Abstract
To investigate the genomic aberrations that are involved in lung tumorigenesis and therefore may be developed as biomarkers for lung cancer diagnosis, we characterized the genomic copy number changes associated with individual genes in 14 tumors from patients with primary non small cell lung cancer (NSCLC). Six squamous cell carcinomas (SQCAs) and eight adenocarcinomas (ADCAs) were examined by high-resolution comparative genomic hybridization (CGH) analysis of cDNA microarray. The SQCAs and ADCAs shared common frequency distributions of recurrent genomic gains of 63 genes and losses of 72 genes. Cluster analysis using 57 genes defined the genomic differences between these two major histologic types of NSCLC. Genomic aberrations from a set of 18 genes showed distinct difference of primary ADCAs from their paired normal lung tissues. The genomic copy number of four genes was validated by fluorescence in situ hybridization of 32 primary NSCLC tumors, including those used for cDNA microarray CGH analysis; a strong correlation with cDNA microarray CGH data emerged. The identified genomic aberrations may be involved in the initiation and progression of lung tumorigenesis and, most importantly, may be developed as new biomarkers for the early detection and classification of lung cancer.
Keywords: Genomic copy number changes, primary non small cell lung cancer, comparative genomic hybridization, cDNA microarray
Introduction
Lung cancer is the most common cause of cancer death in North America. The unsatisfactory cure rate and poor prognosis of affected patients support efforts for better risk assessment and early detection. Non small cell lung cancer (NSCLC), the predominant form of lung cancer, has two major histologic subtypes: squamous cell carcinoma (SQCA) and adenocarcinoma (ADCA) [1].
Lung tumorigenesis is a heterogeneous process that arises after a series of clonal molecular genetic alterations, including genomic gains and losses, particularly deletion of tumor-suppressor genes (TSGs) and amplification of oncogenes [2]. Therefore, defining these genomic aberrations may help us identify tumor-specific signatures involved in the initiation and progression of lung cancer and thus help produce genomic biomarkers for the early detection of lung tumors. Cytogenetic karyotypes have shown that NSCLCs display multiple numeric and structural chromosomal alterations [3]. Loss of heterozygosity (LOH) analysis further disclosed major differences in patterns of allelic imbalances between ADCA and SQCA [3–6]. Metaphase comparative genomic hybridization (CGH) studies detected genomewide copy number changes in lung cancers [7–10]. CGH analysis of the microarray bacterial artificial chromosomal (BAC) clones that cover a limited fraction of the human genome was also used to analyze NSCLCs and showed a clear pattern of genomic changes for SQCAs [11]. However, the low resolution of all the techniques makes it difficult to identify the causal genes whose structural alteration is critical for biologic behavior.
Most recently, CGH analysis of cDNA microarrays has provided high-resolution maps of genomic locations of single genes because it uses cDNA and expressed sequence tag clones as targets [12,13]. This technique has been proven to define genomic copy number gains and losses of individual genes in human cancer [12–14]. With sufficient genetic representation in cDNA microarrays, CGH resolution can be substantially improved to provide important genetic information underlying complex chromosomal rearrangements and genomic imbalances leading to tumorigenesis.
In this study, we used cDNA microarray CGH analysis to characterize, in detail, genomic aberrations associated with single genes in 14 primary NSCLC tumors and their paired normal tissues. The results demonstrate that NSCLC tumors share common frequency distributions of recurrent genomic gains and losses of sets of genes. Our study also defines the genomic difference between the two most common lung tumor subtypes, SQCA and ADCA, and provide a clear genomic profile of primary ADCA, which shows a distinct difference from paired normal lung tissues by a cluster of genomic aberrations. Validation of some of these genomic signatures raises the possibility of using the findings as new biomarkers for early detection of lung cancer.
Materials and Methods
Clinical Samples
For cDNA microarray CGH and metaphase CGH analysis, surgical specimens were obtained from 14 patients with stage I NSCLC between March 1, 2002 and June 28, 2003 at The University of Texas M. D. Anderson Cancer Center. All patients had a smoking history ranging from 32 to 95 pack years. Six SQCAs and eight ADCAs had been definitively resected by either a lobectomy or a pneumonectomy. None of the patients had received preoperative adjuvant chemotherapy or radiotherapy. Tissues samples had been routinely dissected intraoperatively from the surrounding lung parenchyma; paired normal lung tissues had also been obtained from the same patients at an area distant from their tumors. Tissue acquisition was approved by the institutional review board at our institution. Tissue sections (4 µm thick) were stained with hematoxylin and eosin, and reviewed to confirm the diagnosis and to verify the presence of greater than 70% tumor cells. For fluorescence in situ hybridization (FISH) analysis, touch imprints were made from surgical specimens obtained from 32 patients with stage I NSCLC (16 SQCAs and 16 ADCAs, including those used for the cDNA microarray CGH analysis) and then fixed in methanol and acetic acid (3:1).
Cancer cell lines BT474 and H358 were purchased from the American Tissue Culture Collection (Rockville, MD) and maintained in RPMI medium supplemented with 10% fetal bovine serum. Genomic DNA was extracted from cell lines, surgical tissues, and normal human lymphocytes using a DNA tissue kit (QIAGEN, Inc., Valencia, CA) following the manufacturer's instructions.
cDNA Microarray CGH
cDNA microarrays contained a total of 8000 cDNA clones (Research Genetics; Invitrogen, Huntsville, AL). Of these clones, 6894 represented known genes, and the remainder corresponded to uncharacterized expressed sequence tags. The preparation of array slides was performed essentially as described previously [12,13]. Chromosomal assignments of clones were determined from the July 2003 freeze of the assembled human genome available through the UCSC Genome Browser (http://genome.cse.ucsc.edu). CGH experiments on cDNA microarrays were performed as described previously [12,13]. Briefly, 20 µg of genomic DNA from cancer cell lines, tissue specimens, and normal human lymphocytes was digested for 14 to 18 hours with AluI and RsaI (New England Biolaboratories, Beverly, MA) and purified by phenol-chloroform extraction. Six micrograms of tested DNA was labeled with Cy3 by Bioprime labeling kit (Invitrogen, Carlsbad, CA), and normal lung tissue DNA was labeled with Cy5-dUTP (Invitrogen). Hybridization and postwashes were performed as described previously [12,14]. A laser confocal scanner (Agilent Technologies, Palo Alto, CA) was used to measure the fluorescence intensities at the target locations using DEARRAY software. After background subtraction, the average intensity of each clone in the test hybridization was divided by the average intensity of the corresponding clone in the control hybridization. For the copy number analysis, the ratios were normalized on the basis of the distribution of ratios of all targets on the array based on 126 housekeeping genes, which were spotted four times in the array. The distributions of fluorescence ratios were used to define cutpoints for increased or decreased copy number. Only clones that exhibited a log2 hybridization ratio of either >-1 or <-1 were considered completely amplified or deleted candidates, respectively.
Conventional CGH
Chromosomal CGH experiments were carried out as described in our previous publications [15,16]. Briefly, genomic DNA was labeled by nick translation using a nick translation labeling system (Vysis, Downers Grove, IL). Tumor samples were directly labeled with SpectrumGreen (Vysis) and hybridized with SpectrumRed-labeled reference DNA (Vysis). Samples were counterstained with 4,6-diamino-2-phenylindole (DAPI). Each CGH experiment included at least one normal human lymphocyte DNA as a negative control. Images were analyzed with CGH analysis software (Applied Imaging, Clara, CA). A gain of DNA sequence copy number was defined as a tumor-to-reference ratio >1.2 on both standard and inverse hybridizations. A copy number decrease was defined as a tumor-to-reference ratio of <0.8 on both hybridizations.
