement with acid-mediated activation profiles of PKD1L3 and PKD2L1 in our current studies using heterologous cells (Figs. 4 and 5). However, it is worth noting that mice appear to prefer citric acid over water at concentrations below 1.0 mM (40), raising the possibility that PKD1L3 and PKD2L1 may mediate “bad” sour taste and there may be more than one neural mechanism transmitting sour sensation. Generation and analyses of PKD1L3 and PKD2L1 gene knockout animals will help define the roles of PKD1L3 and PKD2L1 in vivo.
Methods
In Situ Hybridization and RT-PCR.
Procedures of in situ hybridization were performed as described previously (41, 42). For RT-PCR, taste tissues were derived from tongue regions, including circumvallate and foliate papillae. Total RNA was extracted and reverse-transcribed into cDNA by using oligo dT primer. Approximately 500-bp coding regions encompassing multiple exons of PKD1L3 and PKD2L1 were amplified from each cDNA for 30, 35, or 40 cycles at an annealing temperature of 50°C. The volume of PCR products for PKD1L3 and PKD2L1 loaded on the agarose gel was approximately normalized by using the PCR products of GAPDH.
Immunohistochemistry.
Rabbits were immunized with peptide corresponding to residue 731–749 of PKD2L1 (Open Biosystems, Huntsville, AL). CD-1 or C57BL6 adult mice or Sprague–Dawley rats were used for immunostaining. Fresh frozen sections (16 μm thick) were fixed with 4% paraformaldehyde, permeabilized with ice-cold methanol, blocked with PBS containing 5% skim milk, incubated with anti-PKD2L1 antiserum (1/500 dilution) and anti-IP3R-3 antibody (1/1,000 dilution; BD Pharmingen, San Diego, CA) followed by incubation with Cy3-conjugated anti-rabbit IgG (Jackson Immunologicals, West Grove, PA) and Alexa Fluor 488-conjugated anti-mouse IgG (Invitrogen, Carlsbad, CA).
Immunoprecipitation and Cell Surface Biotinylation Assay.
Immunoprecipitations were performed essentially as described (41). Protease inhibitor mixture (Sigma, St. Louis, MO) was used. An HA tag was inserted at the C terminus of PKD1L3 (M1594–Y2151) in pCI (Promega, Madison, WI). Putative mature protein (R25-Y2151) from PKD1L3 was inserted into pDisplay to generate N-terminal HA-tagged proteins. A Flag tag was inserted at the N terminus of the ORF of PKD2L1 in pCI. The cell surface biotinylation assay was performed with a FluoReporter Cell-Surface Biotinylation kit (Invitrogen) according to the manufacture’s protocol. Cells were transfected, incubated for 20 h, washed, and resuspended at a concentration of 2.5 × 107 cells per ml in PBS. After incubating cells with a 0.5 μg/ml solution of biotin-XX SSE for 30 min on ice, cells were washed and lysed. The lysis was incubated with streptavidin, immobilized on Agarose CL-4B (Sigma) overnight at 4°C, and washed. The samples were then eluted and subjected to SDS/PAGE and Western blot analysis.
Calcium Imaging.
Expression vectors for PKD1L3 and PKD2L1 were constructed by subcloning coding regions into pDisplay (Invitrogen) and pCI, respectively. Cells were loaded with 4 μM Fluo-4 (Invitrogen) and 7 μM Fura red (Invitrogen) for 45 min at room temperature. For calcium imaging, cells were seeded on glass-bottom plates with poly-d-lysine-coated coverslips (MatTek) and plasmid DNA was transfected with Lipofectamine 2000 (Invitrogen) and incubated for 30–42 h before dye loading. Transfection efficiency was consistent (≈50%), as judged by blue fluorescent protein expression. We used a Leica (Wetzlar, Germany) confocal microscope (excitation, 488 nm; emission, 500–560 nm for Fluo-4 and 605–700 nm for Fura red). We used the live imaging mode of Leica confocal software for data acquisition. Data were collected at 3-s intervals. Cells were exposed to a constant flow of bath solution (Hanks’ buffer containing 10 mM Hepes; Invitrogen). Tastant solutions were applied to cells for ≈15 s by changing the bath solution with a peristaltic pump (Rabbit; Rainin Instrument, Oakland, CA). Data analyses were done with Microsoft (Redmond, WA) Excel and Prizm4. A detailed description of procedures, reagents, and data analysis may be found in Supporting Methods, which is published as supporting information on the PNAS web site.
