Abstract
The NOVH protein belongs to the emerging CCN [Connective tissue growth factor (CTGF), Cyr61/Cef10, nephroblastoma overexpressed gene] family of growth regulators sharing a strikingly conserved multimodular organization but exhibiting distinctive functional features. Two members of the family (CYR61 and CTGF) are positive regulators of cell proliferation, whereas NOVH and two other members (ELM1 and RCOP-1) exhibit features of negative regulators of growth. The multimodular structure of these proteins suggests that their biological role(s) may depend on interactions with several factors as well as proteins constitutive of the extracellular matrix. To gain insight into the functionality of these domains, we have used a two-hybrid system to identify proteins interacting with NOVH. We report here that the C-terminal domain confers on the full-length NOVH protein the capacity to bind fibulin 1C, a protein of the extracellular matrix that interacts with several other regulators of cell adhesion. Furthermore, we show that a natural N-truncated isoform of NOVH produced by cells expressing the full-length NOVH protein also binds fibulin 1C with a high affinity, and we hypothesize that the production of truncated isoforms of NOVH (and probably of other CCN proteins) may be a critical aspect in the modulation of their biological activity. These results set the stage for a study of NOVH–fibulin 1C interactions and their potential significance in cell-adhesion signaling in normal and pathological conditions.
Control of cell proliferation and differentiation requires the interplay of many signaling factors, among which extracellular-matrix proteins play a central role. The existence of a new class of cell growth regulators has been uncovered recently. This emerging family of proteins, often referred to as CCN [Connective tissue growth factor (CTGF), CYR61/CEF10, nephroblastoma overexpressed gene; ref. 1], comprise both positive and negative regulators of cell growth (CYR61/CEF10; CTGF/FISP12; NOV; ELM1; and RCOP-1) sharing a common multimodular organization. The chicken cef10 and murine cyr61 genes were first identified as immediate early genes induced by pp60v-src oncogene and serum growth factors, respectively (2, 3). The CYR61 protein has been shown to promote cell adhesion, migration, and proliferation—probably through the potentialization of platelet-derived growth factor and basic fibroblast growth factor activities (4). The expression of human cyr61 is associated with normal differentiation of mesenchyme cells into chondrocytes (5) and is differentially regulated in brain-derived tumor cell lines (6). The human ctgf also was identified as an immediate early gene encoding a CTGF (7) showing mitogenic activity for human umbilical vein endothelial cells (HUVECs) and fibroblasts in culture (8). On the other hand, the murine elm1 (Expressed in Low-Metastatic cells) gene, which is expressed in low-metastatic but not in high-metastatic K-1735 mouse melanoma cells, exhibits cell growth-inhibitory properties and suppresses the tumorigenic potential of mouse melanoma cells (9). Similarly, the expression of the murine rCop-1 gene is completely abolished after fibroblast transformation, and retrovirus-driven expression of rCop-1 had a dramatic cytotoxic effect on transformed cells but not on untransformed counterparts (10).
The nov gene has been initially characterized as an integration site for the myeloblastosis-associated virus MAV (11), which induces kidney tumors representing a unique model of the Wilms tumor (12). In several human nephroblastomas, the levels of wt1 and novH RNAs are inversely correlated (13) probably as a result of the negative regulation of nov expression by WT1 (14). In other types of human and animal tumors, the expression of the nov gene was found to be altered either positively or negatively (15, 16). Considering that the expression of nov was associated with cell quiescence (17) and that retrovirus-driven expression of a full-length NOV was inhibitory for cell growth (11), the elevated expression of nov in several types of tumors (including Wilms) appeared paradoxical until it was established that novH encodes a secreted 48-kDa protein whose expression is tightly associated to heterotypic blastemal differentiation in Wilms tumors (18) and to normal differentiation of central nervous system (19) and kidney (18). It therefore appears that under normal circumstances, the expression of NOV is associated or required for mesenchyme cells to undergo differentiation. Its accumulation in fully differentiated structures such as glomeruli and axons of nerve cells (18) argues for a role of NOV also being important in the maintenance of their differentiated state.
The presence of an insulin-like growth factor binding protein (IGFBP)-like motif at the N terminus of NOVH and the extensive homology of NOV, CTGF, CYR61, ELM1, RCOP-1, and IGFBP3 at their 5′ terminus raised the possibility that NOV and other related proteins may be acting in the IGF signaling pathway (11, 12). Similarly, the deletion of the N-terminal sequences of NOV was reported to confer oncogenic properties on the resulting NOV-truncated protein (11). However, recent results indicated that IGFs did not bind to recombinant NOV protein secreted by baculovirus-infected Sf9 cells (18) and suggested that NOV and IGFBPs may share common signaling activities through IGF-independent pathways, as recently reviewed for IGFBP3 (20, 21). Two motifs sharing identity with (i) Von Willebrand type C domain likely responsible for oligomerization and (ii) thrombospondin type 1 repeat responsible for interaction with extracellular-matrix proteins have been recognized in the NOV protein and other members of the CCN family. Although the biological activity of these motifs remains to be established, their conservation in all members of the family argues for their biochemical or structural importance. The C-terminal module of NOV, which was proposed to represent a dimerization domain (1), contains a cystine-knot motif present in, and involved in the dimerization of, several growth factors such as nerve growth factor, transforming growth factor β2 (TGFβ2), and platelet-derived growth factor BB. This motif is not represented in the RCOP-1 protein (10).
