Abstract
p73, a member of the p53 family, is overexpressed in many cancers. To understand the mechanism(s) underlying this overexpression, we have undertaken a detailed characterization of the human p73 promoter. The promoter is strongly activated in cells expressing exogenous E2F1 and suppressed by exogenous Rb. At least three functional E2F binding sites, located immediately upstream of exon 1 (at -284, -155 and -132) mediate this induction. 5′ serially deleted promoter constructs and constructs harboring mutated E2F sites were analyzed for their response to exogenously expressed E2F1 or Rb to establish functionality of these sites. Authenticity of E2F sites was further confirmed by electrophoretic mobility shift assay (EMSA) using E2F1/DP1 heterodimers synthesized in vitro, followed by competition assays with unlabeled wild-type or mutant oligonucleotides and supershift analysis using anti-E2F1 antibodies. In vivo binding of E2F1 to the p73 promoter was demonstrated using nuclear extracts prepared from E2F1-inducible Saos2 cells. The region conferring the highest promoter activity was found to reside between -113 to -217 of the p73 gene. Two of the three functional E2F sites (at -155 and -132) reside within this region. Our results suggest that regulation of p73 expression is primarily mediated through binding of E2F1 to target sites at -155 and -132.
Keywords: p73, E2F1, promoter analysis, gene regulation, cancer
Introduction
p53 is mutated in about half of all cancers. p73, a member of the p53 gene family, not only shows significant sequence conservation with p53 [1] but also exhibits some functional overlap. p73 can induce p21Waf1/Cip1, a cyclin-dependent kinase inhibitor, and several p53-responsive genes such as BAX, 14-3-3σ, GADD45, and HDM2 [2–4,5,6]. Although p53 and p73 induce similar sets of genes, the level of response can differ [2,4,5]. p73 also promotes apoptosis when overexpressed in vitro [1,2,7–9,10]. p73 is regulated by HDM2 (the human homolog of MDM2) by an auto-feedback regulatory mechanism but in a manner quite distinct from that of p53 [3,11,12]. Some viral proteins that inactivate p53, such as the adenoviral protein E1A, can also inhibit p73 function [13]. The human p73 gene is located at 1p36.33 [1], a region that is frequently lost in neuroblastomas, melanomas, oligodendrogliomas, breast, liver, and lung cancers. These similarities raised the possibility that p73 could be a tumor suppressor gene.
There are, however, differences between p73 and p53. p73 exists in at least six carboxy-terminal isoforms (α, β, γ, δ, ɛ, ζ) that arise by alternative splicing [1,14–16]. At least, two N-terminal isoforms of p73 encompass all of these carboxy splicing variants [17] — ΔNp73, which lacks the transactivation (TA) domain, and TA-p73 (full-length [FL] p73); the former, transcribed from an intron 3 promoter, is a dominant-negative version of p73 and is the predominant form expressed in murine tissues [17]. The interaction of p73 with HDM2 suppresses the efficient transcription of the p73 gene and its ability to transactivate target genes [11,18] whereas a similar interaction with p53 targets p53 for degradation. Viral oncoproteins such as adenovirus E1B, SV40 large T antigen, and HPV E6 inactivate p53 by protein-protein interactions but do not appear to interact with p73 [13,19,20]. DNA-damaging agents such as γ-irradiation and cisplatin, but not UV irradiation or actinomycin D, induce p73 [21–23]. Thus, p53 and p73 respond to certain DNA-damaging agents through different mechanisms. The p73 gene also appears to be imprinted in certain tissues, unlike p53, but with data showing considerable inter- and intraindividual variations, the tissue specificity of the imprinting process remains unclear at present [10,17,24].
Extensive analyses of the p73 gene in various cancers indicate that it is rarely mutated [10,25,26]. On the contrary, overexpression has been observed in a variety of cancers such as those of the lung [27,28], bladder [29,30], and breast [15] and has been attributed, in part, to biallelic expression involving the reactivation of the normally imprinted silent allele. However, this is not always the case [15,30]. Thus, other mechanisms must account for p73 overexpression. Overall, the lack of mutations and its overexpression in cancer tissues indicate that p73 is unlikely to be a tumor suppressor gene.
A knowledge of p73 gene regulation is, thus, critical to our understanding of p73 function in both normal and tumor tissues. Recently, we and others demonstrated that ectopically expressed E2F1 could induce p73 gene expression leading to apoptosis [31,32]. Here, we provide a more detailed analysis of the p73 promoter and provide functional evidence for its regulation by E2F1.
Materials and Methods
Materials
HeLa cells (CCL 2.3) were obtained from ATCC (Manassas, VA). Saos2 cell subclones in which E2F1 cDNA is under the control of a doxycycline-inducible promoter have been previously described [31]. The p73 positive BAC, 190O18, was from Research Genetics (Huntsville, AL). All other reagents, unless otherwise indicated, were from Promega (Madison, WI), Gibco Life Technologies (Rockville, MD), or Fisher (Hanover Park, IL). Oligonucleotide synthesis and automated nucleotide sequencing were carried out at the Mayo Core Facility, Mayo Clinic.
