Abstract
Cancers originating from epithelial cells are the most common malignancies. No common expression profile of solid tumors compared to normal tissues has been described so far. Therefore we were interested if genes differentially expressed in the majority of carcinomas could be identified using bioinformatic methods. Complete data sets were downloaded for carcinomas of the prostate, breast, lung, ovary, colon, pancreas, stomach, bladder, liver, and kidney, and were subjected to an expression analysis using SAM. In each experiment, a gene was scored as differentially expressed if the q value was below 25%. Probe identifiers were unified by comparing the respective probe sequences to the Unigene build 155 using BlastN. To obtain differentially expressed genes within the set of analyzed carcinomas, the number of experiments in which differential expression was observed was counted. Differential expression was assigned to genes if they were differentially expressed in at least eight experiments of tumors from different origin. The identified candidate genes ADRM1, EBNA1BP2, FDPS, FOXM1, H2AFX, HDAC3, IRAK1, and YY1 were subjected to further validation. Using this comparative approach, 100 genes were identified as upregulated and 21 genes as downregulated in the carcinomas.
Keywords: Cancer, tumor, microarray, comparative analysis, FOXM1
Introduction
Solid tumors are the second most common cause of death in the western world. Besides very few successes in rare solid tumors such as testicular cancer, the survival rate of most of these tumors is still low. However, many of these tumors are characterized by inactivating mutations of common tumor-suppressor genes, such as p53, p16, or Rb, and activation of oncogenes such as Her2/neu [1,2]. Therefore one might guess that not only these genes are differentially expressed within a broad range of tumors, but that there are also other undescribed genes that are differentially expressed in the majority of tumors.
Large-scale gene expression analysis by means of microarrays has yielded large sets of class 2 cancer genes [3]. This enables us to compare the expression profiles of various tumors and to generate sets of common differentially expressed genes. These genes would then represent a pool of interesting candidates used to give new insights into tumor development, and are candidates for new targets for therapy and diagnostics in a variety of cancers. Attempts have been made to compare the gene expression profiles of tumors of a single entity and to assign differentially expressed genes [4–9]. In addition, gene expression differences of tumors from diverse organs have been analyzed with various approaches [10–20].
Gene expression profiles are obtained by different techniques such as DDRT-PCR, SAGE, expressed sequence tag (EST) sequencing, or microarrays [21,22]. Moreover, within the field of microarrays, there are at least three different rival technologies. The most often used techniques, Affymetrix GeneChips and cDNA on glass microarrays, generate different data types and are accompanied by their own challenges within data preprocessing, leading to sets of gene expression data that are not easy to compare [23].
To identify common differentially expressed genes in solid tumors and to overcome these limitations, we have analyzed the differential expression of genes within each experiment by a single method with defined thresholds for 11 different carcinomas. We identified a set of 121 differentially expressed genes and validated eight of these using dot blots containing the transcriptome of different solid cancers.
Materials and Methods
Data Sets
Only data sets containing values for tumors and normal tissues were selected. Data sets for gene expression profiles were obtained by downloading the expression values in tabular form from the Stanford microarray database (http://genome-www5.stanford.edu//), GEO (http://www.ncbi.nlm.nih.gov/geo/), and the supplementary information of published manuscripts (Table 1). All data sets were used as provided and were normalized if needed by median centering. To unify the used gene identifiers, all probe sequences were compared against the Unigene database version 155 using the BlastN program [24]. Sequences were assigned to a Unigene cluster if their homology was below an e value of e-100, or if their score was higher than 1.5 per base analyzed. Data from the following tumors have been acquired: prostate, breast, lung, ovary, colon, pancreas, stomach, bladder, liver, and kidney.
Table 1.
