Abstract
BACKGROUND
Molecular predictors for the effectiveness of adjuvant chemotherapy in colorectal cancer are of considerable clinical interest. To this aim, we analyzed the serine threonine receptor-associated protein (STRAP), an inhibitor of TGF-β signaling, with regard to prognosis and prediction of adjuvant 5-FU chemotherapy benefit.
METHODS
The gene copy status of STRAP was determined using quantitative realtime polymerase chain reaction in 166 colorectal tumor biopsies, which had been collected from a randomized multicenter trial of 5-fluorouracil (5-FU)/mitomycin C (MMC) adjuvant chemotherapy of the Swiss Group for Clinical Cancer Research (SAKK).
RESULTS
Amplification of STRAP was found in 22.8% of the tumors. When left without adjuvant chemotherapy, patients bearing tumors with a STRAP amplification had a significantly better prognosis (hazard ratio for death: 0.26; P = .004). Interestingly, these patients, when receiving adjuvant treatment, had a worse survival (hazard ratio for death: 3.48; P = .019) than without chemotherapy, whereas patients carrying tumors with diploidy or deletion of STRAP benefited from the treatment (hazard ratio for death: 0.44; P = .052). This suggests the amplification of STRAP as a strong predictor of an unfavorable effect of 5-FU-based adjuvant chemotherapy.
CONCLUSION
If confirmed, the STRAP gene copy status might provide a parameter to decide about the use of 5-FU-based adjuvant chemotherapy.
Keywords: TGF-β, SMAD, STRAP, colorectal cancer, predictive, prognostic markers
Introduction
The benefit of adjuvant chemotherapy in colorectal cancer has been assessed in large phase III trials for which patients had been selected with regard to tumor stage as the strongest indicator of prognosis. In stage III carcinoma of the colon, adjuvant chemotherapy with 5-fluorouracil (5-FU) and leucovorin has become a standard treatment because a significant increase of time to relapse and a 10% to 12% absolute improvement in overall survival (OS) have been shown. The benefit of adjuvant chemotherapy in stage II patients who have a better prognosis is less clear and remains controversial [1]. However, even in patients with stage III disease, approximately 40% can be cured by surgery alone, and around 50% will die within 5 years despite adjuvant treatment. Therefore, molecular markers predicting the benefit of adjuvant chemotherapy in an individual patient are urgently needed.
Generally, predictive markers are molecules involved in the metabolism of cytotoxic agents or in signaling pathways leading to cell proliferation, growth control, and apoptosis. As potential targets of 5-FU-containing regimens, thymidilate synthase (TS), dihydropyrimidine dehydrogenase (DPD), and thymidilate phosphorylase (TP), as well as p53, BCL-2 (B-cell CLL/lymphoma 2), and microsatellite instability (MSI) have been analyzed [2]. High expression levels of TS [3], DPD [4], and TP [5] mRNA were associated with resistance to 5-FU treatment in some—but not all—studies and, similarly, the role of p53 as a predictive marker in the clinical situation is still unclear [6]. High expression of BCL-2, an inhibitor of apoptosis, correlates with improved survival, and early reports suggested that BCL-2 expression is a predictive marker for a 5-FU response [7]. However, confirmatory studies are needed before BCL-2 expression can be used to guide decisions on adjuvant chemotherapy of colorectal cancer. Finally, MSI, a marker of functional deficiency of mismatch repair genes, has been associated with an increased likelihood of 5-FU benefit in one report [8], whereas others [9] did not confirm this finding. In summary, none of the potential predictive molecular markers is sufficiently evaluated to be used in the decision for or against adjuvant chemotherapy of colorectal cancer.
Sporadic colorectal cancer develops as a result of an accumulation of genetic alterations [10], including the inactivation of the adenomatous polyposis coli (APC)/β-catenin pathway, mutation of the k-RAS gene, deletion of chromosome 17p (the genetic locus of the p53 gene), and loss of chromosome 18q harboring deleted in colorectal cancer (DCC) and the TGF-β signal-transducing molecules SMAD2 [mothers against decapentaplegic (MAD), Drosophila, homolog 2], SMAD4, and SMAD7 [11]. The importance of the TGF-β pathway in colorectal tumorigenesis is underlined by the finding of inactivating mutations of the TGF-β receptor II, SMAD2, SMAD4, and TGF-β itself [12–14]. It therefore has been speculated that most colorectal cancers carry an alteration in at least one component of the TGF-β signaling pathway [15].
The serine threonine receptor-associated protein (STRAP) is a TGF-β pathway inhibitor interacting at the receptor level. It associates with the TGF-β I and II receptors [16], and overexpression has been shown to inhibit TGF-β-mediated activation of transcription. Specifically, STRAP synergizes with SMAD7, an antagonistic SMAD, in the inhibition of TGF-β signaling by preventing the access of SMAD2 and SMAD3 to the receptor [17].