Isolation of the p73 Promoter
Multiplex restriction site polymerase chain reaction was performed [33] with 5% DMSO, using two antisense primers made to exon 1 (p73as1: 5′-CCGTCGCAGCCCCGGGCA and a nested primer p73as2: 5′-GCGTCCGTCCCGGCTGGCC) and the p73 positive BAC DNA. A distinct PCR band was obtained that was sequenced to affirm its authenticity. An antisense primer (p73as3: 5′-AGCCCGGCGCGCGGGAAGGCAG) was then designed toward the 5′ end of this sequence and used for direct sequencing of the BAC DNA. The resulting sequence data was used for the synthesis of a third antisense primer (p73as4: 5′-GACGCCGGTGCCGACTCTGTG) for direct sequencing. A total of 930 bp of sequence, inclusive of exon 1, was thus obtained. An intact fragment harboring this sequence was finally obtained from the BAC DNA by PCR using p73as1 and a sense primer (p73s5: 5′-GATCCAGAGCCCGAGCCCACA) and was cloned into pGEM-T Easy vector (Promega). This plasmid, p73 pGEM-T, was used for the construction of various reporter constructs described below.
p73 Promoter Constructs
5′ Deletion constructs A luciferase reporter vector, pGL3 Basic (Promega), was used to clone the FL 930-bp promoter fragment by digesting p73 pGEM-T with NcoI, filling in with Klenow polymerase, then digesting with SalI and cloning the resultant fragment into SmaI-XhoI digested pGL3 Basic vector. Various 5′ deletion constructs were then made by utilizing unique restriction sites located upstream in the p73 promoter (BstI, PstI, PvuII, NotI, and AvrII). All deletion constructs terminate at nt +71 of exon 1 where +1 represents the first nt of exon 1 [1].
E2F1 mutant constructs The PvuII construct (Figure 1, A and B) was chosen as a parent plasmid to introduce mutations at the five putative E2F binding sites (E2F1-189, -155, -143, -132, and -6) using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). In general, a 2 nt substitution (AT) was incorporated at the conserved G/C core of these sites (Table 1) as this substitution has been shown to eliminate E2F binding [34]. The E2F1-155/-132 double mutant was created using the mutant primers for E2F1-155 on a plasmid carrying the mutant E2F1-132 site. All mutant plasmids were sequenced to confirm the mutations.
Figure 1.
(A) 5′ Deletion analysis of the p73 promoter. Constructs harboring various 5′ deletions of the p73 promoter were cloned into pGL3 Basic (Promega). Deletion constructs were made using unique restriction enzyme sites present in the promoter, as shown. The consensus E2F motif at -284 (E2F1) and the TATA box at -77 (TATAA) are indicated. The downward arrows indicate putative E2F motifs. Transfections were carried out in HeLa cells with (+E2F1) and without (-E2F1) E2F1 cotransfection. Normalized luciferase activity, expressed as a percentage of the FL construct without exogenous expression of E2F1, is shown. FL, full-length promoter construct; vector, pGL3 Basic. (B) The minimal promoter of the p73 gene. The region conferring the highest promoter activity resides within a 104-bp fragment located between the PvuII (-217) and NotI sites (-113). The five putative E2F elements (E2F1-189, E2F1-155, E2F1-143, E2F1-132, and E2F1-6) identified within this region, a TATA box and the various restriction sites are indicated. A construct that deletes sequences upstream of -165 was made by introducing a KpnI site at -165 (shown) by altering the sequence GGTGGC to GGTACC and then digesting with KpnI. Nucleotide positions are numbered every 50 nts (underlined) with reference to the first nt of exon 1 [1].
Table 1.
Mutation of Putative E2F Sites in the p73 Promoter.
| Mutation | Position | Sequence Change (>AT) |
| Mut-189 | -189 to -182 | CTTGGCCC |
| Mut-155 | -155 to -148 | CTTCCCGC |
| Mut-143 | -143 to -136 | CGGGCTAA |
| Mut-132 | -132 to -125 | GGCGCTAA |
| Mut-6 | -7 to +2 | CCGCGAAG |
All mutations were incorporated using the Quickchange site-directed mutagenesis kit (Stratagene). The putative E2F sites and the nucleotides changed (underlined) are highlighted. An AT dinucleotide was substituted for the underlined nucleotides. The double mutant (Mut-155/-132) incorporates the mutational changes described for Mut-155 and Mut-132.
Kpn constructs Because the PvuII-NotI region is critical for p73 expression, a KpnI site was introduced between nts -170 and -165 in the PvuII construct by altering two nucleotides (Figure 1B, see legend) through site-directed mutagenesis (QuickChange, Stratagene), and deleting all sequences upstream of this site. Further deletions at ∼10-bp intervals were made in an identical manner extending downstream to the NotI site at -113.
Electrophoretic Mobility Shift Assays (EMSA)
Thirty-mer oligonucleotides corresponding to the wild-type and mutant putative E2F binding sites were synthesized, annealed to complementary synthetic oligonucleotide in vitro and radiolabeled with [α-32P]dCTP and dGTP by using Klenow DNA polymerase (New England Biolabs, MA). E2F1/DP1 heterodimers were produced by in vitro translation with the TNT translation system (Promega). DNA binding reactions were performed in 20 µl containing 2 µl of the indicated protein in vitro translation product (or unprogrammed reticulocyte lysate labeled as mock), 10 µl of 2x DNA binding buffer (50 mM Tris, 7.5/40% glycerol/100 mM KCl/100 mM DTT/2 mg/ml BSA/0.2% Triton X-100), 1 ng of labeled probe, and 300 ng of sonicated salmon sperm DNA. Forty nanograms of unlabeled oligonucleotides corresponding to wild-type or mutated (Table 1) p73 E2F binding sites or the known E2F binding sites of the DHFR promoter were added as specific competitors. Antibody supershift experiments were performed by adding 1 µl of anti-E2F1 (Santa Cruz, Santa Cruz, CA — sc 193) or isotype matched control anti-myc antibody (Santa Cruz — sc 789). Binding reactions were preincubated for 10 minutes at room temperature, labeled probe was added and incubated for an additional 15 minutes and then resolved by 4% PAGE at 4°C in 0.5x Tris-Borate-EDTA buffer (2 to 3 hours at 200 V). Gels were dried and exposed to film at -70°C.