Electrophysiology.
Transfected HEK 293T cells were incubated for 6–9 h and were seeded on glass coverslips. Whole-cell patch-clamp recordings were carried out at 30–36 h after transfection in voltage-clamp mode using an Axopatch 200B amplifier and pClamp 8.2 software (Axon instruments, Foster City, CA). Standard bath solution for whole-cell recordings contained 140 mM NaCl, 5 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 10 mM Hepes, and 10 mM glucose, pH 7.4 (adjusted with NaOH). Standard pipette solution contained 140 mM KCl (or 120 mM Cs aspartate, 10 mM CsCl, 1 mM MgCl2 for current–voltage analysis), 5 mM EGTA, and 10 mM Hepes, pH 7.4 (adjusted with KOH or CsOH). The dose–response curves were fit with the Hill equation, Fraction open = [H+]n/([H+]n + K0.5n), by using Origin software, where K0.5 is the proton concentration that causes half the channels to open. Experiments were carried out at room temperature (22–24°C).
Supplementary Material
Acknowledgments
We thank Kelvin Kwan and David Corey (both at Harvard Medical School, Boston, MA) for probes; Renzhi Zhan (Duke University Medical Center) for materials; Wenqin Luo and Larry Katz (both at Duke University Medical Center) for materials and valuable advice; Sid Simon, Dan Tracey, Natasha Thorn, Steve Bray, Ben Arenkiel, Steve Shea, Ian Davidson, and Rich Roberts for critical readings of the manuscript; and Qiuyi Chi for expert technical assistance. This work was supported by National Institutes of Health Grant DC05782 (to H.M.) and a fellowship from Japan Society for the Promotion of Science (to Y.I.).
Glossary
Abbreviations
- TRP
transient receptor potential
- TRPM5
TRP-melastatin 5
- IP3R-3
inositol 1,4,5-trisphosphate receptor 3
- ASIC
acid-sensing ion channel
- HCN
hyperpolarization-activated cyclic nucleotide-gated.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lindemann B. Physiol. Rev. 1996;76:718–766. doi: 10.1152/physrev.1996.76.3.719. [DOI] [PubMed] [Google Scholar]
- 2.Drayna D. Annu. Rev. Genomics Hum. Genet. 2005;6:217–235. doi: 10.1146/annurev.genom.6.080604.162340. [DOI] [PubMed] [Google Scholar]
- 3.Margolskee R. F. J. Biol. Chem. 2002;277:1–4. doi: 10.1074/jbc.R100054200. [DOI] [PubMed] [Google Scholar]
- 4.Montmayeur J. P., Matsunami H. Curr. Opin. Neurobiol. 2002;12:366–371. doi: 10.1016/s0959-4388(02)00345-8. [DOI] [PubMed] [Google Scholar]
- 5.Scott K. Curr. Opin. Neurobiol. 2004;14:423–427. doi: 10.1016/j.conb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Miyamoto T., Fujiyama R., Okada Y., Sato T. Prog. Neurobiol. 2000;62:135–157. doi: 10.1016/s0301-0082(99)00072-6. [DOI] [PubMed] [Google Scholar]
- 7.Ugawa S., Minami Y., Guo W., Saishin Y., Takatsuji K., Yamamoto T., Tohyama M., Shimada S. Nature. 1998;395:555–556. doi: 10.1038/26882. [DOI] [PubMed] [Google Scholar]