The multimodular structure of the NOV and other CCN proteins raises interesting questions as to the participation of each individual module on conferring the biological properties to the full-length proteins. Either the biochemical functions of the IGFBP, Von Willebrand type C, thrombospondin, and C-terminal modules contained in these proteins are indeed conserved and add up in the full length protein or the presence of each module confers on the whole protein specific biological function(s) that may substitute or add those of individual modules.
As a first step in our study of the NOVH biological function(s) and of the putative role of the different modules that it contains, we have used a two-hybrid system strategy to search for factors that interact with NOVH. To avoid selecting targets that may only interact with isolated modules, we used in the initial screening the full-length NOV protein as a bait. We report here that the secreted NOVH protein physically interacts with the C-proximal part of fibulin 1C (fib1C), an extracellular matrix protein that interacts with several signaling factors. We also show that a natural N-truncated isoform of the NOVH protein exhibits a high affinity for fib1C and that the C-terminal domain of NOVH is critical for this interaction.
MATERIALS AND METHODS
Yeast Two-Hybrid Assays.
Screening of libraries. The Matchmaker (CLONTECH) yeast two-hybrid screening system was used. The 5′-CTGCGGATCCTTACATTTTCCCTCTGGT-3′ and 5′-TATTGAATTCGTCGCTGCGACTCAGCG-3′ primers were used with the full-length novH cDNA to PCR-amplify a DNA fragment encoding NOV codons 29–357, which was inserted into pGBT9 in-frame with the DNA-binding domain of Gal4. The resulting E2NOV clone drove the expression of a recombinant GAL4/NOVprotein that contained the entire NOVH coding sequence deprived of signal peptide (exons 2–5) (data not shown). The GAL4/NOVE2 protein was used as a bait to screen a HeLa cell cDNA library fused to the Gal4 transcriptional activation domain in pGADGH. The pGBT9 bait and pGADGH plasmids were sequentially introduced into Saccharomyces cerevisiae yeast strains HF7c and Y190. Double transformants were selected on synthetic minimal medium with dextrose lacking tryptophan, leucine, and histidine but containing 5 mM (for HF7c strain) or 25 mM (for Y190 strain) 3-amino-1,2,4-triazole (3AT). Colonies were restreaked and tested for β-galactosidase activity by using a filter assay (under conditions recommended in the Matchmaker manual). Yeast cotransfected with the NOVE2 bait plasmid and pGADGH parental plasmid were negative for β-galactosidase activity. Library plasmids were isolated from yeast by using the QIAamp tissue kit (Qiagen, Chatsworth, CA) and amplified with the use of 5′-GAAATTGAGATGGTGCACGATGCAC-3′ and 5′-CTATTCGATGATGAAGATACCCCACC-3′ primers and which are complementary to pACTII regions localized 87 nt upstream and 113 nt downstream to the cloning sites, respectively. The size of the amplified DNA fragments estimated by 1% agarose gel electrophoresis ranged between 600 and 2500 bp. After further purification with QIAquick PCR purification kit, the amplified inserts were sequenced with an Applied Biosystems automated sequencer according to the manufacturer’s instructions.
Construction of NOV deletion mutants and CTGF expression plasmids.
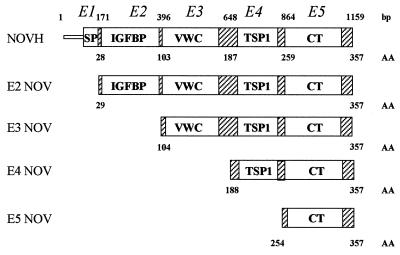
The pGBT9 E3, E4, and E5 NOV clones correspond to deletion constructs lacking NOVH sequences represented in exons 1–2, 1–3, and 1–4 respectively (Fig. 1). The reverse primer 5′-CTGCGGATCCTTACATTTTCCCTCTGGT-3′ was used in combination with forward primers 5′-TGCAGAATTCGCGGTAGAGGGAGATAAC-3′; 5′-CTTAGAATTCGCTTACAGGCCAGAAGCC-3′; and 5′-CCGTGAATTCGAGCAGCCAACAGATAAG-3′; to generate E3 NOV, E4 NOV, and E5 NOV, respectively.
Figure 1.
Schematic representation of the NOVH full-length and truncated proteins expressed by the pGBT9 plasmids. Numbers above the structural motifs indicate the nucleotide position of NOVH exons boundaries. Numbers below the motifs indicate the amino acid boundaries of NOVH exons. VWC, Von Willebrand-type C motif; TSP1, thrombospondin type 1 motif.
To construct the pGBT9–CTGF clone, a full-length ctgf cDNA (22) was used with 5′-CGCTGGATCCTCATGCCATGTCTCCGTA-3′ and 5′-TAATGAATTCGCCGTCGGCCAAACTGCA-3′ primers to PCR-clone CTGF exons 2–5 in-frame with the yeast GAL4 DNA-binding domain sequences of pGBT9.
To obtain pACTII-NOV, the sequences spanning NOVH exons 2–5 were PCR-amplified with the 5′-CTGGGATCCTTACATTTTCCCTCTGGT-3′ and 5′-TATTCCATGGAGGTCGCTGGCGACTCAGC-3′ primers and cloned in-frame with the yeast GAL4 transactivation domain of pACTII. PCR cloning of CTGF exons 2–5 of pACTII in-frame with the GAL4 transactivation domain (to generate pACTII–CTGF) was performed, with the 5′-CGCTGGATCCTCATGCCATGTCTCCGTA-3′ and 5′-TAATCCATGGCCGTCGGCCAGAACTGCA-3′ primers. In all cases, the coding region of recombinant plasmids was checked with DNA sequencing in the presence of [α-35S]dATP and T7 polymerase (Pharmacia). Plasmid DNA transformations were performed as described in the Matchmaker manual, and double transfectants were selected by plating on synthetic minimal medium with dextrose plates deprived of leucine and tryptophan.