EMSA was also performed using nuclear extracts prepared, using NE-PER nuclear and cytoplasmic extraction reagents (Pierce, Rockford, IL), from Saos2 cells treated with doxycycline (2 µg/ml) for 48 hours. For DNA-binding reactions, 2 µl (3 µg) of nuclear extract was preincubated for 15 minutes in a 20-µl reaction with 10 µl of 2x DNA binding buffer, 300 ng of sonicated salmon sperm DNA, and BSA (final concentration, 10 mg/ml) in the presence or absence of unlabeled oligonucleotides and antibodies, after which 1 ng of the labeled probe was then added as described above.
Transfection Experiments
HeLa cells (1x105 cells/well), grown in serum-free RPMI media (ATCC) containing 2% TCH (Celox, St. Paul, MN), were cotransfected with the luciferase reporter plasmid (1 µg) and 100 ng of pRL-TK (Promega), a control plasmid expressing Renilla luciferase. For E2F1 cotransfection, 1 µg of the reporter plasmid, 100 ng of pRL-TK, and 1 µg of the E2F1 expression plasmid, pRC-CMV HA E2F1, were used. The amount of DNA was kept constant by the addition of an appropriate empty vector. Transfections were done in six-well plates using Fugene 6 reagent (Boehringer Mannheim, Indianapolis, IN) and were carried out 24 hours after seeding. Cells were harvested 48 hours later using the Dual-Luciferase assay system (Promega). Firefly and Renilla luciferase activities were measured in a Monolight 2010 (Analytical Luminescence Laboratory, San Diego, CA). All values, from experiments performed at least thrice with duplicates, were normalized and expressed as percent of the activity of the appropriate construct described in the legends to each figure.
Transfection of HeLa cells with Rb was essentially as described above using 1 µg of the FL construct, 100 ng of pRL-TK, and increasing amounts (0 to 2 µg) of the Rb-expressing plasmid, pCMV Rb 379-928, supplemented with appropriate amounts of the empty vector to maintain constant amount of DNA. For transfections examining the effect of Rb on E2F1-induced promoter activity of the FL, deletion and mutant constructs, 1 µg of the reporter construct, 100 ng of the Renilla plasmid, 0.5 µg of the E2F1 expression plasmid, and either 0.5 or 2 µg of the Rb-expressing plasmid were used. The total DNA for all reactions was kept the same by the addition of the empty vector.
Results
Identification and Characterization of the p73 Promoter
The 5′ flanking sequence of p73 that we have characterized extends from +71 (in exon 1) to -857 and matches perfectly the sequence derived by Ding et al. [35]. +1 refers to the first nt of exon 1 [1]. Despite numerous attempts, we were unable to identify the transcriptional start site presumably due to the high GC content in this region, a problem evidently shared by others [32,35]. Salient features of the flanking sequence include the high GC content (76%) and the high proportion of CpGs (1 per 8.2 bp), consistent with the gene residing within a CpG island, and the presence of a consensus E2F binding site (TTTTGGCGC at -284) and a canonical TATA box (TATAA, at -77) (Figure 1B) identified by computer analysis programs (MatInspector V2.2 [36], TFSEARCH [37] and FindPatterns [www.gcg.com]). Putative binding sites for several other factors such as Sp1, MAZ1, AP-2 and EGR-1 are also found to be present (data not shown).
Analysis of the various deletion constructs (Figure 1A; -E2F1) indicates that the PvuII construct harbors the minimal promoter of this gene as it is the smallest construct with a promoter activity (∼112%) comparable to that of the FL construct (set at 100%). Deletions further downstream to the NotI or AvrII sites result in a steep drop in activity to about 13% to 16% (Figure 1A). Thus, the 104-bp region located between the NotI (-113) and PvuII (-217) sites plays a critical role in p73 promoter function (Figure 1, A and B).
E2F Responsiveness of the p73 Promoter
The identification of a consensus E2F site at -284 prompted us to evaluate if the FL promoter fragment was E2F regulated. Because Rb forms complexes with E2F thereby reducing the pool of free E2F, we used Rb to assess its inhibitory effect on the promoter activity of the p73 gene. As shown in Figure 2A, the promoter activity of the FL construct can be suppressed with increasing concentrations of a plasmid expressing Rb, suggesting that E2F regulates the basal promoter activity in HeLa cells. As expected, ectopic expression of E2F1 significantly stimulates the basal activity of the FL construct (Figure 2B); this induced activity is inhibited in a dose-dependent manner by Rb with ∼80% inhibition observed at 2 µg, the highest concentration used. These results suggest that the p73 promoter is E2F1 regulated.
Figure 2.
(A) Rb represses p73 promoter activity in HeLa cells. A p73 reporter construct harboring sequences from -857 to +71 of the p73 gene was cotransfected with increasing amounts (0, 0.5, 1.0, 1.5, and 2 µg) of a plasmid expressing Rb. Normalized luciferase activity was determined and the activity of the reporter construct without cotransfection of the Rb expressing plasmid was taken as 100%. The experiment was repeated twice with duplicates. (B) Effect of Rb on E2F1-induced promoter activity in HeLa cells. The experiments were as in (A) but 0.5 µg of an E2F1 expressing plasmid was added to HeLa cells. Increasing amounts of a plasmid expressing Rb (0.5 to 2 µg) was cotransfected with E2F1 to assess suppression of E2F1-induced activity.