- 8.Richter T. A., Dvoryanchikov G. A., Roper S. D., Chaudhari N. J. Neurosci. 2004;24:4088–4091. doi: 10.1523/JNEUROSCI.0653-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevens D. R., Seifert R., Bufe B., Muller F., Kremmer E., Gauss R., Meyerhof W., Kaupp U. B., Lindemann B. Nature. 2001;413:631–635. doi: 10.1038/35098087. [DOI] [PubMed] [Google Scholar]
- 10.Richter T. A., Caicedo A., Roper S. D. J. Physiol. 2003;547:475–483. doi: 10.1113/jphysiol.2002.033811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin W., Burks C. A., Hansen D. R., Kinnamon S. C., Gilbertson T. A. J. Neurophysiol. 2004;92:2909–2919. doi: 10.1152/jn.01198.2003. [DOI] [PubMed] [Google Scholar]
- 12.Richter T. A., Dvoryanchikov G. A., Chaudhari N., Roper S. D. J. Neurophysiol. 2004;92:1928–1936. doi: 10.1152/jn.00273.2004. [DOI] [PubMed] [Google Scholar]
- 13.Clapham D. E. Nature. 2003;426:517–524. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]
- 14.Montell C. Sci. STKE. 2005;2005(272):re3. doi: 10.1126/stke.2722005re3. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Hoon M. A., Chandrashekar J., Mueller K. L., Cook B., Wu D., Zuker C. S., Ryba N. J. Cell. 2003;112:293–301. doi: 10.1016/s0092-8674(03)00071-0. [DOI] [PubMed] [Google Scholar]
- 16.Delmas P., Padilla F., Osorio N., Coste B., Raoux M., Crest M. Biochem. Biophys. Res. Commun. 2004;322:1374–1383. doi: 10.1016/j.bbrc.2004.08.044. [DOI] [PubMed] [Google Scholar]
- 17.Nauli S. M., Zhou J. BioEssays. 2004;26:844–856. doi: 10.1002/bies.20069. [DOI] [PubMed] [Google Scholar]
- 18.Ramsey I. S., Delling M., Clapham D. E. Annu. Rev. Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez-Perrett S., Kim K., Ibarra C., Damiano A. E., Zotta E., Batelli M., Harris P. C., Reisin I. L., Arnaout M. A., Cantiello H. F. Proc. Natl. Acad. Sci. USA. 2001;98:1182–1187. doi: 10.1073/pnas.98.3.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanaoka K., Qian F., Boletta A., Bhunia A. K., Piontek K., Tsiokas L., Sukhatme V. P., Guggino W. B., Germino G. G. Nature. 2000;408:990–994. doi: 10.1038/35050128. [DOI] [PubMed] [Google Scholar]
- 21.Barr M. M., Sternberg P. W. Nature. 1999;401:386–389. doi: 10.1038/43913. [DOI] [PubMed] [Google Scholar]
- 22.Corey D. P., Garcia-Anoveros J., Holt J. R., Kwan K. Y., Lin S. Y., Vollrath M. A., Amalfitano A., Cheung E. L., Derfler B. H., Duggan A., et al. Nature. 2004;432:723–730. doi: 10.1038/nature03066. [DOI] [PubMed] [Google Scholar]
- 23.Damak S., Rong M., Yasumatsu K., Kokrashvili Z., Perez C. A., Shigemura N., Yoshida R., Mosinger B., Jr, Glendinning J. I., Ninomiya Y., Margolskee R. F. Chem. Senses. 2006;31:253–264. doi: 10.1093/chemse/bjj027. [DOI] [PubMed] [Google Scholar]
- 24.Perez C. A., Huang L., Rong M., Kozak J. A., Preuss A. K., Zhang H., Max M., Margolskee R. F. Nat. Neurosci. 2002;5:1169–1176. doi: 10.1038/nn952. [DOI] [PubMed] [Google Scholar]
- 25.Li A., Tian X., Sung S. W., Somlo S. Genomics. 2003;81:596–608. doi: 10.1016/s0888-7543(03)00048-x. [DOI] [PubMed] [Google Scholar]