Used Gene Expression Profiling Data Sets.
| Reference | Tissue | Type of tumor | Type of microarray | Number of tumor | Number of normal | Number of probes/probesets | Source of data |
| Dyrskjot et al. [30] | Bladder | Carcinoma | Affymetrix | 10 | 4 | 8,793 | GEO |
| Sorlie et al. [19] | Breast | Carcinoma | cDNA | 68 | 4 | 8,102 | SMD |
| Notterman et al. [18] | Colon | Adenocarcinoma | Affymetrix | 18 | 18 | 7,129 | Supplement |
| Boer et al. [11] | Kidney | Renal cell carcinoma | cDNA | 32 | 32 | 31,500 | GEO |
| Higgins et al. [15] | Kidney | Renal cell carcinoma | cDNA | 24 | 3 | 22,648 | SMD |
| Chen et al. [12] | Liver | Hepatocellular carcinoma | cDNA | 82 | 75 | 23,075 | SMD |
| Garber et al. [14] | Lung | Adenocarcinoma and squamous cell carcinoma | cDNA | 36 (A) and 13 (S) | 5 | 8,102 | SMD |
| Bhattacharjee et al. [10] | Lung | Adenocarcinoma and squamous cell carcinoma | Affymetrix | 127 (A) and 21 (S) | 17 | 12,625 | Supplement |
| Welsh et al. [20] | Ovary | Adenocarcinoma | Affymetrix | 27 | 5 | 8,793 | Supplement |
| Iacobuzio-Donahue et al. [16] | Pancreas | Adenocarcinoma | cDNA | 11 | 5 | 44,500 | SMD |
| Dhanasekaran et al. [13] | Prostate | Adenocarcinoma | cDNA | 10 | 9 | 9,948 | Supplement |
| Luo et al. [31] | Prostate | Adenocarcinoma | cDNA | 16 | 9 | 6,500 | Supplement |
| Singh et al. [32] | Prostate | Adenocarcinoma | Affymetrix | 52 | 50 | 12,625 | Supplement |
| Welsh et al. [33] | Prostate | Adenocarcinoma | Affymetrix | 25 | 9 | 8,793 | Supplement |
| Leung et al. [17] | Stomach | Adenocarcinoma | cDNA | 89 | 23 | 44,500 | SMD |
Identification of Differentially Expressed Genes
All experiments were analyzed using the SAM package (http://www-stat.stanford.edu/∼tibs/SAM/index.html) with a cutoff of 25% as q value. To the probes identified by this approach, we assigned the value of 1 if they were overexpressed in the tumor, or -1 if they were underexpressed. If a probe was not differentially expressed, we assigned 0; if it was not analyzed in a given experiment, we assigned a blank field. For all probes, we counted the number of differential expression. To identify genes differentially expressed in the majority of carcinomas, we called only those genes as differentially expressed, which displayed a differential expression within eight of the analyzed 11 cancer entities (73%). For the cancer entities of the kidney, lung, and prostate, we obtained more than one data set. For these, differentially expressed genes were identified by each experiment as mentioned above. Subsequently, within a type of cancer (e.g., prostate cancer), the differentially expressed genes were compared and scored as differential only if they were identified within at least 50% of the experiments. Thus, for prostate cancer, genes were counted as differentially expressed if they had been identified within at least two experiments—for kidney and lung cancer, if they had been identified within at least one experiment. Such genes were then counted only once for the comparison between the different cancer entities.
Validation of Differential Expression
CDNA clones for ADRM1 (accession no. BM913272), EBNA1BP2 (BU541488), FDPS (BQ877587), FOXM1 (BQ691509), H2AFX (BG757479), HDAC3 (BM468317), IRAK1 (BE250451), and YY1 (BE746736) were obtained from the RZPD (www.rzpd.de) and prepared using the GFX Micro Plasmid Prep kit according to the manufacturer's recommendation (Amersham Pharmacia Biotech, Freiburg, Germany). The identity of the inserts of the obtained clones was confirmed by sequencing of the clones with an ABI 3700 sequencer and BigDye terminators (Applied Biosystems, Weiterstadt, Germany). For the derived sequences, we analyzed their identity against the clones using the program BlastN and the Unigene database. The inserts of the clones were prepared by digesting the DNA with the appropriate restriction enzymes and purification from agarose gels. The fragments were labelled with α-32P-dCTP (∼ 6.000 Ci/mmol) using the Rediprime II kit (Amersham Pharmacia Biotech) and hybridized to a cancer profiling array II (CPA II) using the ExpressHyb hybridization solution according to the manufacturer's recommendations (Clontech, Heidelberg, Germany).
Autoradiographs were obtained by using a BAS-II scanner (Raytest, Eggenfelden, Germany). For further analysis, intensities were normalized to β-actin and divided by the normalized intensities of the corresponding normal tissues. These data were subjected to visualization using Treeview (http://rana.lbl.gov/EisenSoftware.htm).