Analysis of Potential E2F Binding Sites in the p73 Promoter
Analysis of deletion constructs indicates that the consensus E2F site at -284 (Figure 1A) is not necessary for p73 induction by E2F. To identify potential E2F motifs that may be modulating p73 regulation, we scanned sequences downstream of the PvuII site (-217) because the PvuII construct is the smallest construct exhibiting maximal E2F1-induced activity (Figure 1A). Five potential sites showing similarity to the E2F consensus binding site [38], TT(G/C)(G/C)CG(G/C), were identified at -189, -155, -143, -132 and -6 (Figure 1B). All but the one at -6 occur in close proximity to each other within the 104-bp critically important PvuII-NotI region.
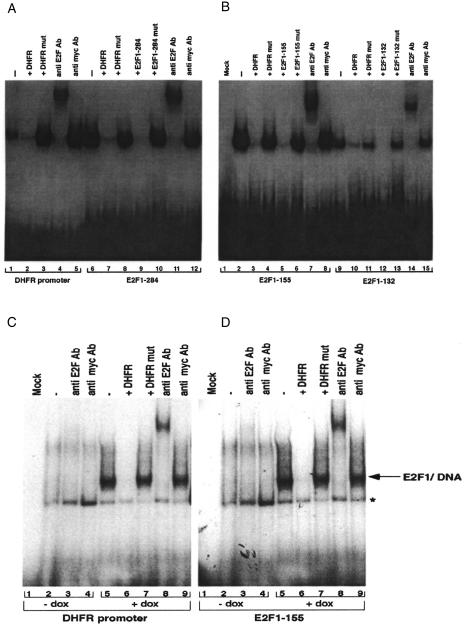
Electrophoretic mobility shift analysis (EMSA) All the six putative E2F sites, including that at -284, were subjected to gel-shift analysis. Although all six sites show variable levels of binding in vitro, only three sites (at -284, -155 and -132) exhibit specific binding to E2F1/DP1 heterodimers. It should be noted that DP1 is an obligate binding partner of E2F1 and is necessary for the binding of E2F1 to target binding sites [39,40]. Figure 3 shows binding of E2F1/DP1 heterodimers to a DHFR E2F element, used as positive control (A, left panel), and to the E2F sites at -284 (A, right panel), -155 (B, left panel) and -132 (B, right panel). Binding to these sites can be effectively competed by unlabeled oligonucleotides containing these sites or by an oligonucleotide containing the DHFR E2F site. Mutant versions of these oligonucleotides do not compete. Further confirmation comes from supershift analysis using anti-E2F1 antibody. A high molecular weight antibody complex is formed with all three sites whereas none is seen with the nonspecific antibody. Site 132 shows reduced affinity for E2F1 presumably because the nt at position 3 of this site varies from the E2F binding consensus sequence [34,38].
Figure 3.
(A, B) Binding of E2F1 to specific E2F binding sites in the p73 promoter. E2F1/DP1 heterodimers produced by in vitro translation were coincubated with radiolabeled probes containing the p73 promoter E2F binding sites at -284 (A, right panel), -155 (B, left panel), and -132 (B, right panel) in the absence or presence of unlabeled competitor oligonucleotides corresponding to wild-type or mutant (Table 1) versions of these sites. Oligonucleotides containing the wild-type and mutant E2F binding sites of the DHFR promoter were used as positive and negative controls, respectively (A, left panel). In addition, supershift analysis using anti-E2F1 antibody or the isotype matched control anti-myc antibody was performed. E2F1/DP1 heterodimers bind E2F site - 132 with reduced affinity because the nt at position 3 of this site varies from the E2F binding consensus sequence [34,38]. (C, D) Cellular E2F1 binds specific E2F binding sites in the p73 promoter. Nuclear extracts from subclones of Saos2 cells, which stably express E2F1 under the control of a doxycycline-inducible promoter, were prepared following no treatment (lanes 1 to 4) or treatment with doxycycline for 48 hours (lanes 5 to 9). Extracts were coincubated with radiolabeled probes containing the E2F binding sites of the DHFR promoter (C) or the E2F site at - 155 (D) of the p73 promoter in the absence or presence of unlabeled competitor oligonucleotides corresponding to the wild-type or mutant versions of the DHFR promoter. Supershift analysis using anti-E2F1 antibody and anti-myc antibody was performed. Extracts from cells induced to express E2F1 demonstrate specific binding to the E2F site at -155. In contrast, extracts from cells not induced to express E2F1 do not demonstrate a measurable level of binding. The asterisk indicates a nonspecific complex.
Specificity of E2F binding was further confirmed by performing gel shift analyses using [32P]-labeled mutant versions of oligonucleotides containing E2F-284, E2F-155 or E2F-132. These mutant versions (Table 1), are not bound by E2F1/DP1 heterodimers (data not shown). In vivo binding of E2F1 to the p73 promoter was demonstrated using nuclear extracts prepared from Saos2 cells that overproduce E2F1 following treatment with doxycycline. Induced extracts demonstrated binding to an oligonucleotide containing the E2F binding site in the DHFR promoter (Figure 3C) as well as the E2F-DNA binding sites of the p73 promoter (Figure 3D and data not shown).