To test whether STRAP is of prognostic value or predictive for chemotherapy benefit, we analyzed a collection of tumor specimens from a randomized trial of the Swiss Group for Clinical Cancer Research (SAKK 40/81) evaluating the effect of 5-FU-based adjuvant chemotherapy in stage II and stage III colorectal cancer [18]. The gene dosage of STRAP in the tumors was determined by quantitative real-time polymerase chain reaction (PCR) and the gene copy status was correlated with survival data.
Patients and Methods
Patients
Paired primary tumor and normal tissue biopsies were obtained from patients involved in a randomized trial on the benefit of 5-FU/MMC adjuvant chemotherapy versus surgery alone (SAKK study 40/81) [18]. The adjuvant treatment consisted of an immediate postoperative infusion with 5-FU (500 mg/m2 per day) for 7 days and one single dose of mitomycin C (MMC; 10 mg/m2) on day 1. The SAKK 40/81 trial population comprised 533 patients younger than 75 years with a median of 62 years. About 63.4% of the patients had colonic carcinomas and 36.6% had rectal carcinomas. A total of 62.4% were node-negative (stage II) and 31.1% were node-positive (stage III); in 6.5%, the nodal status was not assessed. Details of the trial have been described previously [18]. The subpopulation of which tumor specimens were successfully analysed for STRAP gene copy status (Table 1, first column) was very similar to the whole SAKK 40/81 trial population with respect to age, stage, and tumor location. The study was approved by the local ethics committees of the participating institutions.
Table 1.
Clinical Characteristics of Colorectal Cancer Patients from the SAKK 40/81 Trial Whose Tumors Were Evaluated for the STRAP Gene Copy Status.
| All patients (n = 166) | Amplification (n = 38) | Deletion (n = 75) | Normal Diploidy (n = 53) | P (Fisher's Exact Test) | |
| Median age (years) | 63 | 61 | 63 | 63 | |
| Sex [n, (%)] | |||||
| Male | 79(48) | 20 (53) | 37 (49) | 22 (42) | .56 |
| Female | 87 (52) | 18 (47) | 38 (51) | 31 (58) | |
| Lymph node status [n, (%)] | |||||
| Positive | 57 (34) | 14 (37) | 22 (29) | 21 (40) | .69 |
| Negative | 102 (62) | 23 (61) | 50 (67) | 29 (55) | |
| Missing information | 7 (4) | 1 (3) | 3 (4) | 3 (6) | |
| Tumor size [n, (%)] | |||||
| Locally advanced (T stage ≥ 3) | 125 (75) | 31 (82) | 55 (73) | 39 (74) | .60 |
| Locally not advanced (T stage < 3) | 41 (25) | 7 (18) | 20 (27) | 14 (26) | |
| Tumor location [n, (%)] | |||||
| Rectum | 45 (27) | 13 (34) | 16 (21) | 16 (30) | .29 |
| Colon | 121 (73) | 25 (66) | 59 (79) | 37 (70) | |
| Adjuvant chemotherapy [n, (%)] | |||||
| Yes | 65 (39) | 17 (45) | 24 (32) | 24 (45) | .23 |
| No | 101 (61) | 21 (55) | 51 (68) | 29 (55) | |
Methods
Genomic DNA was extracted from formalin-fixed and paraffin-embedded tumor and normal tissue biopsies derived from the same patients using the Nucleospin C+T kit (Macherey-Nagel, Dürren, Germany), according to the manufacturer's instructions.
The gene copy status of STRAP was established by gene dosage with real-time quantitative PCR using the TaqMan system on an ABI Prism 7700 sequence detector (PE Applied Biosystems, Foster City, USA), as described previously [19]. The protocol for gene dosage is given in Boulay et al. [19]. To identify the human STRAP gene (AB 024327: MAWD, Homo sapiens pt-wd mRNA for WD-40 repeat protein), the following primers were selected: STRAPf, cgcgacccgtggttga; STRAPr, aagcgctgattaagaaatacccata; probe: STRAP, ttggccttcagtggcatcacgc.
For each individual, the Ct value obtained for the autosomal reference gene 36B4 (calculated by the built-in software) on normal tissue was subtracted from that of the tumor tissue, thus defining ΔCt 36B4. An analogous calculation was made for STRAP. The gene copy status is indicated by the ΔCt value (ΔCt 36B4 - ΔCt STRAP) as follows: -0.45 < ΔCt < 0.45: normal diploidy; ΔCt < -0.55: deletion; ΔCt > 0.55: amplification. Molecular analysis was performed blinded to the clinical data.
Statistical Analysis
Two multiple Cox regression models were performed to test the prognostic and predictive values of STRAP status separately on disease-free survival (DFS) and OS. Main effects of STRAP status were represented by two covariates: amplification and deletion, with no change at the reference level. Interaction effects between STRAP status and treatment were represented by two interaction terms: treatment x amplification and treatment x deletion. In addition to covariates of STRAP and treatment indicator, five known prognostic factors (i.e., patients' age at trial entry, sex, lymph node status, tumor size, and tumor location) were also included in the regression analysis as confounding variables to control for their influence. The Cox regressions were performed on 159 of 166 STRAP-informative patients because the clinical data were incomplete for seven of them (Table 1).