FISH
By searching the genome sequence database (http://www.ncbi.nlm.nih.gov/BLAST) using the BLAST algorithm, we identified the following BAC clones: 2320O4 for Skp2, 307C12 for Cks1, 391M1 for Gc20/Sui1, and 506M13 for SFTPA1. These clones were used as FISH probes. BAC DNA was prepared using the DNA Maxi Kit (QIAGEN, Inc.). Dual-color FISH was performed as described in our previous publication [17]. Briefly, 1 µg of BAC DNA was labeled with SpectrumGreen (Vysis). Tissue imprint slides were denatured in 70% formamide and 2 x SSC for 5 minutes at 72°C, dehydrated in graded ethanol, and incubated with a hybridization mixture consisting of 60% formamide, 2 x SSC, CotI DNA, and 100 ng of both a Spectrum-Green-labeled BAC DNA probe and a SpectrumOrangelabeled corresponding chromosomal centromeric probe (Vysis). After overnight incubation at 37°C, the slides were washed at 45°C in 50% formamide and 2 x SSC for 10 minutes and counterstained with antifade solution containing DAPI. Two hundred cells on each slide were counted using Leica microscopes equipped with appropriate filter sets (Leica Microsystems, Buffalo, NY). Greater or lesser copy numbers of the tested probes compared with copy numbers of reference indicated gain and loss of the gene, respectively. The cutoff value was calculated from normal tissue samples using the mean number (plus 3 SD) of cells having an abnormal FISH signal pattern.
Statistical Analyses
To analyze the cDNA microarray CGH data, clustering analysis was performed with cluster analysis software and in the TreeView program written by Michael Eisen [18]. Before the clustering algorithm was applied, the fluorescence ratio for each spot was first log-transformed (log2), and then the data for each sample were median-centered to remove experimental biases. To distinguish differences in the copy number of genes between SQCAs, ADCAs, and normal tissues, we measured 7668 clones because these clones were present in more than 12 of 14 specimens. Each clone was assessed by computing a two-sample t statistic with equal variances. The P value for each test was determined using a permutation method to calculate the ability of individual clones to distinguish between the subtypes of lung cancer. This procedure was repeated 10,000 times. P values less than .05 were considered significant, and the clones associated with these significant values were thought to have the power to distinguish between any two groups of tissues.
A Wilcoxon ranks sum test was applied to compare the number of genomic alterations detected by conventional CGH between different histologic subtypes, and the Student's t test was used to evaluate the relationships between genomic copy number changes detected by FISH in the different histologic subtypes. Chi-square analysis was performed to examine the results of correlation between cDNA microarray CGH and FISH regarding the genomic copy number of the genes. A P value of less than .05 was considered statistically significant.
Results
cDNA Microarray CGH Is Sensitive in Defining Genomic Aberrations of Genes
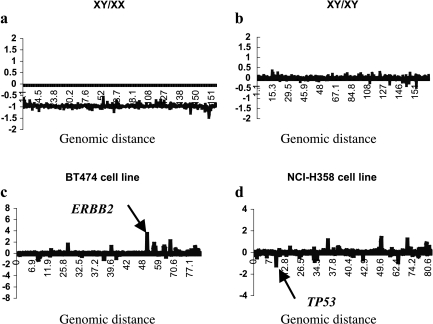
To assess the sensitivity of the cDNA microarray in detecting the genomic copy numbers, we first tested its ability to measure single-copy chromosomal changes by cohybridizing male DNA labeled with Cy5 and female DNA labeled with Cy3 in the cDNA microarrays. The average log2 Cy5:Cy3 hybridization ratio for X chromosome genes was -1, which compares to an ideal log2 value of 1 for a 2:1 female-to-male X chromosome ratio. We then tested the ability of the cDNA microarray to detect the genomic gain of single gene by hybridizing breast cancer cell line BT474 genomic DNA, in which the genomic copy number of the ERBB2 gene is approximately 10. When one third the amount of this DNA was compared with the normal reference DNA, the log2 hybridization ratio for ERBB2 genes was 3.2, suggesting that the ERBB2 copy number was approximately 3 (Figure 1).
Figure 1.
The sensitivity of the cDNA microarray CGH analysis in detecting genomic copy number. Evidence of genes that increased in copy number (log2 hybridization ratio >0) appears above the x-axis, whereas evidence of those that decreased in copy number (log2 hybridization ratio >0) appears below the x-axis. (a) Chromosome X DNA copy number profile for male DNA compared with female DNA. The average log2 Cy5:Cy3 hybridization ratio for X chromosome genes was -1, suggesting a 2:1 female-to-male X chromosome ratio. (b) Chromosome X DNA copy number profile for male DNA cohybridized with male DNA. The average log2 Cy5:Cy3 hybridization ratio for X chromosome genes was 0, suggesting no X chromosome copy number change. (c) Chromosome 17 DNA copy number profile for one third the amount of the BT474 DNA used compared with the normal reference DNA; the log2 hybridization ratio for the ERBB2 gene was 3.2, suggesting that the ERBB2 copy number was approximately 3 (arrow). (d) Chromosome 17 DNA copy number profile for DNA from NCI-H358 lung cancer cells cohybridized with normal reference DNA. The average log2 hybridization ratio for TP53 genes was -1.2 (arrow), suggesting the ability of the cDNA microarray CGH to detect the losses of a single gene.
To assess the ability of the array to detect the deletion of a single gene, we cohybridized DNA from NCI-H358 lung cancer cells, which have homozygous deletion of the TP53 genome, with normal reference DNA. The average log2 hybridization ratio for TP53 genes was -1.2 (Figure 1).
Our study also allowed a direct comparison of the sensitivity of the cDNA array CGH with that of the metaphase CGH because the same samples were applied to the two methods simultaneously. As illustrated in Figure 2, all of the imbalances identified by metaphase CGH were confirmed by microarray CGH, whereas the copy number imbalances at 2p, 2q, 4q, 7p, 7q, 6p, 10q, 14q, 15q, 16q, and Xq detected by microarray CGH were not identified by metaphase CGH. In addition, the genomic aberrations identified by metaphase CGH were delineated by microarray CGH to a much smaller regions, including gains at 1q21-22, 5p13, 8q22.1-23.1, 11q13, 19q13.1, 20q13.3, and 22q11.23, and losses at 5q23-32, 8p21-22, 9p21, 19p13.1, and 21q22.3. There was no statistically significant relationship between smoking pack year and certain genomic aberrations.
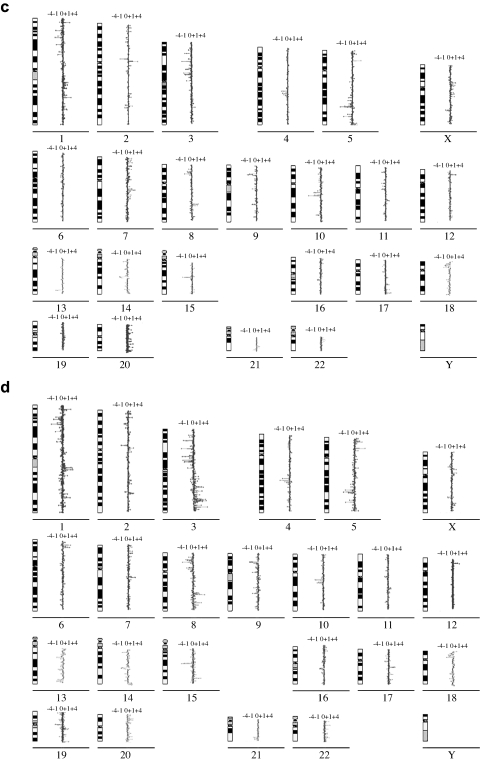
Figure 2.