Transfection analysis of various deletion constructs The promoter activities of the various deletion reporter constructs were also examined after E2F1 cotransfection to identify constructs that would significantly respond to E2F1 stimulation, thereby limiting the E2F1-responsive region (Figure 1A; +E2F1). The FL construct exhibited 250% activity in the presence of exogenous E2F1 (compared to its unstimulated state). Inducibility of the BstI (375%), PstI (288%), and PvuII (298%) constructs were as high as the FL construct whereas very little induction was observed with either the NotI (26%) or the AvrII construct (75%). This again suggests that the PvuII construct must account for most, if not all, of the potential E2F binding element(s) and that these sites should lie within the PvuII-NotI region (-217 to -113). The reason for the consistently higher luciferase activity with the BstI construct (compared to the larger FL construct) and the milder E2F1 induction (75%) with the smallest AvrII construct (compared to the larger NotI construct) is presumably due to the removal of potential negative elements.
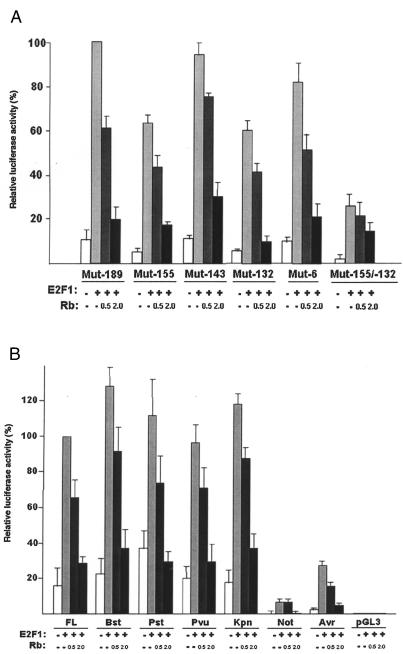
Transfection analysis of E2F mutant constructs Because the PvuII construct harbors five of the putative E2F motifs, respective mutations (Mut-189, Mut-155, Mut-143, Mut-132 and Mut-6) were introduced at these sites (Table 1) and their effect on promoter activity was assessed with and without E2F1, and with E2F1 cotransfected with Rb (0.5 and 2 µg). Additionally a plasmid doubly mutant at sites -155 and -132 (Mut-155/-132) was constructed and similarly analyzed. E2F1-induced Mut-189 promoter activity was taken as 100% because the response of this construct was similar to the unmutated parent PvuII construct (data not shown). From Figure 4A, it is clear that E2F sites at -155 and -132 are the most critical for promoter regulation because mutation of either site significantly reduces basal activity (to ∼5%) compared to Mut-189 (∼10%) whereas activities of the reporters with mutant sites at -143 and -6 are comparable to that of Mut-189. A steep drop in activity (to ∼1%) is observed when sites -155 and -132 are mutated together. Cotransfection with E2F1 enhances the relative activity of all mutant constructs: Mut-189 (set at 100%), Mut-155 (∼62%), Mut-143 (∼95%), Mut-132 (∼60%), and Mut-6 (∼82%) with the lowest induction observed with Mut-132 and Mut-155. The double mutant exhibited only ∼25% induction. The observation that all of the single mutant reporter plasmids display residual E2F1 induction presumably reflects the fact that they all possess at least one, if not two, intact E2F binding sites at -155 and -132. Although E2F1 induction of the double mutant is considerably diminished, it is not totally abrogated. We presume that some of this residual activity arises from binding to cryptic sites, or to an indirect effect of E2F1. We conclude that binding sites at -155 and -132 play important roles in p73 regulation consistent with our EMSA studies and that both sites are required for maximal induction of promoter activity. E2F1-induced promoter activity of all constructs (except for the double mutant) shows dose-dependent suppression with ectopically expressed Rb where 2 µg of Rb is able to suppress ∼75% to 80% of the induced activity.
Figure 4.
(A) Effect of mutations at putative E2F binding sites on p73 promoter activity. The five putative E2F sites at -189, -155, -143, -132, and -6 were mutated individually by site-directed mutagenesis (Table 1) in the context of the PvuII construct and a construct harboring a double mutant at sites -155/-132 was also included. Constructs were transfected into HeLa cells with and without E2F1 cotransfection. To assess suppression of E2F1-induced activity, E2F1 transfected cells were cotransfected with 0.5 or 2 µg of a plasmid expressing Rb. Normalized luciferase activity is expressed as a percentage of the E2F1-induced promoter activity of Mut-189, which has been found to respond like the unmutated parent PvuII construct (data not shown). The experiments were repeated thrice with duplicates. (B) Effect of Rb on E2F1-induced promoter activity of the various 5′ deletion constructs. A total of 0.5 µg of a E2F1 expressing plasmid was added to induce promoter activity of the various 5µ deletion constructs of the p73 gene. A Kpn construct was created and included to further narrow down the region harboring key regulatory elements. Inhibition of induced activity was assessed in the presence of exogenously expressed Rb (0.5 or 2 µg). Normalized luciferase activity is expressed as percentage of the FL construct induced with E2F1. Experiments were repeated thrice with duplicates.