The test for prognostic effects was based on the statistical significance of the coefficients for the terms gene amplification and gene deletion, and the test of predictive effects on the treatment x amplification or treatment x deletion interaction terms. A significant treatment x amplification interaction would mean that the risk for patients with amplification relative to patients without change at the STRAP locus differs between untreated and treated patients. Analogously, it also would mean that the relative risk of treated versus untreated patients differs between patients with STRAP amplification and those with no change. The same logic applies for the treatment x deletion interaction. Survival curves were constructed with the Kaplan-Meier method.
Following regression, diagnostics was employed to check the quality of our Cox models [20]. For each covariate, we graphically checked the scaled Schoenfeld residuals and ran a test to check proportional hazard assumption that underlies the use of Cox regressions; deviance residuals were plotted to discover patients who are poorly predicted by our statistical models; score residuals were plotted to discover individual patients who have a large influence on the models.
Results
To determine the prevalence of STRAP amplifications and STRAP deletions in early colorectal cancer, we analyzed 182 paired tumor and non-neoplastic DNA specimens collected from patients of the SAKK 40/81 trial. Of the 182 specimens, 166 (91%) were informative for the STRAP gene copy status. Complete or partial allelic loss of STRAP was found in 75 (45.2%), normal diploidy in 53 (32%), and gene amplification in 38 (22.8%) tumors, respectively. We did not find any statistically significant association between STRAP gene copy status (amplification, deletion, and normal diploidy) and either gender, tumor size, lymph node status, tumor location, or received treatment (Table 1). These clinical characteristics were similar for all three subgroups. A summary table with these clinical characteristics of individual patients further illustrates the absence of an association of STRAP amplification with any specific combination of them (Table 2).
Table 2.
Clinical Characteristics of Individual Patients with STRAP Amplification of Their Tumors.
| Patient ID Number | Age (years) | Sex | Lymph Node Status | Tumor Size | Tumor Location |
| 8 | 66 | Female | Positive | Locally advanced | Colon |
| 9 | 71 | Female | Positive | Locally advanced | Colon |
| 17 | 54 | Male | Positive | Locally advanced | Colon |
| 34 | 46 | Female | Negative | Locally advanced | Colon |
| 36 | 65 | Male | Negative | Locally advanced | Colon |
| 43 | 55 | Male | Negative | Locally advanced | Colon |
| 54 | 57 | Male | Negative | Locally advanced | Colon |
| 83 | 49 | Female | Positive | Locally advanced | Colon |
| 150 | 60 | Male | Negative | Locally advanced | Colon |
| 164 | 70 | Male | Negative | Locally advanced | Colon |
| 271 | 72 | Male | Negative | Locally advanced | Colon |
| 290 | 73 | Male | Negative | Locally not advanced | Colon |
| 357 | 60 | Female | Negative | Locally advanced | Colon |
| 359 | 64 | Male | Negative | Locally advanced | Colon |
| 365 | 60 | Female | Negative | Locally advanced | Colon |
| 372 | 64 | Female | Negative | Locally advanced | Colon |
| 379 | 44 | Female | Negative | Locally not advanced | Colon |
| 383 | 39 | Female | Negative | Locally advanced | Colon |
| 384 | 62 | Male | Positive | Locally advanced | Colon |
| 393 | 59 | Female | Negative | Locally advanced | Colon |
| 418 | 66 | Female | Negative | Locally advanced | Rectum |
| 443 | 54 | Male | Negative | Locally advanced | Rectum |
| 452 | 50 | Male | Positive | Locally not advanced | Rectum |
| 453 | 74 | Male | Positive | Locally advanced | Rectum |
| 455 | 59 | Male | NA | Locally not advanced | Rectum |
| 456 | 48 | Female | Negative | Locally not advanced | Rectum |
| 478 | 59 | Female | Positive | Locally advanced | Rectum |
| 482 | 59 | Male | Positive | Locally not advanced | Rectum |
| 488 | 71 | Female | Negative | Locally advanced | Rectum |
| 529 | 74 | Male | Negative | Locally advanced | Colon |
| 550 | 68 | Female | Negative | Locally advanced | Rectum |
| 560 | 66 | Female | Positive | Locally advanced | Colon |
| 564 | 62 | Female | Positive | Locally advanced | Colon |
| 567 | 44 | Male | Negative | Locally advanced | Colon |
| 573 | 70 | Male | Positive | Locally not advanced | Colon |
| 636 | 63 | Female | Positive | Locally advanced | Rectum |
| 652 | 60 | Male | Negative | Locally advanced | Rectum |
| 655 | 65 | Male | Positive | Locally advanced | Rectum |
NA = not assessed.