Comparison of metaphase CGH (a and b) and cDNA microarray CGH (c and b) for determining genomic copy number changes in 14 primary NSCLC tumors consisting of eight ADCAs and six SQCAs. Panels (a) and (b) are summaries of relative DNA sequence copy number changes detected by metaphase CGH in ADCAs and SQCAs, respectively. The vertical lines to the right of the chromosomal ideograms indicate copy number gains; those to the left indicate losses. Panels (c) and (d) are summaries of the cDNA microarray CGH profile of genomic imbalances for the ADCA and SQCA specimens tested, respectively, in (a) and (b). Evidence of genes that increased in copy number (log2 hybridization ratio >0) appears to the right of the vertical line next to each chromosomal ideogram, and evidence of genes that decreased in copy number (log2 hybridization ratio <0) appears to the left of the vertical line next to each chromosomal ideogram. The profile for chromosome Y is not shown because of the low representation of genes from this chromosome in the microarray.
Genomic Signatures of Primary NSCLCs Defined by cDNA Microarray CGH
Consistent with the results of conventional CGH, those of cDNA microarray CGH showed that primary SQCAs and ADCAs share common frequency distributions of recurrent gains and losses of genes (Figure 2). The genomic aberrations involved several known oncogenes and TSGs within 1q, 3q, 5p, 8q, 16p, 17q, 19p, 19q, and 20q for gains and 1p, 3p, 5q, 8p, 9p, 11p, 11q, 13q, and 18q for losses. The total numbers of genes with genomic aberrations are 228 in SQCAs and 194 in ADCAs. Furthermore, using clustering analysis of cDNA microarray CGH data, we identified 25 genes with a high number of genomic copy number changes and another 63 genes with a high number of genomic copy number losses in both SQCAs and ADCAs. Notably, as Table 1 shows, molecular genetic alteration of some of the genes has been previously described in lung tumors [19–49].
Table 1.
Genes with Genomic Amplification or Deletion in Both ADCAs and SQCAs.
| Gene Name | Gene Description | Chromosomal Location | Prevalence* | Reference |
| Genes with amplified genomic copy number | ||||
| RPL37 | Ribosomal protein L37 | 5p13 | 12 | [19] |
| LAPTM4B | Lysosome-associated protein transmembrane 4β | 8q22.1 | 12 | |
| ENO1 | Enolase 1 (α) | 1p36.3 | 12 | [20] |
| YWHAZ | Tyrosine 3-monooxygenase | 8q23.1 | 12 | |
| COL1A2 | Collagen, type I, α2 | 7q22.1 | 11 | [21] |
| RPL30 | Ribosomal protein L30 | 8q22 | 11 | |
| RPS2 | Ribosomal protein S2 | 16p13.3 | 11 | |
| RPS4X | Ribosomal protein S4 | Xq13.1 | 10 | |
| GNB2l1 | Guanine nucleotide binding protein, β polypeptide 2-like 1 | 5q35.3 | 10 | |
| SYN1 | Synapsin I | Xp11.23 | 10 | |
| SSR4 | Signal sequence receptor, δ | Xq28 | 10 | |
| JTB | Jumping translocation breakpoint | 1q21 | 10 | |
| RPS21 | Ribosomal protein S21 | 20q13.3 | 9 | |
| MIF | Macrophage migration-inhibitory factor | 22q11.23 | 9 | [22–27] |
| BASP1 | Brain-abundant, membrane-attached signal protein 1 | 5p15.1 | 9 | |
| RPS10 | Ribosomal protein S10 | 6p21.31 | 9 | |
| EIF3S9 | Eukaryotic translation initiation factor 3, subunit 9 | 7p22.3 | 8 | [28] |
| SNRPE | Small nuclear ribonucleoprotein polypeptide E | 1q32 | 8 | |
| RPL38 | Ribosomal protein L38 | 17q23 | 8 | |
| FGF4 | Fibroblast growth factor 4 | 11q13.3 | 8 | [29] |
| IGKC | Immunoglobulin κ constant | 2p12 | 7 | |
| PLP2 | Proteolipid protein 2 | Xp11.23 | 7 | |
| CRABP2 | Cellular retinoic acid binding protein 2 | 1q21.3 | 6 | |
| CCT5 | Chaperonin-containing TCP1, subunit 5 (epsilon) | 5p15.2 | 6 | |
| PFN2 | Profilin 2 | 3q25.1 | 6 | |
| Genes with deleted genomic copy number | ||||
| ACTA2 | Actin, α2, smooth muscle, aorta | 10q23.3 | 13 | |
| GPX3 | Glutathione peroxidase 3 (plasma) | 5q23 | 13 | [30] |
| TNA | Tetranectin (plasminogen binding protein) | 3p22 | 13 | [29–31] |
| ANXA2 | Annexin A2 | 15q21 | 13 | |
| CDKN2B | Cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 9p21 | 13 | [32] |
| GNAI2 | Guanine nucleotide binding protein (G protein) | 3p21 | 13 | [31] |
| DUSP1 | Dual specificity phosphatase 1 | 5q34 | 12 | |
| ELAVL2 | Embryonic lethal, abnormal vision, Drosophila-like 2 | 9p21 | 12 | |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A | 9p21 | 12 | [33,34] |
| SFTPC | Surfactant, pulmonary-associated protein C | 8p21 | 12 | [31] |
| UBC | Ubiquitin C | 12q24.3 | 12 | |
| GC20/SUI1 | Translation factor sui1 homolog | 3p21.3 | 12 | |
| HYAL2 | Hyaluronoglucosaminidase 2 | 3p21.3 | 12 | [35] |
| TGFBR2 | Transforming growth factor β receptor II | 3p22 | 12 | [31] |
| CD74 | Invariant polypeptide of major histocompatibility complex, II | 5q32 | 12 | [36] |
| NEDD4L | Neural precursor cell expressed, developmentally downregulated 4 | 18q21 | 12 | |
| FCN3 | Ficolin (collagen/fibrinogen domain containing) 3 | 1p35 | 12 | [31] |
| SFTPA | Surfactant, pulmonary-associated protein A | 10q22.2 | 12 | [30,31] |
| COL4A2 | Collagen, type IV, α2 | 13q34 | 11 | |
| MACF1 | Microtubule actin cross-linking factor 1 | 1p32 | 11 | |
| SFTPB | Surfactant, pulmonary-associated protein B | 2p12 | 11 | [19,30,37] |
| CACNA2D2 | Calcium channel, voltage-dependent, α2/δ subunit 2 | 3p21.3 | 11 | [30,38,39] |
| APM2 | Adipose-specific 2 | 10q23.2 | 11 | |
| SPARCL1 | SPARC-like 1 | 4q22.1 | 11 | [40] |
| ARHA | Ras homolog gene family, member A | 3p21.3 | 11 | [21,36] |
| PSAP | Prosaposin | 10q21 | 11 | |
| RAB31 | Member RAS oncogene family | 18p11.3 | 11 | [21] |
| B2M | β2-microglobulin | 15q21 | 11 | [41] |
| TU3A | TU3A protein | 3p21.1 | 11 | |
| PPAP2B | Phosphatidic acid phosphatase type 2B | 1pter | 11 | |