Effect of Rb on E2F1-Induced Promoter Activity of the Deletion Constructs
The above results clearly implicate the 104-bp PvuII-NotI region, which contains the E2F sites at -155 and -132, as being critical for p73 promoter function and E2F1 responsiveness. To further narrow down the region harboring key regulatory elements, a plasmid (a Kpn construct) that removed all sequences upstream of nt -165 was created. This plasmid, which truncates by half the 104-bp region at the 5′ end and removes the putative E2F element at -189 (Figure 1B) retains most of the promoter activity of the FL construct (Figure 4B) confirming the minimal role of E2F1-189 in promoter regulation. E2F1-induced promoter activity is repressible in a dose-dependent fashion for the Kpn and all larger constructs and is brought down to near-normal levels (∼80% inhibition) with Rb. These results suggest that functional E2F1 binding site(s) are localized within the 52-bp fragment spanning nts -165 and -113 (KpnI and NotI) and do not reside downstream of -113. Thus, E2F1-induced activity of the p73 gene must be contributed primarily by the two sites at -155 and -132.
Discussion
The data presented here provides a more detailed analysis of E2F1 activation of the human p73 promoter [31,32]. The activity of a promoter construct carrying sequences from -857 to +71 of the p73 gene is significantly stimulated by the exogenous addition of E2F1 and repressed by ectopically expressed Rb. Repression by Rb presumably occurs by depleting the pool of free E2F and formation of pRb-E2F repressor complexes. Association of Rb with E2F1 is a key step in determining cell-cycle progression. Hyperphosphorylation of Rb or loss of Rb function, as seen in various cancers, releases E2F, which activates target genes involved in DNA replication and cell-cycle progression [38,41,42]. The observation that E2F1 induces p73 expression suggests a key role for p73 in cellular function and is consistent with an apoptotic role for p73 [7,8,21–23,31,32].
Gel shift analyses identified three sites within the promoter that specifically bind E2F1/DP1 heterodimers: E2F-284, E2F-155 and E2F-132. In vivo binding of E2F1 to these binding sites has also been demonstrated using nuclear extracts prepared from E2F1-inducible Saos2 cell lines, thus validating the binding specificities of the E2F1/DP1 heterodimers to the p73 promoter both in vitro and in vivo. Deletion of the most upstream site (-284) has negligible effect on the overall promoter activity (Figure 1A) but mutations in either of the two remaining sites reduce activity by more than 50% suggesting that both these sites are required for basal promoter activity (Figure 4A), and by more than 90% when these two sites are simultaneously mutated. Ectopic expression of E2F1 induces promoter activity of the Mut-155 and Mut-132 plasmids to only about 60% and the Mut-155/-132 double mutant to about 25%, relative to that of the unmutated parent construct. These results indicate that binding of E2F1 to both these sites is necessary for maximal induction of the p73 promoter. The two sites are located within the 104-bp region found critical for p73 promoter regulation. Indeed, shorter truncations of the 104-bp minimal region narrow it down to a 52-bp segment (-165 to -113), containing these two E2F binding sites, as harboring much of the p73 promoter activity (Kpn construct; Figure 4B). Stiewe and Pützer [32], who analyzed a larger segment of the p73 promoter (-4040 to +3033), also conclude that the major sites of E2F1 induction are localized between -705 and +537. However, whereas they used larger DNA fragments (105 to 605 bp) in GST-E2F1 pull-down assays to determine regions of binding and then identified putative motifs contained therein, we have used defined oligonucleotides spanning each motif to characterize specific binding and validated their binding specificity by use of competition assays and antibody supershifts. Although our studies are focussed on a promoter region that is immediately proximal to exon 1, E2F sites in intron 1 or those located further upstream in the 5′ flanking region may also bind E2F factors [32]. The observation that the p73 promoter is significantly induced by E2F1 and is brought down to basal levels by Rb underscores the importance of E2F1 in the regulation of p73.
Interestingly, the minimal promoter region described here is quite different from that observed by Ding et al. [35] who identified a 138-bp sequence immediately downstream from ours, spanning nts -119 and +19, as conferring maximal basal activity. A human breast carcinoma cell line, MCF-7, was used in their study whereas we used a HeLa (present study) and Saos2 (osteosarcoma) cell lines (results not shown), both of which provide similar results. Thus, tissue-specific factor(s) may play a role in p73 regulation. Tissue-specific regulation may underlie some of the differential aspects of p73 overexpression observed in many, but not all, cancers [27–29,30,43,44]. A Northern analysis of normal tissues indicates a discrete expression pattern for p73, with maximal expression in heart, liver, and pancreas [45].
Whereas the apoptotic role of TA-p73 is well recognized, an anti-apoptotic role has been assigned to ΔN-p73 in mouse neuronal cells [46]. ΔN-p73 lacks the N-terminal TA domain and represents a dominant-negative version of the FL form [17]. The regulation of these two functionally opposite N-terminal p73 isoforms from different promoters of the same gene must, therefore, be tightly controlled. Identifying signals that modulate each of these promoters will provide significant insights not only into their mode of regulation but also to understanding the mechanisms underlying p73 overexpression in various cancers. In keeping with the above findings, we have recently observed a direct correlation between elevated E2F1 levels and p73 overexpression in lung and bladder cancer samples (manuscript in preparation). Whereas one should await the characterization of the human ΔN-promoter to further understand p73 regulation, the characterization of the TA-promoter marks a step in that direction.
Acknowledgement
The authors sincerely thank W. Krek, Basel, Switzerland, for the kind gift of pRC-CMV HA E2F1.
Footnotes
This work was funded by Mayo Clinic/Mayo Foundation and by the award of a grant by The Robert Black Charitable Foundation to W. L.