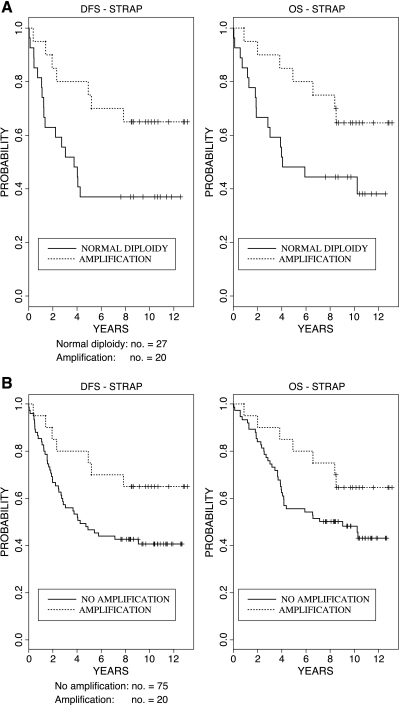
In patients who did not receive adjuvant treatment (control arm of the SAKK 40/81 trial), the STRAP gene copy status was prognostic for DFS and OS. Compared to patients with normal diploidy at the STRAP locus, amplification of STRAP significantly reduced the relative risk of recurrence (P = .005) and the relative risk of death (P = .004) (Table 3). Patients with STRAP amplification in their tumors showed a much better DFS and OS than patients harboring tumors with normal diploidy of STRAP (Figure 1A). Contrary to STRAP amplification, STRAP deletion did not have any statistically significant association with either DFS or OS (Table 3). Because there was no significant association of STRAP deletion and survival, we pooled patients bearing tumors with normal diploidy or deletion of STRAP (“no amplification”) and compared them to patients with tumors harboring a STRAP amplification. This analysis, including all patients who did not receive adjuvant chemotherapy, allows testing whether STRAP amplification separates a specific prognostic subgroup from the rest of the patients. In fact, a significant prognostic effect of STRAP amplification was confirmed with respect to DFS (P = .011) and OS (P = .020) (Table 3 and Figure 1B). Because our multivariable Cox regressions contain sex, tumor size, lymph node stage, and tumor location as covariates, the hazard ratio for STRAP amplification is adjusted for these variables (i.e., the results for STRAP amplification are independent of any effects of sex, size, lymph node stage, and tumor location). These findings suggest a role of STRAP amplification as an independent favorable prognostic marker in early colorectal cancer.
Table 3.
Prognostic Relevance of STRAP Gene Copy Status in Patients Without Adjuvant Chemotherapy.
| Markers | Relative Risk of Recurrence (95% CI) | P | Relative Risk of Death (95% CI) | P |
| Amplification (n = 20) versus normal diploidy (n = 27) | 0.27 (0.11–0.68) | .005 | 0.26 (0.10–0.70) | .004 |
| Deletion (n = 48) versus normal diploidy (n = 27) | 0.71 (0.35–1.36) | .280 | 0.60 (0.25–1.05) | .130 |
| Amplification (n = 20) versus no amplification (n = 75) | 0.34 (0.15–0.79) | .011 | 0.37 (0.16–0.85) | .020 |
The relative risk has been calculated using a multivariate Cox proportional hazard model. The relative risk of patients with normal diploidy of STRAP is 1.
Figure 1.
Prognostic effect of STRAP amplification on DFS and OS in the untreated patient group. (A) Kaplan-Meier curves for DFS and OS of patients without adjuvant chemotherapy who are harboring tumors with STRAP amplification versus tumors with normal diploidy of STRAP. In the absence of adjuvant chemotherapy, the patients with tumors displaying STRAP amplification had a significantly longer DFS and higher OS [relative risk of recurrence: 0.27 (95% confidence interval: 0.11–0.68, P = .005) and relative risk of death: 0.26 (95% confidence interval: 0.10–0.70, P = .004)]. (B) Kaplan-Meier curves for DFS and OS of all patients without adjuvant chemotherapy. Patients harboring tumors with STRAP amplification versus pooled patients with tumors containing diploidy or deletion of STRAP (“;no amplification”).
Among patients who received adjuvant 5-FU/MMC chemotherapy, we also found a significant association of the STRAP gene copy status with prognosis. But surprisingly, chemotherapy-treated patients bearing tumors with STRAP amplification had a significantly shorter DFS [HR for relapse 2.61 (1.05-6.48), P = .039] and a trend toward worse OS [HR for death 2.05 (0.79-5.37), P = .140] than patients bearing tumors with normal diploidy at the STRAP locus. Thus, the effect of the STRAP gene copy status differed between the treatment groups.