| TMSB4X | Thymosin, β4, X-linked | Xq21.3 | 10 | |
| DHCR24 | 24-Dehydrocholesterol reductase | 1p33 | 10 | |
| ANXA11 | Annexin A11 | 10q23 | 10 | |
| CDW52 | CDW52 antigen (CAMPATH-1 antigen) | 1p36 | 10 | |
| ASAH1 | N-acylsphingosine amidohydrolase (acid ceramidase) 1 | 8p22 | 10 | |
| GPRK5 | G protein-coupled receptor kinase 5 | 10q24 | 10 | |
| ARHGEF7 | ρ-Guanine nucleotide exchange factor (GEF7) | 13q33.3 | 10 | |
| GPC3 | Glypican 3 | Xq26.1 | 10 | [30] |
| EPB41L3 | Erythrocyte membrane protein band 4.1-like 3 | 18p11.32 | 10 | |
| STOM | Stomatin | 9q34.1 | 9 | |
| SLIT2 | Slit homolog 2 | 4p15.2 | 9 | [30,31] |
| VAT1 | Vesicle amine transport protein 1 homolog | 17q21 | 9 | |
| TNFAIP3 | Tumor necrosis factor, α-induced protein 3 | 6q23 | 9 | |
| PTPRM | Protein tyrosine phosphatase, receptor type, M | 18p11.2 | 9 | |
| ADH1A | Alcohol dehydrogenase 1A (class I), α polypeptide | 4q21 | 9 | [30] |
| C2 | Complement component 2 | 6p21.3 | 9 | |
| ORM1 | Orosomucoid 1 | 9q31 | 9 | [42,43] |
| PCDHGC3 | Protocadherin gamma subfamily C, 3 | 5q31 | 9 | |
| DLGAP1 | Discs, large (Drosophila) homolog-associated protein 1 | 18p11.3 | 9 | [44] |
| NSG-X | Brain and nasopharyngeal carcinoma susceptibility protein | 7q31 | 8 | |
| ID1 | Inhibitor of DNA binding 1 | 20q11 | 8 | [45] |
| CYP4B1 | Cytochrome P450, family 4, subfamily B, polypeptide 1 | 1p34 | 8 | [30,46] |
| COL6A2 | Collagen, type VI, α2 | 21q22.3 | 8 | |
| HBB | Hemoglobin, β | 11p15.5 | 8 | |
| GSN | Gelsolin | 9q33 | 8 | [47–49] |
| SCGB1A1 | Secretoglobin, family 1A, member 1 (uteroglobin) | 11q12.3 | 8 | |
| CD14 | CD14 antigen | 5q31.1 | 8 | |
| TEK | TEK tyrosine kinase, endothelial | 9p21 | 8 | [21] |
| VAMP3 | Vesicle-associated membrane protein 3 | 1p36.22 | 8 | |
| MT1G | Metallothionein 1G | 16q13 | 8 | |
| FHL1 | Four and a half LIM domains 1 | Xq26 | 7 | |
| FTH1 | Ferritin, heavy polypeptide 1 | 11q13 | 7 | |
| VIPR1 | Vasoactive intestinal peptide receptor 1 | 3p22 | 7 | |
| PGC | Progastricsin (pepsinogen C) | 6p21.3 | 7 | |
| SH3BP5 | SH3 domain binding protein 5 (BTK-associated) | 3p24.3 | 7 | |
Genes with amplified genomic copy number refer to genes that are greater than four times the normal copy number.
Number of the tumor specimens in which genomic copy number change of the genes was observed.
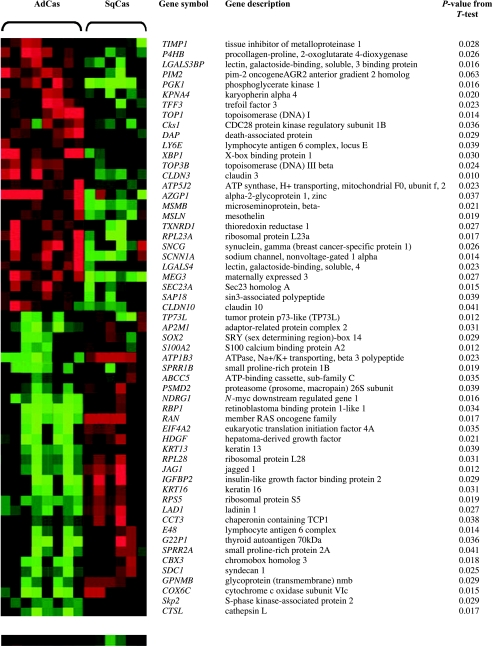
Genomic differences associated with individual genes between lung SQCA and ADCA subgroups can be deduced from Figure 2, c and d. A paired t-test performed on the logtransformed cDNA microarray CGH data identified the 57 most informative genes that allowed accurate discrimination between SQCAs and ADCAs (Figure 3). Furthermore, a permutation t-test using 18 genes with genomic changes was able to show a significant difference between ADCAs and their paired normal lung tissues (Figure 4).
Figure 3.
The 57 identified most informative gene associated with a P value of less than .05 in a paired t-test comparing eight ADCAs and six SQCAs. The heat map was made using TreeView [18]. The histologic type was given on top of the heat map.
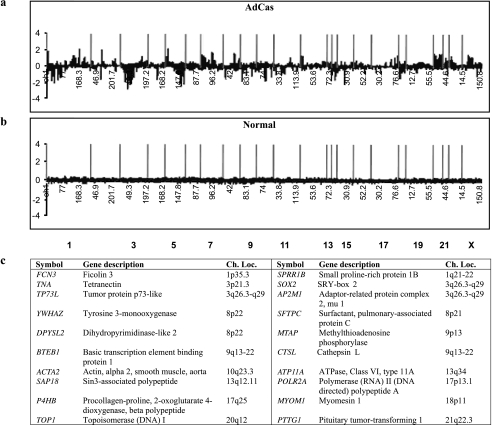
Figure 4.
Summary of the relative DNA sequence copy number changes detected by cDNA microarray CGH analysis in eight primary ADCAs and their paired normal lung tissues. Primary ADCAs have a clear genomic aberration profile of genes (a), which shows distinct differences from that of normal lung tissues (b). The vertical red bars represent the separation of chromosomes. Panel (c) shows the results of a permutation t test using 18 genes that have genomic changes and shows a significant difference between ADCAs and their paired normal lung tissues.
Validation of Genomic Signatures by FISH
To confirm whether the genomic signatures detected by our cDNA microarray CGH analysis reflected the real frequencies of the gene alterations in primary NSCLCs and have the potential to correctly identify the two major histology types of lung tumor, two genes (Gc20/Sui1 and SFTPA1) with deletions in both the SQCAs and ADCAs and two genes (Skp2 and Cks1) with genomic gains in either SQCAs or ADCAs were selected. The copy number aberrations of the genes were detected by FISH in a set of lung tissue specimens, including those used for the cDNA microarray CGH analysis. As shown in Table 2, there was complete concordance between the cDNA microarray CGH and FISH results. The genomic deletion of Gc20/Sui1 and SFTPA1 is common for both subtypes of NSCLCs. Genomic amplification of Skp2 is specific for SQCA, whereas Cks1 amplification is more common for ADCA (Table 2, Figure 5).
Table 2.
Comparison of Results Obtained for Four Selected Genes From cDNA Microarray CGH and FISH Analyses*
| General Chromosomal Location | % Positive Cases in ADCAs by cDNA | Microarray CGH % Positive Cases in SQCAs by cDNA Microarray CGH | % Positive Cases in ADCAs by FISH | % Positive Cases in SQCAs by FISH | cDNA Microarray CGH Versus FISH† |
| Sui1 (3p21.33) | 88 (7/8) | 83 (5/6) | 94 (15/16) | 87 (14/16) | <0.003 |
| Skp2 (5p13.2) | 13 (1/8) | 67 (4/6) | 19 (3/16) | 56 (9/16) | <0.01 |
| SFTPA1 (10q22) | 100 (8/8) | 100 (6/6) | 84 (14/16) | 94 (15/16) | <0.001 |
| Cks1 (1q21) | 75 (6/8) | 17 (1/6) | 69 (11/16) | 19 (3/16) | <0.01 |
Specimens containing a two-fold or higher increase in the number of test Skp2 or Cks1 probe signals, compared with corresponding centromere signals, in at least 10% of the tumor cells were considered to be amplified, whereas specimens containing fewer Sui1 or SFTPA1 probe signals than the corresponding centromeric probe signals in at least 10% of the tumor cells were considered to be deleted.