Cloning and Preparation of Glutathione S-Transferase (GST) Fusion Proteins.
The pGEX-4T vectors (Pharmacia) were used to construct fusion proteins in-frame with the GST. A cDNA fragment encoding the NOVH polypeptide deprived of the signal peptide was PCR-amplified in the presence of the 5′-GACTGGATCCGTCGCTGCGACTCAGCGCTGC-3′ and 5′-GGCCGGAATTAGCTTGGCTGC-3′ primers and cloned in pGEX-4T-1. The 1.1-kb cDNA fragment corresponding to nucleotides 1164–2156 of the fib1C gene (see Results) was excised from pGADGH plasmid by an EcoRI–XhoI digestion and cloned in pGEX-4T-3. Escherichia coli BL21 competent cells were transformed with either the GST–NOVH or the GST–fib1C-COOH plasmids, and fusion proteins were produced as recommended by Pharmacia. The apparent molecular mass of the recombinant GST–NOVH and GST–fib1C-COOH proteins estimated after SDS/PAGE was 62 and 55 kDa, respectively.
GST Pull-Down Assays.
Serum-free conditioned TC100 medium (GIBCO) was obtained from either Sf9/82 cells infected with recombinant baculovirus expressing the NOVH protein (18) or Sf9/4 cells infected with recombinant baculovirus expressing the CTGF protein. The conditioned media were concentrated and dialyzed by using PM10 Centricon (Amicon) and adjusted to 1 ml of lysis buffer (50 mM Hepes/250 mM NaC/10 mM MgCl2/1% Triton X-100/0.5 mM phenylmethylsulfonyl fluoride/2 mM benzamidine). Conditioned minimal essential medium also was obtained from of MDCK cells stably transfected with pCMV–novH in the sense (54.14) and antisense (51.2) orientations (18). To avoid nonspecific binding during the pull-down assays, the different conditioned media were first incubated for 30 min at 4°C with glutathione–Sepharose beads and after centrifugation with GST, GST–fib1C-COOH or GST–NOVH-Sepharose beads for 14 hr at 4°C. Treated beads were washed several times with the lysis buffer, and proteins bound to the recombinant-protein beads were eluted with SDS sample buffer. Eluted proteins were separated by using SDS/PAGE and transferred onto poly(vinylidene difluoride) membrane (Amersham) for immunochemical detection. The NOVH and CTGF proteins were detected by the enhanced chemiluminescence method according to the manufacturer’s instructions (Amersham) by using anti-NOVH (K19M) or anti-CTGF (K28T) antibodies at a 1:500 dilution.
Antibodies.
The K19M anti-NOV polyclonal antibody has been described (18). The anti-CTGF (K28T) antibody was raised against a 28-aa synthetic peptide corresponding to CTGF positions 232–259. The specificity of this antibody has been confirmed with serum-free conditioned medium from Sf9 cells infected with recombinant baculoviruses in which the CTGF cDNA has been cloned either in the sense (Sf9/4) or in the antisense orientation (Sf9/3) (see Results).
Cell Cultures.
MDCK 54.14 and 54.2 subclones are cells stably transfected with pCMV vectors driving the expression of NOV in the sense [pCMV–novH(S)] and antisense [pCMV–novH(AS)] orientations (8). HeLa cells, MDCK 54.14 and 54.2, rhabdomyosarcoma-derived cells (RD), and glioblastoma-derived cell lines (G59 and G22) (23) were grown in DMEM (GIBCO/BRL); 293 cells (American Type Culture Collection CRL1573) were grown in Eagle’s minimal essential medium (GIBCO/BRL). All culture media were supplemented with 10% fetal calf serum.
RNA Purification and Northern Blot Analysis.
Procedures for RNA purification from cell cultures and Northern blotting were described (24). Nytran+ membrane (Schleicher and Schuell) blots were sequentially hybridized with the 1.9-kb EcoRI–novH probe (13, 18), the 1.1-kb EcoRI–XhoI fragment derived from the pGADGH–fib1C–COOH subclone (see Results), and the human glyceraldehyde-3-phosphate dehydrogenase probe (CLONTECH).
N-Terminal Microsequencing.
Recombinant NOVH protein expressed by Sf9/82 baculovirus-infected cells was purified to homogeneity from cell culture supernatant by affinity chromatography (C.M., Y. Blouquit, and B.P., unpublished results) and concentrated on PM10 Centricon before being subjected to 12% SDS/PAGE. Proteins were transferred onto a poly(vinylidene difluoride) membrane for 18 hr at 16 V in 50 mM Tris base/50 mM boric acid buffer. Localization of the purified 44-kDa NOVH and 25-kDa proteins was performed by staining the membrane with amido black in 40% methanol, 1% acetic acid. Both protein bands were excised and subjected to N-terminal amino sequencing on an Applied Biosystems 473A sequencer.
RESULTS
The C-Terminal Domain of NOVH Protein Mediates Interaction with Fib1C in a Yeast Two-Hybrid System.