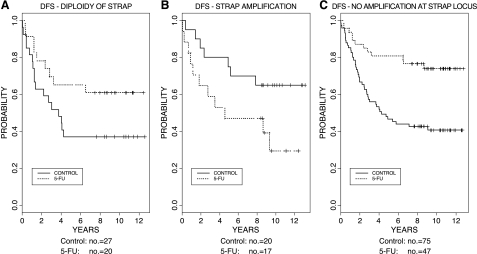
Because the patients from whom tumor specimens were collected were randomized to receive perioperative adjuvant 5-FU/MMC chemotherapy or no adjuvant treatment [18], we were able to test whether the gene copy status of STRAP was correlated with the treatment effect. With regard to DFS, the significant interaction of STRAP amplification and treatment (treatment x amplification interaction term: hazard ratio for relapse = 9.58; P = .001) (Table 4) showed that the STRAP status can predict the effect of treatment. Specifically, among patients with tumors having normal diploidy of STRAP, those receiving adjuvant treatment had a relative risk of relapse of .42 (P = .041) compared with those without adjuvant therapy (i.e., they derive a significant benefit from the treatment) (Table 5). On the other hand, among patients bearing tumors with an amplification of STRAP, those receiving adjuvant treatment had a 4.07 times higher relative risk of relapse (P = .006) than those without adjuvant therapy (Table 5). These results are illustrated by Figure 2, A and B, which shows a better DFS for treated patients in the group with normal diploidy of STRAP, but a worse DFS for treated patients in the group with STRAP amplification. Similarly, there was a significant interaction between treatment and STRAP amplification with regard to OS (P = .003). Treatment with 5-FU/MMC reduced the relative risk of death to .44 (P = .052) in patients bearing tumors with normal diploidy of STRAP, whereas it increased the relative risk of death 3.48-fold (P = .019) in patients carrying a tumor with STRAP amplification.
Table 4.
Predictive Value of STRAP Amplification.
| STRAP | Relative Risk of Recurrence (95% CI) | P | Relative Risk of Death (95% CI) | P |
| STRAP amplification (n = 37) versus normal diploidy (n = 50) | 9.58 | .001 | 7.95 | .003 |
| STRAP amplification (n = 37) versus no amplification (n = 122) | 12.79 | <.001 | 10.37 | <.001 |
The amplification x treatment interaction in the multivariate proportional hazard model is highly significant for DFS and OS in comparison to either patients with diploidy of strap in their tumors, or to pooled patients with tumors containing diploidy or deletion of strap (no amplification).
Table 5.
Influence of STRAP Gene Copy Status on Treatment Effect.
| Markers | Relative Risk of Recurrence (95% CI) | P | Relative Risk of Death (95% CI) | P |
|
STRAP normal diploidy Treated (n = 23) versus untreated (n = 27) |
0.42 (0.19–0.96) | .041 | 0.44 (0.19–1.01) | .052 |
|
STRAP amplification Treated (n = 17) versus untreated (n = 20) |
4.07 (1.51–11.00) | .006 | 3.48 (1.23–9.85) | .019 |
|
STRAP no amplification Treated (n = 48) versus untreated (n = 80) |
0.32 (0.17–0.62) | <.001 | 0.33 (0.17–0.66) | .002 |
The relative risk of patients who did not receive adjuvant chemotherapy is 1.
Figure 2.
Predictive effect of STRAP amplification. (A) Effect of 5-FU/MMC adjuvant treatment on DFS in patients with tumors bearing an amplification at STRAP locus (amplification) compared with patients with diploidy of STRAP. Patients with diploidy of STRAP in their tumors who had received adjuvant treatment had a relative risk of relapse of 0.42 (P = .041) compared with those without adjuvant therapy. (B) However, among patients bearing tumors with an amplification of STRAP, those receiving adjuvant treatment had a 4.07 times higher relative risk of relapse (P = .006) compared with those without therapy. (C) Pooled patients (no amplification) having tumors with either normal diploidy of STRAP or deletion of STRAP have a relative risk of relapse of 0.32 when they receive 5-FU/MMC treatment compared to patients without adjuvant treatment.
Contrary to STRAP treatment x amplification interaction, STRAP treatment x deletion interaction did not show a statistically significant association with either DFS or OS (DFS: treatment x deletion: HR = 0.38, P = .190; OS: treatment x deletion: HR = 0.28, P = .140). This allows us to compare patients with tumors harboring a STRAP amplification to pooled patients with normal diploidy or deletion of STRAP (“no amplification”). The results of this analysis were highly significant (P < .001), confirming the strong predictive relevance of STRAP amplification (Table 4). The survival curves of all patients without amplification of STRAP (“no amplification” group) illustrate the high benefit they derive from 5-FU/MMC adjuvant therapy (Figure 2C) similar to the patients with tumors containing a normal diploidy of STRAP (Figure 2A).
The analysis of Schoenfeld residuals did not show any evidence of departure from the proportional hazards assumption. Deviance residuals did not identify any patients who were poorly predicted by our models. No single observations showed a large influence on the multivariate models. Because all our multivariable Cox regressions contain gender, age at study entry, site, and stage of disease as covariates, the hazard ratios for treatment, amplification, and their interaction are adjusted for these confounding parameters. Thus, our findings strongly support a role of STRAP amplification as an independent negative predictive marker for 5-FU-based chemotherapy benefit in early colorectal cancer patients.