Concordance between cDNA array CGH and FISH results was determined by chi-square analysis.
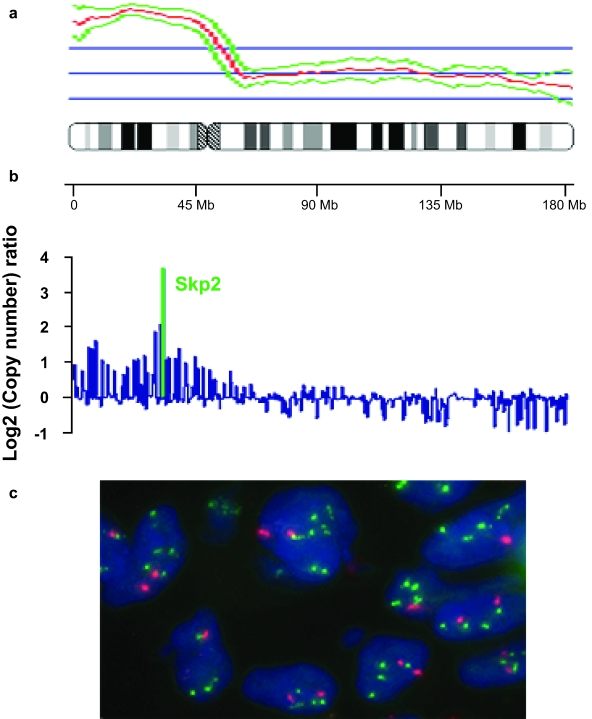
Figure 5.
Validation of Skp2 genomic copy number changes in a SQCA specimen by metaphase CGH, cDNA microarray CGH analyses, and FISH. Metaphase CGH (a) shows a large amplicon in chromosome 5p, which is confirmed and precisely defined by cDNA microarray CGH and includes the Skp2 oncogene (b). The increased Skp2 copy number (c) was validated in the specimens by FISH using a Skp2 probe (green) and a centromeric probe (red), which was used as a reference. SQCA cells contain a two- to four-fold increase in the number of Skp2 probe signals compared with the number of the reference, indicating Skp2 amplification.
Discussion
Chromosomal aberrations reflect the selective retention of genomic fragments housing driver genes, whose abnormality contributes to tumorigenesis. The metaphase CGH assay has been used for the identification of novel driver genes, and its profiles correspond well to the chromosomal location of some known or suspected oncogenes and TSGs in lung tumors [7–10]. However, for other frequently observed aberrations, no specific driver genes have yet been implicated because the method has relatively low resolution. We demonstrated in our study that the use of cDNA microarray CGH analysis may address this issue because cDNA micro-arrays represent high-resolution maps (in our study, one clone every 376 kb through the human genome), an approximately 20-fold higher mapping resolution than that attained by metaphase CGH. With the completion of the human genomic database, cDNA microarray CGH can map genomic gains and losses by their gene position rather than their chromosomal band, and therefore can immediately provide a list of candidate genes that occur within the region of interest.
The genomic copy number imbalances identified by our cDNA microarray CGH analysis appear comparable to those found in a recent study of lung cancer that used a BAC array CGH technique [11]. Furthermore, because the cDNA microarray has a much higher mapping resolution (376 kb) than that achieved by BAC array CGH (1.4 Mb), our study restricted the larger fragment of genomic copy number changes to a small focal point of copy number aberrations of individual genes in primary lung tumors. For example, we determined two peaks of genomic gain at 5p13 and 5p35.3 in both the SQCAs and ADCAs that were not detected by the BAC array CGH, suggesting that the use of cDNA microarrays for analysis of DNA copy number variation has marked advantages over the use of large genomic DNA clone array-based CGH methods. Moreover, we determined the pattern of composite genomic losses of variable regions on several chromosomal loci in lung cancer, which was in keeping with the complex pattern of chromosomal rearrangements observed for deletions discovered by LOH [4,5]; however, our results defined a narrower region and even identified the individual genes with genomic deletion. Thus, our cDNA microarray CGH analysis has a higher resolution than other methods and can be used to detect a small region or individual genes of amplification or deletion, and, finally, define the unique genomic signatures associated with lung tumorigenesis.
The results of our cDNA microarray CGH analysis also imply that primary SQCAs and ADCAs share common recurrent DNA copy number gains and losses of certain gene clusters, confirming previous findings that lung tumors involve a series of clonal molecular-genetic alterations [11,50]. We also showed that substantial genomic differences exist between SQCAs and ADCAs. For example, when using metaphase CGH, we detected genomic gains in both ADCAs and SQCAs; the frequency of the genomic copy number of 3q was higher in SQCAs (100%, 6/6) than in ADCAs (50%, 4/8) (P < .001). Correspondingly, when conducting cDNA array analysis of the same specimens, gene amplification of chromosome 3q was more frequent in SQCAs than in ADCAs. ADCAs tended to have a more heterogeneous gene transcript pattern and, in some cases, to exhibit a genomic profile in 3q more similar to that of nonneoplastic parenchymal lung tissues. The genomic copy number difference between primary SQCAs and ADCAs suggests that they may differ in the level of genomic instability or mechanisms by which they initiate and progress; the genomic aberrations of specific genes in the genomic phenotype of each of the histologic subtypes reflect their different genomic clonal evaluations and appear as different diseases at the molecular level [11]. Most importantly, these unique genomic abnormalities may be developed as predictor sets of biomarkers for the early detection and classification of lung cancers.
Previous reports using the serial analysis of gene expression, oligonucleotides, or cDNA array analysis have described sets of genes overexpressed or downregulated in primary SQCAs and ADCAs [21,22,30,31,36,51]. In contrast to SQCAs, which always showed clusters as a distinct tumor group, ADCAs tended to have a more heterogeneous gene transcript pattern and, in some cases, to exhibit a profile more similar to that of nonneoplastic parenchymal lung tissues. Both of these facts make it difficult to molecularly classify ADCA using transcript signatures [51]. In our study, cDNA microarray CGH analysis provided a clear genomic profile of genes in primary ADCAs that is distinctly different from that in normal lung tissues. That genes in primary ADCAs had a distinct genomic pattern in our study but no clear transcriptional profile in other study is not surprising for several reason: 1) genomic DNA is a different mixture from the mRNA representation of cells; 2) transcription of genes has different biologic changes and behaviors from their genomic ancestors in lung cancer; and 3) the level of mRNA expression does not completely reflect genomic copy number changes. In addition, the inclusion of some ADCAs with normal lung from the previous reports may due to the profiling of BAC bronchioloalveolar carcinomas. The comparison of ADCAs with normal lung at the genomic DNA level by cDNA microarray CGH analysis should reveal the differences. Future assessment of transcript level and gene copy number changes of the same set of lung tumors in parallel using the same array may define whether genomic structural abnormalities directly affect imbalances of expression in lung tumorigenesis. However, our findings showing that primary ADCAs have a set of genes with a unique genomic profile may be of interests because these minimal gene sets can be used for developing biomarkers for ADCAs. This finding is particularly important because ADCAs have become more prevalent than SQCAs—a trend that is occurring worldwide—and are more difficult to diagnose than SQCAs because they always arise from the smaller airways [1].