Results
Overlap of Data Sets
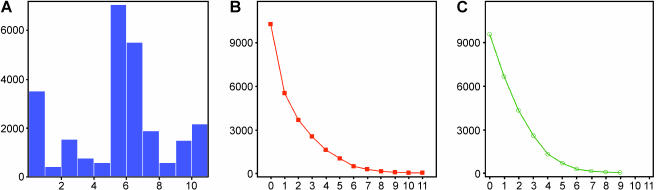
A prerequisite for a comparative analysis of gene expression differences is that a high number of genes are analyzed in more than one experiment. This is especially true for comparisons using microarray data sets with incomplete genome representation. However, within the data sets used, the majority of genes is analyzed by at least six experiments, and more than 4000 genes are interrogated by at least eight experiments (Figure 1A).
Figure 1.
(A) Histogram of the number of genes analyzed by the different experiments. (B and C) Histogram of the number of genes found to be differentially expressed in the different experiments: (B) overexpressed; (C) underexpressed. Interestingly, we did not find a gene that is underexpressed in more than nine experiments, indicating a more heterogeneous gene expression in normal tissues of different origins. X-axis: number of experiments; Y-axis: number of genes.
Identification of Differential Expression
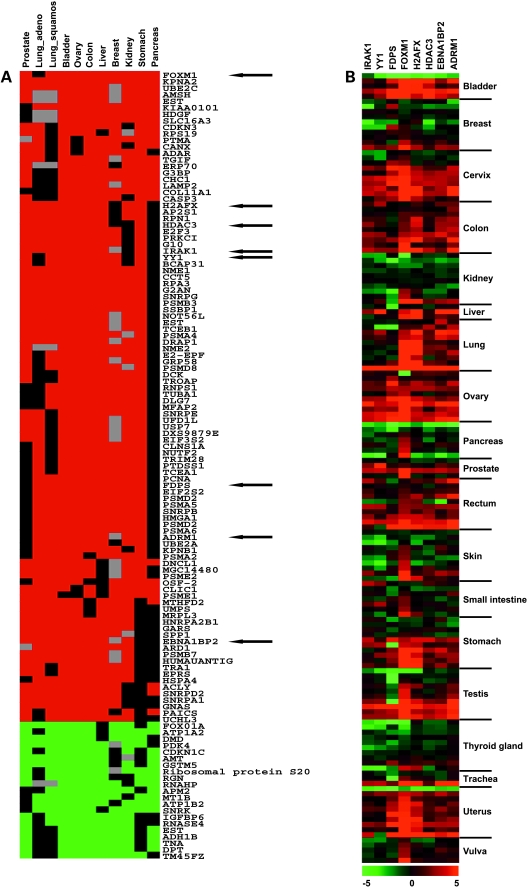
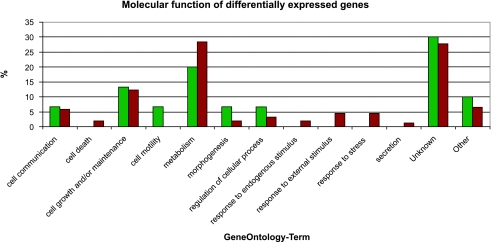
Assignment of significance values is a common procedure in the analysis of gene expression. There are several algorithms known, but the obtained values are rarely corrected for multiple testing. To control the error associated with multiple testing, we used the software SAM (http://wwwstat.stanford.edu/∼tibs/SAM/index.html). We tested several cutoffs for the q value generated by SAM in a comparison of four different prostate experiments. From these results, we concluded that a cutoff of 25% would be sensitive enough to generate a list of common differentially expressed genes because we found an overlap of 50% with the data set generated by Rhodes et al. [25] (data not shown). We also tried to use only those genes with a cutoff below 5%, but failed to identify differentially expressed genes in several studies, probably due to a high data variance, which could in part be attributed to a low sample number of normal tissues in different studies (data not shown). When we analyzed the frequency of classification of a gene as differentially expressed, we observed that most of the genes were never classified as differentially expressed and only a minor number were classified more than five times (Figure 1, B and C). Because we were interested to further analyze genes differentially expressed in the majority of investigated tumors, we chose to examine genes differentially expressed in at least eight tumors. Using this threshold, we identified 100 genes as commonly upregulated and 21 as commonly downregulated in 11 different cancer types from 10 different tissues, as listed in Figure 2A. Within this set, we identified genes such as PCNA and OSF-2, which are well known to be overexpressed within human carcinomas. We then classified the differentially expressed genes according to their purposed function (www.geneontology.org; Figure 3). However, for most of the identified genes, a function has not assigned yet. Of these genes that were characterized by a category, most of them were grouped into the category of metabolism and cell growth/maintenance. Differences in the distribution of the assigned categories between normal and tumor genes could not be observed. Annotation of the genes revealed that many of the genes are either transcriptional regulators, or take part in the degradation of proteins (Figure 2A).