References
- 1.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J-C, Valent A, Minty A, Chalon P, Lelias J-M, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 2.Zhu J, Jiang J, Zhou W, Chen X. The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res. 1998;58:5061–5065. [PubMed] [Google Scholar]
- 3.Fang L, Lee SW, Aaronson SA. Comparative analysis of p73 and p53 regulation and effector functions. J Cell Biol. 1999;147:823–830. doi: 10.1083/jcb.147.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee C-W, La Thangue NB. Promoter specificity and stability control of the p53-related protein p73. Oncogene. 1999;18:4171–4181. doi: 10.1038/sj.onc.1202793. [DOI] [PubMed] [Google Scholar]
- 5.Di Como CJ, Gaiddon C, Prives C. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol Cell Biol. 1999;19:1438–1449. doi: 10.1128/mcb.19.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levrero M, De Laurenzi V, Costanzo A, Gong J, Melino G, Wang JYJ. Structure, function and regulation of p63 and p73. Cell Death Differ. 1999;6:1146–1153. doi: 10.1038/sj.cdd.4400624. [DOI] [PubMed] [Google Scholar]
- 7.Jost CA, Marin MC, Kaelin WG., Jr p73 is a human p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 8.Jost CA, Marin MC, Kaelin WG., Jr Correction: p73 is a human p53-related protein that can induce apoptosis. Nature. 1999;399:817. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 9.Prabhu N, Somasundaram K, Satyamoorthy K, Herlyn M, El-Deiry W. p73β, unlike p53, suppresses growth and induces apoptosis of human papillomavirus E6-expressing cancer cells. Int J Oncol. 1998;13:5–9. doi: 10.3892/ijo.13.1.5. [DOI] [PubMed] [Google Scholar]
- 10.Kaelin WG., Jr The p53 gene family. Oncogene. 1999;18:7701–7705. doi: 10.1038/sj.onc.1202955. [DOI] [PubMed] [Google Scholar]
- 11.Zeng X, Chen L, Jost CA, Maya R, Keller D, Wang X, Kaelin WG, Jr, Oren M, Chen J, Lu H. MDM2 suppresses p73 function without promoting p73 degradation. Mol Cell Biol. 1999;19:3257–3266. doi: 10.1128/mcb.19.5.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lohrum MAE, Vousden KH. Regulation and function of the p53-related proteins: same family, different rules. Trends Cell Biol. 2000;10:197–202. doi: 10.1016/s0962-8924(00)01736-0. [DOI] [PubMed] [Google Scholar]
- 13.Steegenga WT, Shvarts A, Riteco N, Bos J, Jochemsen AG. Distinct regulation of p53 and p73 activity by adenovirus E1A, E1B, and E4orf6 proteins. Mol Cell Biol. 1999;19:3885–3894. doi: 10.1128/mcb.19.5.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Laurenzi V, Costanzo A, Barcaroli D, Terrinoni A, Falco M, Annicchiarico-Petruzzelli M, Levrero M, Melino G. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J Exp Med. 1998;188:1763–1768. doi: 10.1084/jem.188.9.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaika AI, Kovalev S, Marchenko ND, Moll UM. Over expression of the wild type p73 gene in breast cancer tissues and cell lines. Cancer Res. 1999;59:3257–3263. [PubMed] [Google Scholar]
- 16.De Laurenzi V, Catani MV, Terrinoni A, Corazzari M, Melino G, Costanzo A, Levrero M, Knight RA. Additional complexity in p73: induction by mitogens in lymphoid cells and identification of two new splicing variants epsilon and zeta. Cell Death Differ. 1999;6:389–390. doi: 10.1038/sj.cdd.4400521. [DOI] [PubMed] [Google Scholar]
- 17.Yang A, McKeon F. P63 and P73: P53 mimics, menaces and more. Nature Rev Mol Cell Biol. 2000;1:199–207. doi: 10.1038/35043127. [DOI] [PubMed] [Google Scholar]
- 18.Balint E, Bates S, Vousden KH. Mdm2 binds p73 alpha without targeting degradation. Oncogene. 1999;18:3923–3929. doi: 10.1038/sj.onc.1202781. [DOI] [PubMed] [Google Scholar]
- 19.Marin MC, Jost CA, Irwin MS, DeCaprio JA, Caput D, Kaelin WG., Jr Viral oncoproteins discriminate between p53 and the p53 homolog p73. Mol Cell Biol. 1998;18:6316–6324. doi: 10.1128/mcb.18.11.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobbelstein M, Roth J. The large T antigen of simian virus 40 binds and inactivates p53 but not p73. J Gen Virol. 1998;79:3079–3083. doi: 10.1099/0022-1317-79-12-3079. [DOI] [PubMed] [Google Scholar]
- 21.Agami R, Blandino G, Oren M, Shaul Y. Interaction of c-Abl and p73 alpha and their collaboration to induce apoptosis. Nature. 1999;399:809–813. doi: 10.1038/21697. [DOI] [PubMed] [Google Scholar]
- 22.Gong J, Costanzo A, Yang H-Q, Melino G, Kaelin WG, Jr, Levrero M, Wang JYJ. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 23.Yuan Z-M, Shioya H, Ishiko T, Sun X, Gu J, Huang YY, Lu H, Kharbanda S, Weichselbaum R, Kufe D. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature. 1999;399:814–817. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- 24.Nomoto S, Haruki N, Kondo M, Konishi H, Takahashi T, Takahashi T, Takahashi T. Search for mutations and examination of allelic expression imbalance of the p73 gene at 1p36.33 in human lung cancers. Cancer Res. 1998;58:1380–1383. [PubMed] [Google Scholar]