There was no statistically significant relationship between smoking pack year and certain genomic aberrations; one possible reason may be the small sample size of the current study. Currently, we are analyzing a large cohort of clinical specimens in an ongoing study, assessing the concordance of the genomic findings detected by cDNA microarray CGH and correlating these data with smoking history, prognosis, tumor progression, and treatment of the patients.
Although we used only four genes for confirmation in this study, all four showed a strong correlation between FISH analysis and cDNA microarray CGH data for genomic copy number changes, indicating that the genomic signatures discovered by cDNA microarray CGH might be developed as biomarkers for early interventional strategies for lung cancer. Furthermore, our results may suggest that the genomic changes we observed are likely relevant to lung tumorigenesis. In fact, alterations of some of the genes have been previously reported in lung cancer (Table 1). For example, SFTPA1 is a phospholipid-protein complex that lowers the surface tension at the air-liquid interface in the alveoli of the lung and plays a key role in the innate host defenses there. The transcription-level and protein-level aberration of SFTPA1 has previously been observed in lung tumorigenesis [52–54]. The product of GC20/Sui1 is a general monitor of the translational accuracy of proteins through recognition of the protein synthesis initiation codon, and the expression of GC20/Sui1 induced is related to cellular stress and may represent an important adaptive response to genotoxic agents [55]. GC20/Sui1 has been detected in normal liver cells but not in hepatocellular carcinoma cells [56]. We found that the GC20/Sui1 transcript was diminished in 80% of lung cancer cell lines tested by using reverse transcription polymerase chain reaction (RT-PCR) (data not shown). Skp2 displays an S-phase-promoting function in the cell cycle and is implicated in the ubiquitin-mediated degradation of several key regulators of mammalian G1 progression, including p27, a dosage-dependent tumor-suppressor protein. Skp2 protein is overexpressed in oral epithelial carcinomas, and its expression levels correlate positively with prognosis [57]. A positive correlation of an increased relative copy number of Skp2 with a transcriptional level was found in small cell lung cancers cell lines [58]. Cks1 is one of the components of the Skp1-Cullin1-F-box-Roc1 complex [59]. Inui et al. [60] recently found a high expression of Cks1 in ADCAs and suggested that such high expression may be involved in the pathogenesis of the diseases. In agreement with that report, our study's detection of genomic gain of Cks1 by FISH and cDNA microarray CGH analyses was common in ADCAs. However, further characterization of the genes with a genomic aberration identified in our study is needed to evaluate the effects of the genomic aberrations on transcriptional and protein levels in lung cancer.
In summary, we have generated a profile of genomic copy number aberrations in the two major histologic subtypes of primary NSCLC tumors. Our findings may be a step toward defining a new genomic taxonomy of such tumors and demonstrate the potential power of genomic copy number profiling in lung cancer diagnosis. The development and implication of a relevant panel of probes for detecting genomic signatures might be of great value in lung cancer diagnosis and surveillance strategies in a clinical laboratory setting. Nevertheless, double-blind, prospective, confirmatory studies by independent groups are necessary to further validate these findings.
Acknowledgements
We thank Vickie J. Williams and Ellen M. McDonald for editing the manuscript.
Footnotes
This work was supported by an M. Keck Center for Cancer Gene Therapy Award, the University of Texas M. D. Anderson Cancer Center Institutional Research Grant, the Developmental Project/Career Development Award from The University of Texas specialized programs of research excellence in lung cancer, and the M. D. Anderson Cancer Center Tobacco Settlement Fund.
References
- 1.Mountain CT. Revision in the international system for staging of lung cancer. Chest. 1997;111:1710–1717. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Christiani DC, Mark EJ, Nelson H, Wiencke JK, Gunn L, Wain JC, Kelsey KT. Carcinogen exposure, p53 alteration, and K-ras mutation in synchronous multiple primary lung carcinoma. Cancer. 1999;85:1734–1739. [PubMed] [Google Scholar]
- 3.Johansson M, Jin Y, Mandahl N, Hambraeus G, Johansson L, Mitelman F, Heim S, Lukeis R, Irving L, Garson M, Hasthorpe S. Cytogenetics of non-small cell lung cancer: analysis of consistent nonrandom abnormalities. Genes Chromosomes Cancer. 1990;2:116–124. doi: 10.1002/gcc.2870020207. [DOI] [PubMed] [Google Scholar]
- 4.Virmani AK, Fong KM, Kodagoda D, McIntire D, Hung J, Tonk V, Minna JD, Gazdar AF. Allelotyping demonstrates common and distinct patterns of chromosomal loss in human lung cancer types. Genes Chromosomes Cancer. 1998;21:308–319. doi: 10.1002/(sici)1098-2264(199804)21:4<308::aid-gcc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 5.Yoshino I, Fukuyama S, Kameyama T, Shikada Y, Oda S, Maehara Y, Sugimachi K. Detection of loss of heterozygosity by highresolution fluorescent system in non-small cell lung cancer: association of loss of heterozygosity with smoking and tumor progression. Chest. 2003;123:545–550. doi: 10.1378/chest.123.2.545. [DOI] [PubMed] [Google Scholar]
- 6.Girard L, Zochbauer-Muller S, Virmani AK, Gazdar AF, Minna JD. Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small cell lung cancer and non-small cell lung cancer, and loci clustering. Cancer Res. 2000;60:4894–4906. [PubMed] [Google Scholar]
- 7.Petersen I, Bujard M, Petersen S, Wolf G, Goeze A, Schwendel A, Langreck H, Gellert K, Reichel M, Just K, du Manoir S, Cremer T, Dietel M, Ried T. Patterns of chromosomal imbalances in adenocarcinoma and squamous cell carcinoma of the lung. Cancer Res. 1997;12:2331–2335. [PubMed] [Google Scholar]
- 8.Luk C, Tsao MS, Bayani J, Shepherd F, Squire JA. Molecular cytogenetic analysis of non-small cell lung carcinoma by spectral karyotyping and comparative genomic hybridization. Cancer Genet Cytogenet. 2001;125:87–99. doi: 10.1016/s0165-4608(00)00363-0. [DOI] [PubMed] [Google Scholar]
- 9.Michelland S, Gazzeri S, Brambilla E, Robert-Nicoud M. Comparison of chromosomal imbalances in neuroendocrine and non-small-cell lung carcinomas. Cancer Genet Cytogenet. 1999;114:22–30. doi: 10.1016/s0165-4608(99)00035-7. [DOI] [PubMed] [Google Scholar]