Figure 2.
(A) A total of 121 common differentially expressed genes. Red: Overexpressed in tumors; green: underexpressed in tumors; black: not differentially expressed; grey: not investigated in the original study. Within our comparison, values of +1 for overexpression and -1 for underexpression are assigned. Therefore, no graduations of differential expression are shown. (B) Heatmap of expression of the selected genes on the CPA II. Intensity values for the gene of interest were normalized by the value of β-actin of the respective probe and then divided by the normalized value of the corresponding normal tissue.
Figure 3.
Grouping of identified genes into the molecular function categories of gene ontology using Fatigo (http://fatigo.bioinfo.cnio.es/). Red: Genes overexpressed in tumors; green genes underexpressed in tumors.
Validation of Differential Expression
We selected eight upregulated genes based on their degree of differential expression and proposed function (ADRM1, EBNA1BP2, FDPS, FOXM1, H2AFX, HDAC3, IRAK1, and YY1) and investigated their differential expression by hybridization of a gene-specific probe to a CPA II. The CPA II filter contains the transcriptome from 19 different tumor entities. Each tumor is represented by more than one sample, and a corresponding sample of normal tissue from the same donor is provided. For each of the spots on the array, we also obtained the expression values for β-actin as a “housekeeping” control. To evaluate the differential expression of the genes, we divided each value by the corresponding spot value of β-actin. Furthermore, we divided the normalized values of the tumor by the corresponding normal values (Figure 2B). As expected, most of these genes were overexpressed in a number of samples of different tumor entities displaying the validity of our approach, though we could not observe any upregulation of the genes within the kidney tissues analyzed. Also, only sporadic upregulation was seen in the mammary tumor samples. This might argue against our approach although, except for FOXM1, none of the genes was identified as overexpressed in kidney cancer by our comparative analysis and only five (ADRM1, EBNA1BP2, FDPS, FOXM1, and H2AFX) were identified in breast cancer. Also, we were not able to analyze the quality of the samples in detail. We observed also the heterogeneity of gene expression in the different tumor entities. In our comparative analysis, FOXM1 was overexpressed in 10 of 11 different tumors. This is reflected by the high overexpression of FOXM1 in nearly every tumor analyzed. However, overexpression of FOXM1 could not be observed for some samples of different origins.
Discussion
In an attempt to identify genes commonly overexpressed within solid tumors, we have compared gene expression profiling experiments of 11 different tumors from 10 different organs. We identified the differentially expressed genes using a comparative analysis. The major challenge herein was to unify the different identifiers of the experiments. The data sets used were derived from different microarrays platforms with different structures of identifiers. Whereas Affymetrix GeneChip provides a probeset identifier with a target sequence, the obtained data sets from cDNA microarrays provide only an accession number of the used clones. This leads to the need for a large computational effort. Unfortunately, most of the gene expression data have not been published as raw data so far. Therefore, published articles are the prevalent repository for differentially expressed genes. Comparison of these data has its own merit [26], but only analysis of the raw data circumvents the inherent problems of microarrays within the field of data normalization and expression level assignment. Moreover, comparison of published data is hampered by different modes of gene expression analysis, possibly leading to a subjective choice of genes.
Within this set of analyzed genes, most of them have never been assigned as differentially expressed. Also, only a few genes are identified as differentially expressed in more than five experiments (Figure 1, B and C). From those data, we might conclude that a comparison of microarray data leads not to an accumulation of false positives but to subsets of genes that might be worthwhile in further analyses.