- 25.Ikawa S, Nakagawara A, Ikawa Y. p53 family genes: structural comparison, expression and mutation. Cell Death Differ. 1999;6:1154–1161. doi: 10.1038/sj.cdd.4400631. [DOI] [PubMed] [Google Scholar]
- 26.Han S, Semba S, Abe T, Makino N, Furukawa T, Fukushige S, Takahashi H, Sakurada A, Sato M, Shiiba K, Matsuno S, Nimura Y, Nakagawara A, Horii A. Infrequent somatic mutations of the p73 gene in various human cancers. Eur J Surg Oncol. 1999;25:194–198. doi: 10.1053/ejso.1998.0626. [DOI] [PubMed] [Google Scholar]
- 27.Mai M, Yokomizo A, Qian C, Yang P, Tindall DJ, Smith DI, Liu W. Activation of p73 silent allele in lung cancer. Cancer Res. 1998;58:2347–2349. [PubMed] [Google Scholar]
- 28.Tokuchi Y, Hashimoto T, Kobayashi Y, Hayashi M, Nishida K, Hayashi S, Imai K, Nakachi K, Ishikawa Y, Nakagawa K, Kawakami Y, Tsuchiya E. The expression of p73 is increased in lung cancer, independent of p53 gene alteration. Br J Cancer. 1999;80:1623–1629. doi: 10.1038/sj.bjc.6690572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chi S-G, Chang S-G, Lee S-J, Lee C-H, Kim JI, Park J-H. Elevated and biallelic expression of p73 is associated with progression of human bladder cancer. Cancer Res. 1999;59:2791–2793. [PubMed] [Google Scholar]
- 30.Yokomizo A, Mai M, Tindall DJ, Cheng L, Bostwick DG, Naito S, Smith DI, Liu W. Overexpression of the wild type p73 gene in human bladder cancer. Oncogene. 1999;18:1629–1633. doi: 10.1038/sj.onc.1202474. [DOI] [PubMed] [Google Scholar]
- 31.Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH, Kaelin WG., Jr Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 32.Stiewe T, Pützer BM. Role of the p53-homologue p73 in E2F1-induced apoptosis. Nature Genet. 2000;26:464–469. doi: 10.1038/82617. [DOI] [PubMed] [Google Scholar]
- 33.Weber KL, Bolander ME, Sarkar G. Rapid acquisition of unknown DNA sequence adjacent to a known segment by multiplex restriction site PCR. BioTechniques. 1998;25:415–419. doi: 10.2144/98253st02. [DOI] [PubMed] [Google Scholar]
- 34.Farnham PJ, Slansky JE, Kollmar R. The role of E2F in the mammalian cell cycle. Biochim Biophys Acta. 1993;1155:125–131. doi: 10.1016/0304-419x(93)90001-s. [DOI] [PubMed] [Google Scholar]
- 35.Ding Y, Inoue T, Kamiyama J, Tamura Y, Ohtani-Fujita N, Igata E, Sakai T. Molecular cloning and functional characterization of the upstream promoter region of the human p73 gene. DNA Res. 1999;6:347–351. doi: 10.1093/dnares/6.5.347. [DOI] [PubMed] [Google Scholar]
- 36.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, Kaelin WG., Jr The cellular effects of E2F overexpression. Curr Top Microbiol Immunol. 1996;208:79–93. doi: 10.1007/978-3-642-79910-5_4. [DOI] [PubMed] [Google Scholar]
- 39.Adams PD, Kaelin WG., Jr Transcriptional control by E2F. Semin Cancer Biol. 1995;6:99–108. doi: 10.1006/scbi.1995.0013. [DOI] [PubMed] [Google Scholar]
- 40.Lam EW, La Thangue NB. DP and E2F proteins: coordinating transcription with cell cycle progression. Curr Opin Cell Biol. 1994;6:859–866. doi: 10.1016/0955-0674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 41.Sladek TL. E2F transcription factor action, regulation and possible role in human cancer. Cell Proliferation. 1997;30:97–105. doi: 10.1046/j.1365-2184.1997.00085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muller H, Helin K. The E2F transcription factors: key regulators of cell proliferation. Biochim Biophys Acta. 2000;1470:M1–M12. doi: 10.1016/s0304-419x(99)00030-x. [DOI] [PubMed] [Google Scholar]
- 43.Chen CL, Ip SM, Cheng D, Wong LC, Ngan HYS. p73 gene expression in ovarian cancer tissues and cell lines. Clin Cancer Res. 2000;6:3910–3915. [PubMed] [Google Scholar]
- 44.Mihara M, Nimura Y, Ichimiya S, Sakiyama S, Kajikawa S, Adachi W, Amano J, Nakagawara A. Absence of mutation of the p73 gene localized at chromosome 1p36.3 in hepatocellular carcinoma. Br J Cancer. 1999;79:164–167. doi: 10.1038/sj.bjc.6690027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Senoo M, Seki N, Ohira M, Sugano S, Watanabe M, Tachibana M, Tanaka T, Shinkai Y, Kato H. A second p53-related protein, p73L, with high homology to p73. Biochem Biophys Res Commun. 1998;248:603–607. doi: 10.1006/bbrc.1998.9013. [DOI] [PubMed] [Google Scholar]
- 46.Pozniak CD, Radinovic S, Yang A, McKeon F, Kaplan DR, Miller FD. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science. 2000;289:304–306. doi: 10.1126/science.289.5477.304. [DOI] [PubMed] [Google Scholar]