Screening of the HeLa cell library was first performed with the NOVE2 clone encoding the full-length NOVH protein as a bait. A total of 220 (H1-H220) and 105 (Y1-Y105) histidine-positive colonies were obtained after Y190 and HF7c transformations, respectively. From these, 75 and 37 colonies that were highly positive for β-galactosidase activity were selected for further studies.
Among several other clones of interest, Y3 appeared as a good candidate for encoding a potential partner of NOVH because its nucleotide sequence matched the published sequence of the human fib1C over 694 nt. The purified Y3 insert (designated fib1C-COOH) has been used in cotransfection assays to confirm that NOVH and the C-terminal portion of fib1C indeed interacted in the yeast system (Table1). Cotransfections of fib1C-COOH and pGBT9 DNAs did not give rise to β-galactosidase activity, indicating that fib1C-COOH was not exhibiting transactivating properties on its own. We also confirmed that the pGBT9 novH constructs did not give rise to β-galactosidase activity when cotransfected with the pGADGH vector. It is worth noting that the 100% similarity between Y3 and fib1C amino acid sequences extended from residue 385 to 683 at the C terminus of fib1C. This region contains a set of EGF-like repeats that are critical for fibulin 1 self-association and fibulin 1 interactions with fibronectin and calcium (see Discussion).
To establish whether interactions between the fib1C and NOVH require several of the four structural domains of NOVH, two-hybrid system assays were performed with the E3NOV, E4NOV, and E5NOV deletion mutants and fib1C-COOH. The results of cotransfections performed with these clones (Table 1) indicated that the C-terminal domain of NOVH, which contains a cystine knot motif responsible for dimerization of several growth factors (25), is sufficient to mediate the interaction with fib1C-COOH.
Table 1.
Results obtained in B-galactosidase assays performed on yeast cotransfected with plasmids expressing indicated recombinant proteins
| pGBT9 | pGBT9-E2-NOV | pGBT9-E3-NOV | pGBT9-E4-NOV | pGBT9-E5-NOV | pACTII | pACTII-E2-NOV | pACTII-CTGF | pGADGH-fib1C | |
|---|---|---|---|---|---|---|---|---|---|
| pGADGH-fib1C | − | + | + | + | + | ND | ND | ND | ND |
| pGADGH | − | − | − | − | − | ND | ND | ND | ND |
| pACTII-E2-NOV | − | + | + | + | + | ND | ND | ND | ND |
| pACTII-CTGF | − | + | + | + | + | ND | ND | ND | ND |
| pGBT9-CTGF | ND | ND | ND | ND | ND | − | + | − | + |
ND, not done.
The C Termini of NOVH and Fib1C Physically Interact in Vitro. GST pull-down assays were performed to determine whether the C terminus of fib1C was indeed able to physically interact with the NOVH protein in vitro.
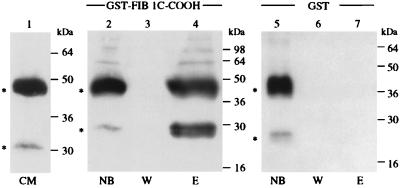
Conditioned medium of Sf9/82 cells infected with a recombinant baculovirus expressing NOVH in the sense orientation was used as a source of NOVH protein. Incubation of GST–fib1C-COOH beads with the NOV-containing conditioned medium resulted in specific binding of two polypeptides (apparent molecular mass 44 and 25 kDa) that could be recovered by elution with lysis buffer followed by SDS/PAGE and that reacted with the K19M anti-NOVH antibody (Fig. 2). Inasmuch as two such polypeptides were detected only with the NOV-containing medium (this work and ref. 18), they were likely to represent NOV proteins. The 44-kDa protein corresponded to the expected full-length recombinant NOV protein expressed in Sf9/82 cells (18). The identity of the 25-kDa polypeptide, which also was detected at a very low level in initial unconcentrated conditioned medium from Sf9/82 cells, was established by N-terminal microsequencing. For this purpose, the conditioned medium from Sf9/82 cells was fractionated by using affinity chromatography and the 44-kDa and 25-kDa proteins purified by SDS/PAGE transferred onto nylon membrane for microsequencing. The N terminus of the 44-kDa protein (TQRC) corresponded to that expected after cleavage of the signal peptide identified in NOV (11). The N terminus of the 25-kDa protein (AYRPE) indicated that it was a NOV-related polypeptide generated by cleavage of the full-length NOV protein at position 187 and that it contained NOVH sequences encoded by exons 4 and 5 (including thrombospondin type 1 and C-terminal domains).
Figure 2.
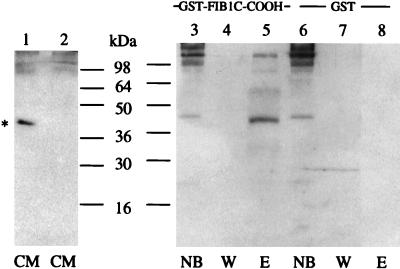
The recombinant NOVH proteins produced by baculovirus-infected cells physically interacts with fib1C-COOH. Three hundred microliters of a 50% GST–fib1C-COOH Sepharose bead slurry (corresponding to 100 μg of recombinant GST–fib1C-COOH protein) were incubated for 14 hr at 4°C with 4 ml of conditioned medium from Sf9/82 cells expressing both the 44-kDa and 25-kDa NOVH proteins (see text). After washing the beads 5 times with 1 ml of lysis buffer, the proteins bound to the column were eluted with 40 μl of sample buffer and analyzed by using SDS/PAGE. The same procedure was used with Sepharose beads coupled to GST alone as a control for nonspecific binding. After protein transfer onto poly(vinylidene) difluoride membranes, the blots were reacted with the K19M NOVH-specific antibody as described (18). CM, serum-free conditioned medium from Sf9/82 cells; NB, proteins not bound to the columns; W, content of the last wash; E, proteins eluted from the columns.