Discussion
Adjuvant chemotherapy of colorectal cancer is impaired by at least two major obstacles: the lack of efficacy of current standard treatments, and our inability to predict which patients will derive benefit from this frequently toxic intervention. Response rates to modern chemotherapy regimens in advanced disease, containing eloxatin and/or CPT-11 [21,22], have given rise to the hope that improved survival might be achievable in patients with early colorectal cancer, but results of phase III trials comparing these regimens to standard 5-FU-based adjuvant treatments are pending.
We have determined the prevalence of STRAP deletions and amplifications in a large number (n = 166) of patients with colorectal cancer. Amplification of STRAP in 22% of the tumors is in line with the published amplification frequency of chromosome 12p12, where STRAP is located, which has been found in 9 of 30 primary colon carcinomas using comparative genomic hybridization [23]. Interestingly, chromosome 12p12 has also been described to be amplified in pancreatic, gastric, oral squamous cell, and testicular germ cell cancers, suggesting a broader role for this molecular alteration in tumorigenesis [24]. In addition to amplification, we have also observed the frequent deletion of the STRAP locus, again in line with findings in several other tumor entities, such as ovarian cancer, childhood acute lymphatic leukemia, and prostate carcinoma [25].
Patients in the control arm of the 40/81 study who did not receive adjuvant therapy allowed us to determine the true survival advantage of patients whose tumors had a STRAP amplification. Although amplified in only around 22% of the tumors, STRAP turned out to be an independent prognostic marker for better DFS (P = .0098) as well as OS (P = .0054) (Table 3). It is tempting to speculate that the positive prognosis associated with amplification of STRAP is explained by the increased formation of the STRAP/SMAD7 complex, one of the physiological pathways of inhibition of TGF-β signaling, and thus tumor cell proliferation. In a recent study, amplification of STRAP was uniformly associated with overexpression of its protein product in the cytoplasm of 21 of 46 breast cancer tissue specimens compared with matched normal breast tissues [26]. Assuming that in colorectal cancer STRAP amplification also leads to protein overexpression, it is conceivable that the TGF-β pathway is inhibited more efficiently by STRAP in tumors carrying an amplification of this gene. Because in late tumor stages TGF-β signaling has been shown to enhance tumor progression [27], amplification of a TGF-β pathway inhibitor should be protective and lead to a more favorable prognosis. Results of preliminary in vitro experiments in our laboratory showing an inhibition of proliferation of tumor cell lines when STRAP is overexpressed (data not shown) are well in line with this hypothesis. Our data are also in accordance with the observation of Markowitz et al. [12], who found a better prognosis of hereditary nonpolyposis colon cancer patients carrying inactivating mutations of the TGF-β II receptor.
By far, the most interesting finding of our study is the deleterious effect of adjuvant chemotherapy on patients with a STRAP-amplified carcinoma. These patients had a nearly three-fold better chance to be alive and free of disease if left untreated compared to the same molecularly defined group of patients after treatment with adjuvant chemotherapy (Tables 4 and 5, Figure 2). Obviously, this leaves patients without STRAP amplification with a correspondingly higher benefit of adjuvant treatment compared with an unselected group of patients with colorectal cancer.
Our data on STRAP amplification as a predictive factor for chemotherapy benefit might help to better select patients for future adjuvant chemotherapies in colorectal cancer. However, this analysis of a molecular marker for chemotherapy benefit is limited by the inherent problem of many similar analyses i.e., that a treatment regimen for which mature clinical follow-up data are available has frequently been replaced by a more modern standard when the molecular analyses are performed. A further limitation of our study relates to the fact that we cannot formally separate the potential effects of the two drugs within this regimen. Although the perioperative 5-FU/MMC treatment regimen used in the SAKK 40/81 trial is not today's clinical standard, it was clearly shown to be an effective treatment in this population [18]. Taking these limitations into consideration, it seems plausible that the predictive value of STRAP amplification we have shown for short-term adjuvant 5-FU/MMC might also be present when the current standard regimen of 6 months of 5-FU/leucovorin or future eloxatin and/or CPT-11-containing regimens is analyzed. This hypothesis, however, needs to be tested in independent large, randomized trials.
How can this provocative finding be explained? Because patients harboring tumors with an amplification of STRAP have a higher relapse and death rate with than without adjuvant chemotherapy, 5-FU resistance cannot explain this finding. It must result from a direct unfavorable effect of 5-FU on tumor biology in this situation. On a purely descriptive level, 5-FU seems to abolish the positive prognostic effect of STRAP amplification because the survival of patients harboring tumors with a STRAP amplification after adjuvant chemotherapy is very similar to the survival of patients with a normal diploidy of STRAP in their tumors when left without adjuvant chemotherapy (Figure 2, A and B).