We were interested in those whose expression is deregulated in a majority of tumors. Therefore, we decided to concentrate on genes that are differentially expressed in 8 of 11 analyzed tumors. We identified 100 genes as commonly overexpressed and 21 as commonly downregulated within carcinomas investigated. Interestingly, we were able to identify more genes as commonly overexpressed than genes commonly underexpressed in tumors. This might be due to a higher variance of the observed gene expression in normal tissues. This variance might result from an inadequate low number of these tissues in individual comparisons (i.e., there were only 4 normal breast tissues and 68 tumor tissues analyzed by Sorlie et al. [19]) (Table 1). As normal tissue has to abide by the precise function of an organ, which is different between organs, a tumor has, first of all, the need to grow. To accomplish this task, a tumor has to circumvent several safeguard functions of cell maintenance and growth limitations [1]. To bypass these, only a few genes have to be modified, which might lead to a common tumor phenotype. Therefore, gene expression of tumors of different organs might probably be more homogenous than the gene expression of normal tissues from the same organs.
Within the set of overexpressed genes, we identified genes from different functional categories. Interestingly, genes from the protein degradation pathway by the ubiquitin ligation/proteasome pathway as well as transcription factors were often found. Those genes have already been described as overexpressed within tumors [1,27], therefore demonstrating the feasibility of our approach. In addition, these genes represent key members for new targets for cancer therapy. Validation of differential expression is a key for successful microarray experiments. This should also refer to comparative or meta-analysis. However, tissue samples of different tumor entities with an adequate number of samples can rarely be obtained. We validated the differential expression of 8 of 100 overexpressed genes and observed an overexpression in most cases. That not all genes were overexpressed in all samples might be attributed to either the heterogeneity of gene expression in tumors or perhaps errors of the sampling of tissues, or that the gene was falsely assigned as differentially expressed by our method.
Within the group of overexpressed genes, we identified FOXM1 and ADAR1. FOXM1 is activated by the hedgehog signalling pathway [28], indicating a common activation of this elemental developmental pathway in cancers. ADAR is a member of a diverse family of proteins involved in the editing of mRNA. Overexpression of this gene might lead to a higher amount of edited RNA and might therefore lead to mutated proteins. A high level of ADAR within cancers therefore might act as a mutator mutation. That enzymes of the RNA editing pathway are capable of acting in this manner has been shown for APOBEC [29].
In conclusion, using a novel approach to compare gene expression data, we identified a set of genes that might be useful in the further analysis of fundamental signal transduction pathways that lead to carcinomas.
Acknowledgement
The authors thank Alfred E. Neumann for his precious comments.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Sherr CJ. Principles of tumor suppression. Cell. 2004;116:235–246. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 3.Sager R. Expression genetics in cancer: shifting the focus from DNA to RNA. Proc Natl Acad Sci USA. 1997;94:952–955. doi: 10.1073/pnas.94.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amatschek S, Koenig U, Auer H, Steinlein P, Pacher M, Gruenfelder A, Dekan G, Vogl S, Kubista E, Heider KH, et al. Tissue-wide expression profiling using cDNA subtraction and microarrays to identify tumor-specific genes. Cancer Res. 2004;64:844–856. doi: 10.1158/0008-5472.can-03-2361. [DOI] [PubMed] [Google Scholar]
- 5.Buckhaults P, Zhang Z, Chen YC, Wang TL, St Croix B, Saha S, Bardelli A, Morin PJ, Polyak K, Hruban RH, et al. Identifying tumor origin using a gene expression-based classification map. Cancer Res. 2003;63:4144–4149. [PubMed] [Google Scholar]
- 6.Dennis JL, Vass JK, Wit EC, Keith WN, Oien KA. Identification from public data of molecular markers of adenocarcinoma characteristic of the site of origin. Cancer Res. 2002;62:5999–6005. [PubMed] [Google Scholar]