As shown in Fig. 2, the molar ratio of the 25-kDa and 44-kDa NOV proteins was increased considerably by selective retention on the GST–fib1C-COOH, therefore suggesting that the C-terminal portion of fib1C exhibits a much higher affinity toward the C-terminal part of NOV. This observation was in agreement with the positive results obtained with the pGBT9 E5NOV clone in the two-hybrid system. It has been confirmed that none of the NOVH protein forms interacted with GST alone (Fig. 2).
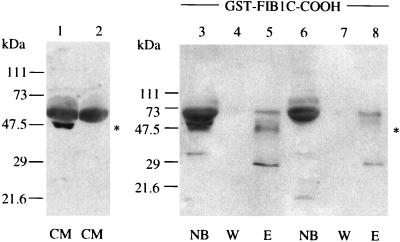
Interaction between NOVH and fib1C-COOH also was confirmed with conditioned medium from MDCK 54.14 cells stably transfected with pCMV-novH(S). As shown in Fig. 3, a 48-kDa and a 28-kDa protein specifically reacted with the K19M antibody in the fraction eluted from the GST–fib1C-COOH beads. Three other bands (64, 50, and 29 kDa) also detected in the samples obtained from MDCK 54.2 (nov antisense) conditioned medium probably result from nonspecific crossreactions occurring with the cellular extracts. The 48-kDa band corresponds to the previously described secreted glycosylated NOV isoform (18). The 28-kDa polypeptide likely corresponds to the 25-kDa truncated NOV protein detected in Sf9/82 conditioned medium (see above). The differences in apparent molecular mass between baculovirus- and MDCK-expressed NOV proteins result from previously described posttranslational modifications of NOV in higher eukaryotic cells (18). Again, the molar ratio of the full-length (48-kDa) and short (28-kDa) NOV proteins was significantly increased by selective retention on the fib1C beads because the 28-kDa band was not detected in the sample of unfractionated medium from MDCK cells.
Figure 3.
The NOVH proteins produced by MDCK transfected cells physically interact with fib1C-COOH. Four-microliter samples of conditioned medium (CM) from MDCK-novH sense (S, lane 1) and antisense (AS, lane 2) were incubated with GST–fib1C-COOH and GST–Sepharose beads and further treated as described in the legend of Fig. 2. Lane abbreviations are as in Fig. 2. Lanes 1, 3, 4, and 5: MDCK-novH (S) expressing NOVH. Lanes 2, 6, 7, and 8: control MDCK-novH(AS). ∗ indicates the position of the 48-kDa NOVH protein.
Expression of NOVH and fib1C in Tumor Cell Lines.
Because there is a fairly good overlap of novH and fib1C expression sites in normal tissues (11, 18, 26, 27), we have compared their expression in various types of tumor cells showing wide variations in their novH RNA content to determine whether any correlation could be drawn between expression of NOVH and fibulin in these cells.
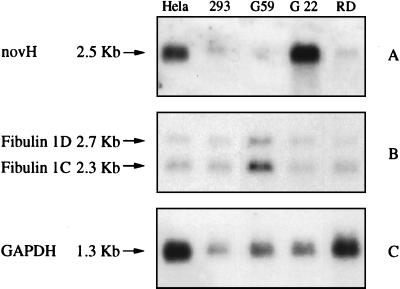
The 3′-proximal region of fib1C (nucleotides 1,164–2,156) was used as a probe on RNA preparations from cells that express nov at high (HeLa, G22), moderate (293, RD), or undetectable (G59) levels. Because of a high degree of homology in their 3′-proximal sequences, both the 2.3-kb fib1C and 2.7-kb fib1D mRNA species are detected by this probe. The results obtained (Fig. 4) established that elevated levels of novH were detected only in cells containing low amounts of fib1C, whereas high levels of fib1C were detected only in the case of G59 cells that do not express novH (16). Athough the 293 and RD cells express comparable levels of both NOVH and fib1C, these observations suggested that an inverse relationship may exist between the expression of NOVH and fib1C in certain types of tumor cells.
Figure 4.
Northern blot analysis of novH and fib1C in human tumor cell lines. Blots containing 20 μg of total RNA derived from human cell lines were successively hybridized with novH (A), fib1C (B) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, C) probes and treated for autoradiography as described in Materials and Methods.
CTGF Interacts with Fib1C-COOH.
Inasmuch as the C-terminal domain represented at the C terminus of NOVH also is present at the C terminus of CTGF, experiments were aimed at establishing whether CTGF can interact with fib1C-COOH.
Cotransfections experiments performed with pGBT9-CTGF and pGADGH-fib1C-COOH also indicated that in the two-hybrid system, CTGF was able to interact with the C-terminal portion of fib1C (Table 1). Under similar conditions, cotransfection of pGBT9-CTGF with pACTII-CTGF did not give rise to β-galactosidase activity, indicating that, as previously reported (8), CTGF did not interact with itself.