- 10.Taguchi T, Cheng GZ, Bell DW, Balsara B, Liu Z, Siegfried JM, Testa JR. Combined chromosome microdissection and comparative genomic hybridization detect multiple sites of amplification DNA in a human lung carcinoma cell line. Genes Chromosomes Cancer. 1997;20:208–212. doi: 10.1002/(sici)1098-2264(199710)20:2<208::aid-gcc13>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 11.Massion PP, Kuo WL, Stokoe D, Olshen AB, Treseler PA, Chin K, Chen C, Polikoff D, Jain AN, Pinkel D, Albertson DG, Jablons DM, Gray JW. Genomic copy number analysis of non-small cell lung cancer using array comparative genomic hybridization: implications of the phosphatidylinositol 3-kinase pathway. Cancer Res. 2002;62:3636–3640. [PubMed] [Google Scholar]
- 12.Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genomewide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 13.Monni O, Barlund M, Mousses S, Kononen J, Sauter G, Heiskanen M, Paavola P, Avela K, Chen Y, Bittner ML, Kallioniemi A. Comprehensive copy number and gene expression profiling of the 17q23 amplicon in human breast cancer. Proc Natl Acad Sci USA. 2001;98:5711–5716. doi: 10.1073/pnas.091582298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark J, Edwards S, Feber A, Flohr P, John M, Giddings I, Crossland S, Stratton MR, Wooster R, Campbell C, Cooper CS. Genomewide screening for complete genetic loss in prostate cancer by CGH onto cDNA microarrays. Oncogene. 2003;22:1247–1252. doi: 10.1038/sj.onc.1206247. [DOI] [PubMed] [Google Scholar]
- 15.Jiang F, Richter J, Gasser T, Sauter G, Mihatsch MJ, Moch H. Chromosomal imbalances in papillary renal cell carcinoma: genetic differences between histological subtypes. Am J Pathol. 1998;153:1467–1472. doi: 10.1016/S0002-9440(10)65734-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richter J, Jiang F, Gorog JP, Gasser TC, Moch H, Mihatsch MJ, Sauter G. Marked genetic differences between stage pTa and stage pT1 papillary bladder cancer detected by comparative genomic hybridization. Cancer Res. 1997;15:2860–2864. [PubMed] [Google Scholar]
- 17.Jiang F, Lin F, Price R, Gu J, Medeiros LJ, Zhang HZ, Xie SS, Caraway NP, Katz RL. Rapid detection of IgH/BCL2 rearrangement in follicular lymphoma by interphase fluorescence in situ hybridization with bacterial artificial chromosome probes. J Mol Diagn. 2002;4:144–149. doi: 10.1016/S1525-1578(10)60695-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang DY, Brown PO, Eisen MB. Visualizing associations between genome sequences and gene expression data using genomemean expression profiles. Bioinformatics. 2001;17:49–55. doi: 10.1093/bioinformatics/17.suppl_1.s49. [DOI] [PubMed] [Google Scholar]
- 19.Nacht M, Dracheva T, Gao Y, Fujii T, Chen Y, Player A, Akmaev V, Cook B, Dufault M, Zhang M, Zhang W, Guo M, Curran J, Han S, Sidransky D, Buetow K, Madden SL, Jen J. Molecular characteristics of non-small cell lung cancer. Proc Natl Acad Sci USA. 2001;26:15203–15208. doi: 10.1073/pnas.261414598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Racz A, Brass N, Hofer M, Sybrecht GW, Remberger K, Meese EU. Gene amplification at chromosome 1pter-p33 including the genes PAX7 and ENO1 in squamous cell lung carcinoma. Int J Oncol. 2000;17:67–73. doi: 10.3892/ijo.17.1.67. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura H, Saji H, Ogata A, Hosaka M, Hagiwara M, Saijo T, Kawasaki N, Kato H. cDNA microarray analysis of gene expression in pathologic Stage IA nonsmall cell lung carcinomas. Cancer. 2003;97:2798–2805. doi: 10.1002/cncr.11406. [DOI] [PubMed] [Google Scholar]
- 22.Wikman H, Kettunen E, Seppanen JK, Karjalainen A, Hollmen J, Anttila S, Knuutila S. Identification of differentially expressed genes in pulmonary adenocarcinoma by using cDNA array. Oncogene. 2002;22:5804–5813. doi: 10.1038/sj.onc.1205726. [DOI] [PubMed] [Google Scholar]
- 23.Kamimura A, Kamachi M, Nishihira J, Ogura S, Isobe H, Dosaka-Akita H, Ogata A, Shindoh M, Ohbuchi T, Kawakami Y. Intracellular distribution of macrophage migration inhibitory factor predicts the prognosis of patients with adenocarcinoma of the lung. Cancer. 2000;89:334–341. [PubMed] [Google Scholar]
- 24.White ES, Strom SR, Wys NL, Arenberg DA. Non-small cell lung cancer cells induce monocytes to increase expression of angiogenic activity. J Immunol. 2001;166:7549–7555. doi: 10.4049/jimmunol.166.12.7549. [DOI] [PubMed] [Google Scholar]
- 25.Tomiyasu M, Yoshino I, Suemitsu R, Okamoto T, Sugimachi K. Quantification of macrophage migration inhibitory factor mRNA expression in non-small cell lung cancer tissues and its clinical significance. Clin Cancer Res. 2002;8:3755–3760. [PubMed] [Google Scholar]
- 26.White ES, Flaherty KR, Carskadon S, Brant A, Iannettoni MD, Yee J, Orringer MB, Arenberg DA. Macrophage migration inhibitory factor and CXC chemokine expression in non-small cell lung cancer: role in angiogenesis and prognosis. Clin Cancer Res. 2003;9:853–860. [PubMed] [Google Scholar]
- 27.Wang MZ, Howard B, Fitzgerald MC, Patz EF., Jr Protein expression profiling identifies macrophage migration inhibitory factor and cyclophilin a as potential molecular targets in non-small cell lung cancer. Cancer Res. 2003;63:1652–1656. [PubMed] [Google Scholar]
- 28.Pincheira R, Chen Q, Zhang JT. Identification of a 170-kDa protein over-expressed in lung cancers. Br J Cancer. 2001;84:1520–1527. doi: 10.1054/bjoc.2001.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, Tyan T, Cedrone E, Savaraj N, Wang N. Detection of 11q13 amplification as the origin of a homogeneously staining region in small cell lung cancer by chromosome microdissection. Genes Chromosomes Cancer. 1996;17:172–178. doi: 10.1002/(SICI)1098-2264(199611)17:3<172::AID-GCC5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 30.Heighway J, Knapp T, Boyce L, Brennand S, Field JK, Betticher DC, Ratschiller D, Gugger M, Donovan M, Lasek A, Rickert P. Expression profiling of primary non-small cell lung cancer for target identification. Oncogene. 2002;21:7749–7763. doi: 10.1038/sj.onc.1205979. [DOI] [PubMed] [Google Scholar]
- 31.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, Meyerson M. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamada K, Kohno T, Kawanishi M, Ohwada S, Yokota J. Association of CDKN2A(p16)/CDKN2B(p15) alterations and homozygous chromosome arm 9p deletions in human lung carcinoma. Genes Chromosomes Cancer. 1998;22:232–240. doi: 10.1002/(sici)1098-2264(199807)22:3<232::aid-gcc9>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 33.Mariatos G, Gorgoulis VG, Zacharatos P, Kotsinas A, Vogiatzi T, Rassidakis G, Foukas P, Liloglou T, Tiniakos D, Angelou N, Manolis M, Veslemes M, Field JK, Kittas C. Expression of p16(INK4A) and alterations of the 9p21-23 chromosome region in non-small-cell lung carcinomas: relationship with tumor growth parameters and ploidy status. Int J Cancer. 2000;89:133–141. doi: 10.1002/(sici)1097-0215(20000320)89:2<133::aid-ijc6>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 34.Lamy A, Sesboue R, Bourguignon J, Dautreaux B, Metayer J, Frebourg T, Thiberville L. Aberrant methylation of the CDKN2a/p16INK4a gene promoter region in preinvasive bronchial lesions: a prospective study in high-risk patients without invasive cancer. Int J Cancer. 2002;100:189–193. doi: 10.1002/ijc.10474. [DOI] [PubMed] [Google Scholar]