We have reported that deletion of another member of the TGF-β signaling pathway, SMAD4, was also associated with reduced benefit of 5-FU/MMC adjuvant chemotherapy in the same collection of tumor specimens [28]. Because SMAD4 is an agonist and STRAP is an antagonist in the TGF-β pathway, it is consistent that deletion of the first and amplification of the latter have the same effect on chemotherapy efficacy. Certainly, these two studies suggest that the effect of 5-FU-based chemotherapy might be dependent on TGF-β signaling. In vitro experiments, which demonstrate that 5-FU inhibits TGF-β-induced expression of collagen type I in fibroblasts through JNK/AP-1 activation [29], support this hypothesis of an interference between 5-FU and TGF-β signaling. In the context of STRAP amplification 5-FU might lead to a severe imbalance in the TGF-β signaling pathway, which provokes an activation of undetectable residual neoplastic cells after removal of the primary tumor.
In conclusion, amplification of STRAP defines a subgroup of colorectal cancer patients with a favorable prognosis, who, according to current clinical standards, would receive adjuvant chemotherapy with a deleterious effect. Limiting treatment to those patients who are likely to benefit might spare a substantial minority of patients the side effects of a useless or even harmful chemotherapy, and thus decrease the ineffective use of resources and, perhaps, improve longterm results. If confirmed, determination of the STRAP gene copy status might be a helpful parameter in the management of patients with stage II and stage III colorectal cancer.
Abbreviations
- DFS
disease-free survival
- OS
overall survival
- STRAP
serine threonine receptor-associated protein
- 5-FU
5-fluorouracil
- MMC
mitomycin C
Footnotes
This work was sponsored by grants from the Swiss Cancer League and Krebsliga Beider Basel.
References
- 1.Wils J, O'Dwyer P, Labianca R. Adjuvant treatment of colorectal cancer at the turn of the century: European and US perspectives. Ann Oncol. 2001;12:13–22. doi: 10.1023/a:1008357725209. [DOI] [PubMed] [Google Scholar]
- 2.Adlard JW, Richman SD, Seymour MT, Quirke P. Prediction of the response of colorectal cancer to systemic therapy. Lancet Oncol. 2002;3:75–82. doi: 10.1016/s1470-2045(02)00648-4. [DOI] [PubMed] [Google Scholar]
- 3.Leichman CG, Lenz HJ, Leichman L, Danenberg K, Baranda J, Groshen S, Boswell W, Metzger R, Tan M, Danenberg PV. Quantitation of intratumoral thymidylate synthase expression predicts for disseminated colorectal cancer response and resistance to protracted-infusion fluorouracil and weekly leucovorin. J Clin Oncol. 1997;15:3223–3229. doi: 10.1200/JCO.1997.15.10.3223. [DOI] [PubMed] [Google Scholar]
- 4.Salonga D, Danenberg KD, Johnson M, Metzger R, Groshen S, Tsao-Wei DD, Lenz HJ, Leichman CG, Leichman L, Diasio RB, et al. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin Cancer Res. 2000;6:1322–1327. [PubMed] [Google Scholar]
- 5.Metzger R, Danenberg K, Leichman CG, Salonga D, Schwartz EL, Wadler S, Lenz HJ, Groshen S, Leichman L, Danenberg PV. High basal level gene expression of thymidine phosphorylase (platelet-derived endothelial cell growth factor) in colorectal tumors is associated with nonresponse to 5-fluorouracil. Clin Cancer Res. 1998;4:2371–2376. [PubMed] [Google Scholar]
- 6.Soong R, Powell B, Elsaleh H, Gnanasampanthan G, Smith DR, Goh HS, Joseph D, Iacopetta B. Prognostic significance of TP53 gene mutation in 995 cases of colorectal carcinoma. Influence of tumour site, stage, adjuvant chemotherapy and type of mutation. Eur J Cancer. 2000;36:2053–2060. doi: 10.1016/s0959-8049(00)00285-9. [DOI] [PubMed] [Google Scholar]
- 7.McKay JA, Lloret C, Murray GI, Johnston PG, Bicknell R, Ahmed FY, Cassidy J, McLeod HL. Application of the enrichment approach to identify putative markers of response to 5-fluorouracil therapy in advanced colorectal carcinomas. Int J Oncol. 2000;17:153–158. doi: 10.3892/ijo.17.1.153. [DOI] [PubMed] [Google Scholar]
- 8.Elsaleh H. The microsatellite instability phenotype in human colorectal carcinoma: relationship to sex, age, and tumor site. Gastroenterology. 2001;121:230–231. doi: 10.1053/gast.2001.26044. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe T, Wu TT, Catalano PJ, Ueki T, Satriano R, Haller DG, Benson AB, III, Hamilton SR. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med. 2001;344:1196–1206. doi: 10.1056/NEJM200104193441603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 11.Chung DC. The genetic basis of colorectal cancer: insights into critical pathways of tumorigenesis. Gastroenterology. 2000;119:854–865. doi: 10.1053/gast.2000.16507. [DOI] [PubMed] [Google Scholar]