To determine whether CTGF could physically interact with fib1C-COOH, conditioned medium from Sf9/4 cells infected with a recombinant baculovirus expressing CTGF was incubated in the presence of GST–fib1C-COOH beads. Proteins selectively adsorbed to the GST–fib1C-COOH were separated by using SDS/PAGE, and the resulting Western blot was reacted with the K28T CTGF antibody. As shown in Fig. 5, the 40-kDa CTGF protein contained in the conditioned medium was found to interact with fib1C-COOH. Interestingly, the CTGF protein bound to the fib1C-COOH beads is slightly smaller than the unbound protein detected in the wash fraction (Fig. 5). This observation suggests that two CTGF proteins with slightly different molecular masses (possibly resulting from posttranslational modifications) compose the 40-kDa band and that posttranslational modifications of CTGF may modulate the binding of CTGF to fib1C.
Figure 5.
Selective retention of CTGF onto GST–fib1C-COOH Sepharose beads. Two ml of Sf9/4 conditioned medium were incubated in the presence of either GST–fib1C-COOH (lanes 3–5) or GST (lanes 6–8) Sepharose beads and analyzed as described in Fig. 2. The blots were reacted with the K28T anti-CTGF antibody. Lane 1, untreated conditioned medium from Sf9/4 cells; lane 2, untreated conditioned medium from Sf9/3 cells; other abbreviations are as in Fig. 2.
Low-Affinity NOVH–NOVH and NOVH–CTGF Interactions Can Be Visualized by the Two-Hybrid Assay.
The presence of the C-terminal domain in both NOVH and CTGF prompted us to test whether NOVH could interact with itself and with CTGF.
To determine whether NOVH and CTGF can interact via their C-terminal domain, yeast were cotransfected with pACTII-CTGF and either pGBT9-E2NOV, pGBT9-E3NOV, pGBT9-E4NOV, or pGBT9-E5NOV. A control cotransfection was performed with pACTII-CTGF and pGBT9 plasmids. As established by the β-galactosidase assays, interactions between CTGF and NOVH were possible with the whole NOVH protein and all of the deletion mutants (Table 1). This observation reinforced the possibility that the cystine knot motif present in the C-terminal domain played a central role in these interactions.
When yeast were cotransfected with pACTII-NOV and pGBT9 E2NOV, E3NOV, E4NOV, or E5NOV, colonies positive for β-galactosidase activity also were obtained. However, when GST pull-down assays were performed with GST-E2NOV beads and conditioned media from either Sf9/82 cells expressing the 44-kDa NOVH protein or Sf9/4 cells expressing the 40-kDa CTGF protein, very low amounts of NOVH and CTGF were selectively retained and recovered after binding (data not shown). These observations suggest that low-affinity NOVH–NOVH and NOVH–CTGF interactions had been visualized by the sensitive two-hybrid assays and were in agreement with the detection of small amounts of high-molecular weight NOVH multimers in the conditioned medium from Sf9/82 cells (Fig. 2).
DISCUSSION
Several lines of evidence indicate that although they share a highly conserved structure, the proteins of the CCN family do not possess fully redundant functions. Two of these proteins (CYR61 and CTGF) appear to be positive regulators, whereas the other three (NOVH, ELM1, and RCOP-1) are likely to represent negative regulators. Understanding the biological diversity of the CCN family of proteins will be eased through characterization of their targets.
The identification of fib1C as a protein that interacts with the secreted 48-kDa NOV protein provides a clue for possible participation of NOV in signaling pathways involving extracellular matrix and cytoskeleton proteins. Fib1C is a 100-kDa member of an emerging family of cystein-rich extracellular matrix and blood proteins comprising fibulin 1A, 1B, and 1C, fibulin 2, and fibulin 3. Fibulin 1 A, B, and C are generated by alternative splicing of the 3′-proximal coding sequences (28). The three isoforms contain two structural domains (1 and 2) sharing identity with anaphylatoxins and EGF. The N-proximal domain 1 (amino acids 36–144) contains three motifs similar to anaphylatoxins (C3a, C4a, and C5a). Domain 2 (amino acids 179–566) contains nine repeats similar to EGF (28). Although the biological function of fibulin 1 remains to be established, its association with matrix structures such as components of the basement membranes and connective tissue matrix fibers (29, 30) is probably related to its ability to bind fibronectin (FN), laminin, nidogen, and fibrinogen (Fg) (31, 32). Sequences responsible for self-association and binding FN and calcium have been localized in domain 2 of fibulin 1 (amino acids 356–440) that contain the EGF-like modules 5 and 6 (33). Our finding that interaction of NOV and fib1C also proceeds through a C-terminal portion (amino acids 385–683) of fib1C containing five of nine EGF repeats raises the possibility that binding of NOV to fib1C may be an important element in mediating the potential role of fib1C in assembly of the extracellular matrix and in cell adhesion.
It is worth noting that expression of fibulin 1 in the endocardial cushion tissue and endocardium of the heart, in the perichondrium and calcifying regions of developing bones, and in gut subepithelium and peripheral nerves of human embryos of gestational weeks 4–10 (27) matches the major sites of NOVH expression at the same developmental stages. Recent studies have established that NOV expression is tightly related to normal central nervous system development of human embryos of gestational weeks 10–32 (19), metanephric differentiation of blastemal cells in normal human kidney (18).
Inasmuch as fibulin was reported to interact with the C-terminal heparin-binding region of fibronectin (31), its interaction with NOVH may provide a basis for the expression of NOV in terminally differentiated structures and during cell quiescence under normal conditions. The interaction of fibulin with fibrinogen and the resulting increased platelet adhesion (34) also are of particular interest in light of the similarity existing between primary structures of NOV and the Von Willebrand factor (11), which plays a central role in homeostasis through its interaction with platelets and factor VIII (35).