- 7.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 8.Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov JP, et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci USA. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su AI, Welsh JB, Sapinoso LM, Kern SG, Dimitrov P, Lapp H, Schultz PG, Powell SM, Moskaluk CA, Frierson HF, Jr, et al. Molecular classification of human carcinomas by use of gene expression signatures. Cancer Res. 2001;61:7388–7393. [PubMed] [Google Scholar]
- 10.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boer JM, Huber WK, Sultmann H, Wilmer F, von Heydebreck A, Haas S, Korn B, Gunawan B, Vente A, Fuzesi L, et al. Identification and classification of differentially expressed genes in renal cell carcinoma by expression profiling on a global human 31,500-element cDNA array. Genome Res. 2001;11:1861–1870. doi: 10.1101/gr.184501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, et al. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13:1929–1939. doi: 10.1091/mbc.02-02-0023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 14.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins JP, Shinghal R, Gill H, Reese JH, Terris M, Cohen RJ, Fero M, Pollack JR, van de Rijn M, Brooks JD. Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray. Am J Pathol. 2003;162:925–932. doi: 10.1016/S0002-9440(10)63887-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq R, et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung SY, Chen X, Chu KM, Yuen ST, Mathy J, Ji J, Chan AS, Li R, Law S, Troyanskaya OG, et al. Phospholipase A2 group IIA expression in gastric adenocarcinoma is associated with prolonged survival and less frequent metastasis. Proc Natl Acad Sci USA. 2002;99:16203–16208. doi: 10.1073/pnas.212646299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001;61:3124–3130. [PubMed] [Google Scholar]
- 19.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welsh JB, Zarrinkar PP, Sapinoso LM, Kern SG, Behling CA, Monk BJ, Lockhart DJ, Burger RA, Hampton GM. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci USA. 2001;98:1176–1181. doi: 10.1073/pnas.98.3.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed FE. Molecular techniques for studying gene expression in carcinogenesis. J Environ Sci Health Part C Environ Carcinog Ecotoxicol Rev. 2002;20:77–116. doi: 10.1081/GNC-120016201. [DOI] [PubMed] [Google Scholar]
- 22.Grutzmann R, Pilarsky C, Staub E, Schmitt AO, Foerder M, Specht T, Hinzmann B, Dahl E, Alldinger I, Rosenthal A, et al. Systematic isolation of genes differentially expressed in normal and cancerous tissue of the pancreas. Pancreatology. 2003;3:169–178. doi: 10.1159/000070087. [DOI] [PubMed] [Google Scholar]
- 23.Kuo WP, Jenssen TK, Butte AJ, Ohno-Machado L, Kohane IS. Analysis of matched mRNA measurements from two different microarray technologies. Bioinformatics. 2002;18:405–412. doi: 10.1093/bioinformatics/18.3.405. [DOI] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Rhodes DR, Barrette TR, Rubin MA, Ghosh D, Chinnaiyan AM. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res. 2002;62:4427–4433. [PubMed] [Google Scholar]
- 26.Grutzmann R, Saeger HD, Luttges J, Schackert HK, Kalthoff H, Kloppel G, Pilarsky C. Microarray-based gene expression profiling in pancreatic ductal carcinoma: status quo and perspectives. Int J Colorectal Dis. 2004 doi: 10.1007/s00384-003-0563-3. In press. [DOI] [PubMed] [Google Scholar]
- 27.Voorhees PM, Dees EC, O'Neil B, Orlowski RZ. The proteasome as a target for cancer therapy. Clin Cancer Res. 2003;9:6316–6325. [PubMed] [Google Scholar]
- 28.Teh MT, Wong ST, Neill GW, Ghali LR, Philpott MP, Quinn AG. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res. 2002;62:4773–4780. [PubMed] [Google Scholar]
- 29.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 30.Dyrskjot L, Thykjaer T, Kruhoffer M, Jensen JL, Marcussen N, Hamilton-Dutoit S, Wolf H, Orntoft TF. Identifying distinct classes of bladder carcinoma using microarrays. Nat Genet. 2003;33:90–96. doi: 10.1038/ng1061. [DOI] [PubMed] [Google Scholar]
- 31.Luo J, Duggan DJ, Chen Y, Sauvageot J, Ewing CM, Bittner ML, Trent JM, Isaacs WB. Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res. 2001;61:4683–4688. [PubMed] [Google Scholar]
- 32.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D'Amico AV, Richie JP, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 33.Welsh JB, Sapinoso LM, Su AI, Kern SG, Wang-Rodriguez J, Moskaluk CA, Frierson HF, Jr, Hampton GM. Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 2001;61:5974–5978. [PubMed] [Google Scholar]