- 12.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 13.Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 14.Tang B, Bottinger EP, Jakowlew SB, Bagnall KM, Mariano J, Anver MR, Letterio JJ, Wakefield LM. Transforming growth factor-beta1 is a new form of tumor suppressor with true haploid insufficiency. Nat Med. 1998;4:802–807. doi: 10.1038/nm0798-802. [DOI] [PubMed] [Google Scholar]
- 15.Grady WM, Myeroff LL, Swinler SE, Rajput A, Thiagalingam S, Lutterbaugh JD, Neumann A, Brattain MG, Chang J, Kim SJ, et al. Mutational inactivation of transforming growth factor beta receptor type II in microsatellite stable colon cancers. Cancer Res. 1999;59:320–324. [PubMed] [Google Scholar]
- 16.Datta PK, Chytil A, Gorska AE, Moses HL. Identification of STRAP, a novel WD domain protein in transforming growth factor-beta signaling. J Biol Chem. 1998;273:34671–34674. doi: 10.1074/jbc.273.52.34671. [DOI] [PubMed] [Google Scholar]
- 17.Datta PK, Moses HL. STRAP and Smad7 synergize in the inhibition of transforming growth factor beta signaling. Mol Cell Biol. 2000;20:3157–3167. doi: 10.1128/mcb.20.9.3157-3167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swiss Group for Clinical Cancer Research (SAKK), author Long-term results of single course of adjuvant intraportal chemotherapy for colorectal cancer. Lancet. 1995;345:349–353. [PubMed] [Google Scholar]
- 19.Boulay JL, Reuter J, Ritschard R, Terracciano L, Herrmann R, Rochlitz C. Gene dosage by quantitative real-time PCR. Biotechniques. 1999;27:228–230. 232. doi: 10.2144/99272bm03. [DOI] [PubMed] [Google Scholar]
- 20.Therneau TaG PM. Modeling Survival Data. In: Springer PM, editor. New York: Springer; 2000. [Google Scholar]
- 21.Giacchetti S, Perpoint B, Zidani R, Le Bail N, Faggiuolo R, Focan C, Chollet P, Llory JF, Letourneau Y, Coudert B, et al. Phase III multicenter randomized trial of oxaliplatin added to chronomodulated fluorouracil-leucovorin as first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2000;18:136–147. doi: 10.1200/JCO.2000.18.1.136. [DOI] [PubMed] [Google Scholar]
- 22.Saltz LB, Cox JV, Blanke C, Rosen LS, Fehrenbacher L, Moore MJ, Maroun JA, Ackland SP, Locker PK, Pirotta N, et al. Irinotecan plus fluorouracil and leucovorin for metastatic colorectal cancer. Irinotecan Study Group. N Engl J Med. 2000;343:905–914. doi: 10.1056/NEJM200009283431302. [DOI] [PubMed] [Google Scholar]
- 23.Aragane H, Sakakura C, Nakanishi M, Yasuoka R, Fujita Y, Taniguchi H, Hagiwara A, Yamaguchi T, Abe T, Inazawa J, et al. Chromosomal aberrations in colorectal cancers and liver metastases analyzed by comparative genomic hybridization. Int J Cancer. 2001;94:623–629. doi: 10.1002/ijc.1522. [DOI] [PubMed] [Google Scholar]
- 24.Henegariu O, Vance GH, Heiber D, Pera M, Heerema NA. Triple-color FISH analysis of 12p amplification in testicular germ-cell tumors using 12p band-specific painting probes. JMolMed. 1998;76:648–655. doi: 10.1007/s001090050262. [DOI] [PubMed] [Google Scholar]
- 25.Dong JT. Chromosomal deletions and tumor suppressor genes in prostate cancer. Cancer Metastasis Rev. 2001;20:173–193. doi: 10.1023/a:1015575125780. [DOI] [PubMed] [Google Scholar]
- 26.Matsuda S, Katsumata R, Okuda T, Yamamoto T, Miyazaki K, Senga T, Machida K, Thant AA, Nakatsugawa S, Hamaguchi M. Molecular cloning and characterization of human MAWD, a novel protein containing WD-40 repeats frequently overexpressed in breast cancer. Cancer Res. 2000;60:13–17. [PubMed] [Google Scholar]
- 27.Cui W, Fowlis DJ, Bryson S, Duffie E, Ireland H, Balmain A, Akhurst RJ. TGFbeta1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell. 1996;86:531–542. doi: 10.1016/s0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- 28.Boulay JL, Mild G, Lowy A, Reuter J, Lagrange M, Terracciano L, Laffer U, Herrmann R, Rochlitz C. SMAD4 is a predictive marker for 5-fluorouracil-based chemotherapy in patients with colorectal cancer. Br J Cancer. 2002;87:630–634. doi: 10.1038/sj.bjc.6600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wendling J, Marchand A, Mauviel A, Verrecchia F. 5-Fluorouracil blocks transforming growth factor-beta-induced alpha 2 type I collagen gene (COL1A2) expression in human fibroblasts via c-Jun NH2-terminal kinase/activator protein-1 activation. Mol Pharmacol. 2003;64:707–713. doi: 10.1124/mol.64.3.707. [DOI] [PubMed] [Google Scholar]