Interaction of the fib1C C terminus with CTGF raises the possibility that fib1C is at a crossroad between signaling pathways involving CTGF and NOVH. It would be interesting to establish whether fib1C’s biological properties are affected under the previously reported transforming growth factor β-stimulated expression of CTGF (36).
The NOVH low-affinity homotypic interactions visualized in the two-hybrid system are in agreement with the existence of the high-molecular weight NOV-related proteins that are detected in the conditioned medium of Sf9/82 baculovirus-infected cells expressing NOVH (Fig. 2). The absence of homotypic interaction of CTGF monomers in the two-hybrid system confirmed previous observations establishing that CTGF did not dimerize (8) and may highlight biochemical differences between CTGF and NOVH.
A salient feature of our results is that an N-truncated isoform of NOVH lacking the IGFBP-related and Von Willebrand type C-related motifs exhibits a high affinity toward the C terminus of fib1C. The existence of a 28- to 30-kDa NOV-related polypeptide detected at various levels in the culture medium of several cell lines is reminiscent of the production of biologically active CTGF-truncated polypeptides in the medium of cultured cells (37). Inasmuch as the N terminus of the recombinant 25-kDa NOV protein detected in the conditioned medium from Sf9/82 cells does not correspond to any consensus proteolytic cleavage site, this peptide could result from digestion of NOVH by an unknown specific protease, the activity of which would vary within different cell types. Secondary structure predictions indicate that the putative cleavage site is localized in the hinge region linking structural domains from exons 2 and 3 to those encoded by exons 4 and 5. Our working hypothesis is that the N-truncated NOVH protein can compete with the secreted NOVH for the binding of fib1C EGF repeats and can therefore play a central role in regulating its association with other signaling factors. Imbalanced expression of nov (such as that observed in tumors) or expression of a truncated nov (such as that resulting from MAV integration in avian nephroblastomas) (11) would compromise the interactions of NOVH and fib1C with their partners and may result in improper control of cell growth and adhesion. Along this line, the absence of the C-terminal domain in RCOP-1 may be relevant. Although it has not been established whether the lack of C-terminal domain in the RCOP-1 protein results from alternative splicing or from transcription of a gene deprived of the CCN family’s exon 5 (10), one can hypothesize that RCOP-1’s negative effects on the growth of transformed cells results from the lack of the C-terminal domain and the lack of capacity to interact with C-terminal targets.
In summary, our results suggest that the C-terminal domain of NOVH and other proteins of the CCN family play a central role in mediating the interactions of these proteins with targets involved in the control of cell adhesion and assembly of the extracellular matrix. The identification of other proteins interacting with NOVH may help in understanding the biochemical properties and conserved structural organization of this emerging family of cell growth regulators.
Acknowledgments
We thank Dr. André Sentenac for his support and advice. The work performed in the Laboratoire d’Oncologie Virale et Moléculaire has been funded by grants from the Ligue Nationale Contre le Cancer (Comités National, du Cher et de l’Indre) and Groupement des Entreprises Françaises dans la Lutte Contre le Cancer. The two-hybrid system screening and preliminary characterization of the clones were performed during a short-term stay of B.P. in the Marjorie Kovler Laboratories (University of Chicago) and funded by a grant to the University of Chicago from the National Cancer Institute (CA71933) of the U.S. Public Health Services. C.M. is on leave from Centre National de la Recherche Scientifique.
ABBREVIATIONS
- fib1C
fibulin 1C
- GST
glutathione S-transferase
- CTGF
connective tissue growth factor
- IGF
insulin-like growth factor
- IGFBP
IGF binding protein
References
- 1.Bork P. FEBS Lett. 1993;327:125–130. doi: 10.1016/0014-5793(93)80155-n. [DOI] [PubMed] [Google Scholar]
- 2.Simmons D L, Levy D B, Yannoni Y, Erikson R L. Proc Natl Acad Sci USA. 1989;86:1178–1182. doi: 10.1073/pnas.86.4.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Brien T P, Yang G P, Sanders L, Lau L F. Mol Cell Biol. 1990;10:3569–3577. doi: 10.1128/mcb.10.7.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kireeva M L, Mo F E, Yang G P, Lau L F. Mol Cell Biol. 1996;16:1326–1334. doi: 10.1128/mcb.16.4.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Brien T P, Lau L F. Cell Growth Differ. 1992;3:645–654. [PubMed] [Google Scholar]