- 35.Junker N, Latini S, Petersen LN, Kristjansen PE. Expression and regulation patterns of hyaluronidases in small cell lung cancer and glioma lines. Oncol Rep. 2003;10:609–616. [PubMed] [Google Scholar]
- 36.Kikuchi T, Daigo Y, Katagiri T, Tsunoda T, Okada K, Kakiuchi S, Zembutsu H, Furukawa Y, Kawamura M, Kobayashi K, Imai K, Nakamura Y. Expression profiles of non-small cell lung cancers on cDNA microarrays: identification of genes for prediction of lymph-node metastasis and sensitivity to anti-cancer drugs. Oncogene. 2003;22:2192–2205. doi: 10.1038/sj.onc.1206288. [DOI] [PubMed] [Google Scholar]
- 37.Borczuk AC, Gorenstein L, Walter KL, Assaad AA, Wang L, Powell CA. Non-small-cell lung cancer molecular signatures recapitulate lung developmental pathways. Am J Pathol. 2003;163:1949–1960. doi: 10.1016/S0002-9440(10)63553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lerman MI, Minna JD. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Consortium. Cancer Res. 2000;60:6116–6133. [PubMed] [Google Scholar]
- 39.Carboni GL, Gao B, Nishizaki M, Xu K, Minna JD, Roth JA, Ji L. CACNA2D2-mediated apoptosis in NSCLC cells is associated with alterations of the intracellular calcium signaling and disruption of mitochondria membrane integrity. Oncogene. 2003;22:615–626. doi: 10.1038/sj.onc.1206134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isler SG, Schenk S, Bendik I, Schraml P, Novotna H, Moch H, Sauter G, Ludwig CU. Genomic organization and chromosomal mapping of SPARC-like 1, a gene down regulated in cancers. Int J Oncol. 2002;8:521–526. doi: 10.3892/ijo.18.3.521. [DOI] [PubMed] [Google Scholar]
- 41.Forbes MA, Cox AM, Cooper EH. A method to detect the in vitro modification of serum beta-2-microglobulin in health and disease. Clin Chim Acta. 1988;177:89–99. doi: 10.1016/0009-8981(88)90311-7. [DOI] [PubMed] [Google Scholar]
- 42.Fan C, Stendahl U, Stjernberg N, Beckman L. Association between orosomucoid types and cancer. Oncology. 1995;6:498–500. doi: 10.1159/000227518. [DOI] [PubMed] [Google Scholar]
- 43.Duche JC, Urien S, Simon N, Malaurie E, Monnet I, Barre J. Expression of the genetic variants of human alpha-1-acid glycoprotein in cancer. Clin Biochem. 2000;33:197–202. doi: 10.1016/s0009-9120(00)00048-5. [DOI] [PubMed] [Google Scholar]
- 44.Fukuhara H, Masvuda M, Yageta M, Fukami T, Kuramochi M, Maruyama T, Kitamura T, Murakami Y. Association of a lung tumor suppressor TSLC1 with MPP3, a human homologue of Drosophila tumor suppressor Dlg. Oncogene. 2003;22:6160–6165. doi: 10.1038/sj.onc.1206744. [DOI] [PubMed] [Google Scholar]
- 45.Eickhoff B, Ruller S, Laue T, Kohler G, Stahl C, Schlaak M, van der Bosch J. Trichostatin A modulates expression of p21waf1/cip1, Bcl-xL, ID1, ID2, ID3, CRAB2, GATA-2, hsp86 and TFIID/TAFII31 mRNA in human lung adenocarcinoma cells. Biol Chem. 2000;381:107–112. doi: 10.1515/BC.2000.015. [DOI] [PubMed] [Google Scholar]
- 46.Czerwinski M, McLemore TL, Gelboin HV, Gonzalez FJ. Quantification of CYP2B7, CYP4B1, and CYPOR messenger RNAs in normal human lung and lung tumors. Cancer Res. 1994;54:1085–1091. [PubMed] [Google Scholar]
- 47.Sagawa N, Fujita H, Banno Y, Nozawa Y, Katoh H, Kuzumaki N. Gelsolin suppresses tumorigenicity through inhibiting PKC activation in a human lung cancer cell line, PC10. Br J Cancer. 2003;88:606–612. doi: 10.1038/sj.bjc.6600739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shieh DB, Godleski J, Herndon JE, II, Azuma T, Mercer H, Sugarbaker DJ, Kwiatkowski DJ. Cell motility as a prognostic factor in Stage I nonsmall cell lung carcinoma: the role of gelsolin expression. Cancer. 1999;85:47–57. doi: 10.1002/(sici)1097-0142(19990101)85:1<47::aid-cncr7>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 49.Dosaka-Akita H, Hommura F, Fujita H, Kinoshita I, Nishi M, Morikawa T, Katoh H, Kawakami Y, Kuzumaki N. Frequent loss of gelsolin expression in non-small cell lung cancers of heavy smokers. Cancer Res. 1998;58:322–327. [PubMed] [Google Scholar]
- 50.Park IW, Wistuba II, Maitra A, Milchgrub S, Virmani AK, Minna JD, Gazdar AF. Multiple clonal abnormalities in the bronchial epithelium of patients with lung cancer. J Natl Cancer Inst. 1999;91:1863–1868. doi: 10.1093/jnci/91.21.1863. [DOI] [PubMed] [Google Scholar]
- 51.Fujii T, Dracheva T, Player A, Chacko S, Clifford R, Strausberg RL, Buetow K, Azumi N, Travis WD, Jen J. A preliminary transcriptome map of non-small cell lung cancer. Cancer Res. 2002;62:3340–3346. [PubMed] [Google Scholar]
- 52.Saitoh H, Shimura S, Fushimi T, Okayama H, Shirato K. Detection of surfactant protein-A gene transcript in the cells from pleural effusion for the diagnosis of lung adenocarcinoma. Am J Med. 1997;103:400–404. doi: 10.1016/s0002-9343(97)00240-4. [DOI] [PubMed] [Google Scholar]
- 53.Betz C, Papadopoulos T, Buchwald J, Dammrich J, Muller-Hermelink HK. Surfactant protein gene expression in metastatic and micrometastatic pulmonary adenocarcinomas and other non-small cell lung carcinomas: detection by reverse transcriptasepolymerase chain reaction. Cancer Res. 1995;19:4283–4286. [PubMed] [Google Scholar]
- 54.Shijubo N, Tsutahara S, Hirasawa M, Takahashi H, Honda Y, Suzuki A, Kuroki Y, Akino T. Pulmonary surfactant protein A in pleural effusions. Cancer. 1992;69:2905–2909. doi: 10.1002/1097-0142(19920615)69:12<2905::aid-cncr2820691207>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 55.Cui Y, Dinman JD, Kinzy TG, Peltz SW. The Mof2/Sui1 protein is a general monitor of translational accuracy. Mol Cell Biol. 1998;18:1506–1516. doi: 10.1128/mcb.18.3.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lian Z, Pan J, Liu J, Zhang S, Zhu M, Arbuthnot P, Kew M, Feitelson MA. The translation initiation factor, hu-Sui1 may be a target of hepatitis B X antigen in hepatocarcinogenesis. Oncogene. 1999;18:1677–1687. doi: 10.1038/sj.onc.1202470. [DOI] [PubMed] [Google Scholar]
- 57.Gstaiger M, Jordan R, Lim M, Catzavelos C, Mestan J, Slingerland J, Krek W. Skp2 is oncogenic overexpressed in human cancers. Proc Natl Acad Sci USA. 2001;98:5043–5048. doi: 10.1073/pnas.081474898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kudo Y, Kitajima S, Sato S, Miyauchi M, Ogawa I, Takata T. High expression of S-phase kinase-interacting protein 2, human F-box protein, correlates with poor prognosis in oral squamous cell carcinomas. Cancer Res. 2001;62:7044–7047. [PubMed] [Google Scholar]
- 59.Harper JW. Protein destruction: adapting roles for Cks proteins. Curr Biol. 2001;5;11(11):431–435. doi: 10.1016/s0960-9822(01)00253-6. [DOI] [PubMed] [Google Scholar]
- 60.Inui N, Kitagawa K, Miwa S, Hattori T, Chida K, Nakamura H, Kitagawa M. High expression of Cks1 in human non-small cell lung carcinomas. Biochem Biophys Res Commun. 2003;303:978–984. doi: 10.1016/s0006-291x(03)00469-8. [DOI] [PubMed] [Google Scholar]