- 26.Murakami M., Ohba T., Xu F., Shida S., Satoh E., Ono K., Miyoshi I., Watanabe H., Ito H., Iijima T. J. Biol. Chem. 2005;280:5626–5635. doi: 10.1074/jbc.M411496200. [DOI] [PubMed] [Google Scholar]
- 27.Max M., Shanker Y. G., Huang L., Rong M., Liu Z., Campagne F., Weinstein H., Damak S., Margolskee R. F. Nat. Genet. 2001;28:58–63. doi: 10.1038/ng0501-58. [DOI] [PubMed] [Google Scholar]
- 28.Matsunami H., Montmayeur J.-P., Buck L. Nature. 2000;404:601–604. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- 29.Hoon M. A., Adler E., Lindemeier J., Battey J. F., Ryba N. J., Zuker C. S. Cell. 1999;96:541–551. doi: 10.1016/s0092-8674(00)80658-3. [DOI] [PubMed] [Google Scholar]
- 30.Miyoshi M. A., Abe K., Emori Y. Chem. Senses. 2001;26:259–265. doi: 10.1093/chemse/26.3.259. [DOI] [PubMed] [Google Scholar]
- 31.Clapp T. R., Stone L. M., Margolskee R. F., Kinnamon S. C. BMC Neurosci. 2001;2:6. doi: 10.1186/1471-2202-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zawistowski J. S., Serebriiskii I. G., Lee M. F., Golemis E. A., Marchuk D. A. Hum. Mol. Genet. 2002;11:389–396. doi: 10.1093/hmg/11.4.389. [DOI] [PubMed] [Google Scholar]
- 33.Wong S. T., Henley J. R., Kanning K. C., Huang K. H., Bothwell M., Poo M. M. Nat. Neurosci. 2002;5:1302–1308. doi: 10.1038/nn975. [DOI] [PubMed] [Google Scholar]
- 34.Gunthorpe M. J., Smith G. D., Davis J. B., Randall A. D. Pflügers Arch. 2001;442:668–674. doi: 10.1007/s004240100584. [DOI] [PubMed] [Google Scholar]
- 35.Ganzevles P. G., Kroeze J. H. Physiol. Behav. 1987;40:641–646. doi: 10.1016/0031-9384(87)90111-9. [DOI] [PubMed] [Google Scholar]
- 36.Mueller K. L., Hoon M. A., Erlenbach I., Chandrashekar J., Zuker C. S., Ryba N. J. Nature. 2005;434:225–229. doi: 10.1038/nature03352. [DOI] [PubMed] [Google Scholar]
- 37.Nauli S. M., Alenghat F. J., Luo Y., Williams E., Vassilev P., Li X., Elia A. E., Lu W., Brown E. M., Quinn S. J., et al. Nat. Genet. 2003;33:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- 38.Chen X. Z., Vassilev P. M., Basora N., Peng J. B., Nomura H., Segal Y., Brown E. M., Reeders S. T., Hediger M. A., Zhou J. Nature. 1999;401:383–386. doi: 10.1038/43907. [DOI] [PubMed] [Google Scholar]
- 39.DeSimone J. A., Lyall V., Heck G. L., Feldman G. M. Respir. Physiol. 2001;129:231–245. doi: 10.1016/s0034-5687(01)00293-6. [DOI] [PubMed] [Google Scholar]
- 40.Bachmanov A. A., Tordoff M. G., Beauchamp G. K. Alcohol Clin. Exp. Res. 1996;20:201–206. doi: 10.1111/j.1530-0277.1996.tb01630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saito H., Kubota M., Roberts R. W., Chi Q., Matsunami H. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 42.Ishimaru Y., Okada S., Naito H., Nagai T., Yasuoka A., Matsumoto I., Abe K. Mech. Dev. 2005;122:1310–1321. doi: 10.1016/j.mod.2005.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.