- 6.Martinerie C, Viegas-Pequignot E, Van-Cong N, Perbal B. Mol Pathol. 1997;50:310–317. doi: 10.1136/mp.50.6.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryseck R P, Macdonald-Bravo H, Mattei M G, Bravo R. Cell Growth Differ. 1991;2:225–233. [PubMed] [Google Scholar]
- 8.Bradham D M, Igarashi A, Potter R L, Grotendorst G R. J Cell Biol. 1991;114:1285–1294. doi: 10.1083/jcb.114.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashimoto Y, Shindo-Okada N, Tani M, Nagamachi Y, Takeuchi K, Shiroishi T, Toma H, Yokota J. J Exp Med. 1998;187:289–296. doi: 10.1084/jem.187.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang R, Averbouk L, Zhu W, Zhang H, Jo H, Dempsey P J, Coffey R J, Pardee A B, Liang P. Mol Cell Biol. 1998;18:6131–6141. doi: 10.1128/mcb.18.10.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joliot V, Martinerie C, Dambrine G, Plassiart G, Brisac M, Crochet J, Perbal B. Mol Cell Biol. 1992;12:10–21. doi: 10.1128/mcb.12.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perbal B. Crit Rev Oncog. 1995;5:589–613. [PubMed] [Google Scholar]
- 13.Martinerie C, Huff V, Joubert I, Badzioch M, Saunders G, Strong L, Perbal B. Oncogene. 1994;9:2729–2732. [PubMed] [Google Scholar]
- 14.Martinerie C, Chevalier G, Rauscher F J, Perbal B. Oncogene. 1996;12:1479–1492. [PubMed] [Google Scholar]
- 15.Perbal B. Bull Cancer (Paris) 1994;81:957–961. [Google Scholar]
- 16.Li W X, Martinerie C, Zumkeller W, Westphal M, Perbal B. Mol Pathol. 1996;49:M91–M97. doi: 10.1136/mp.49.2.m91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scholz G, Martinerie C, Perbal B, Hanafusa H. Mol Cell Biol. 1996;16:481–486. doi: 10.1128/mcb.16.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chevalier G, Yeger H, Martinerie C, Laurent M, Alami J, Kocialkowski S, Schofield P N, Perbal B. Am J Pathol. 1997;152:1563–1575. [PMC free article] [PubMed] [Google Scholar]
- 19.Su B-Y, Cai W-Q, Zhang C-G, Su H-C, Perbal B. C R Acad Sci. 1998;321:883–892. doi: 10.1016/s0764-4469(99)80002-x. [DOI] [PubMed] [Google Scholar]
- 20.Lalou C, Lasserre C, Binoux M. Endocrinology. 1996;137:3206–3212. doi: 10.1210/endo.137.8.8754741. [DOI] [PubMed] [Google Scholar]
- 21.Oh Y, Yamanaka Y, Kim H-S, Vorwerk P, Wilson E, Hwa V, Yang D H, Spagnoli A, Wanek D, Rosenfeld R G. In: Molecular Mechanisms to Regulate the Activities of Insulin-Like Growth Factors. Takano K, Hizuka N, Takahashi S-I, editors. Amsterdam: Elsevier; 1998. pp. 125–133. [Google Scholar]
- 22.Martinerie C, Perbal B. C R Acad Sci Paris. 1991;313:345–351. [PubMed] [Google Scholar]
- 23.Westphal M, Hansel M, Brunken M, Köning A, Köppen JA, Herrmann H D. Exp Cell Biol. 1987;55:152–163. doi: 10.1159/000163411. [DOI] [PubMed] [Google Scholar]
- 24.Perbal B. A Practical Guide to Molecular Cloning. 2nd Ed. New York: Wiley; 1988. [Google Scholar]
- 25.McDonald N Q, Hendrickson W A. Cell. 1993;73:421–424. doi: 10.1016/0092-8674(93)90127-c. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H Y, Timpl R, Sasaki T, Ekblom P. Dev Dyn. 1996;205:348–364. doi: 10.1002/(SICI)1097-0177(199603)205:3<348::AID-AJA13>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 27.Miosge N, Gotz W, Sasaki T, Chu M L, Timpl R, Herken R. Histochemistry. 1996;28:109–116. doi: 10.1007/BF02331415. [DOI] [PubMed] [Google Scholar]
- 28.Argraves W S, Tran H, Burgess W H, Dickerson K. J Cell Biol. 1990;111:3155–3164. doi: 10.1083/jcb.111.6.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spence S G, Argraves W S, Walters L, Hungerford J E, Little C D. Dev Biol. 1992;151:473–484. doi: 10.1016/0012-1606(92)90186-k. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H Y, Kluge M, Timpl R, Chu M L, Ekblom P. Differentiation. 1993;52:211–220. doi: 10.1111/j.1432-0436.1993.tb00633.x. [DOI] [PubMed] [Google Scholar]
- 31.Balbona K, Tran H, Godyna S, Ingham K C, Strickland D K, Argraves W S. J Biol Chem. 1992;267:20120–20125. [PubMed] [Google Scholar]
- 32.Pan T C, Kluge R Z, Mayer U, Timpl R, Chu M L. Eur J Biochem. 1993;215:733–740. doi: 10.1111/j.1432-1033.1993.tb18086.x. [DOI] [PubMed] [Google Scholar]
- 33.Tran H, Van Dusen W J, Argraves W S. J Biol Chem. 1997;272:22600–22606. doi: 10.1074/jbc.272.36.22600. [DOI] [PubMed] [Google Scholar]
- 34.Tran H, Tanaka A, Litvinovich S V, Medved L V, Haudenschild C C, Argraves W S. J Biol Chem. 1995;270:19458–19464. doi: 10.1074/jbc.270.33.19458. [DOI] [PubMed] [Google Scholar]
- 35.Ruggeri Z M. J Clin Invest. 1997;99:559–564. doi: 10.1172/JCI119195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grotendorst G R, Okochi H, Hayashi N. Mol Cell Biol. 1996;16:1326–1334. [Google Scholar]
- 37.Steffen C L, Ball-Mirth D K, Harding P A, Bhattacharyya N, Pillai S, Brigstock D R. Growth Factors. 1998;15:199–213. doi: 10.3109/08977199809002117. [DOI] [PubMed] [Google Scholar]