Abstract
The neurofibromatoses represent two of the most common inherited tumor predisposition syndromes affecting the nervous system. Individuals with neurofibromatosis 1 (NF1) are prone to the development of astrocytomas and peripheral nerve sheath tumors whereas those affected with neurofibromatosis 2 (NF2) develop schwannomas and meningiomas. The development of traditional homozygous knockout mice has provided insights into the roles of the NF1 and NF2 genes during development and in differentiation, but has been less instructive regarding the contribution of NF1 and NF2 dysfunction to the pathogenesis of specific benign and malignant tumors. Recent progress employing novel mouse targeting strategies has begun to illuminate the roles of the NF1 and NF2 gene products in the molecular pathogenesis of NF-associated tumors.
Keywords: neurofibromin, RAS, tumor suppressor gene, ERM proteins, actin cytoskeleton
Introduction
The neurofibromatoses comprise two distinct clinical conditions, neurofibromatosis 1 (NF1) and neurofibromatosis 2 (NF2). Each of these disorders shares the common feature of benign and malignant tumor predisposition; however, the tumor types and clinical manifestations are markedly different [1]. In addition, the genetic basis for NF1 is clearly distinct from that of NF2, in that the NF1 and NF2 genes encode unique and markedly dissimilar gene products, neurofibromin and merlin, located on chromosomes 17 and 22, respectively. Significant progress toward elucidating the molecular pathogenesis of NF1 and NF2 has resulted from the identification of their associated genes; however, our ability to design targeted therapies for these disorders is heavily dependent on the development of relevant and accurate preclinical mouse models of the specific clinical features of NF1 and NF2. This review will focus on the development and refinement of mouse models for NF1 and NF2.
Clinical Disorders
Neurofibromatosis 1
Neurofibromatosis 1 is the most common autosomal dominant disorder affecting the nervous system. Affected individuals are prone to the development of both benign and malignant tumors, including peripheral nerve sheath tumors (neurofibromas and malignant peripheral nerve sheath tumors [MPNSTs]), astrocytomas (gliomas), leukemias, and pheochromocytomas [2]. Aside from the tumor phenotypes, individuals with NF1 also exhibit pigmentary abnormalities, including hyperpigmented skin macules (café-au-lait macules), skinfold freckling, and iris hamartomas (Lisch nodules). In addition, children with NF1 often have skeletal defects (long bone deformities, scoliosis, sphenoid wing dysplasia) and learning disabilities. The most common tumor in NF1 is the benign peripheral nerve sheath tumor (neurofibroma) composed of Schwann cells, fibroblasts, and mast cells [3]. The onset of these tumors typically coincides with the onset of puberty and they continue to appear throughout adulthood. These tumors do not transform into malignant tumors, but carry a tremendous cosmetic burden. Approximately 25% to 30% of individuals with NF1 will develop a more extensive neurofibroma (plexiform neurofibroma) that is thought to represent a congenital lesion. These tumors, although also benign, may grow to enormous proportions, develop a rich blood supply, and transform into aggressive MPNSTs. Within the central nervous system (CNS), the most common tumor is the optic pathway glioma, seen in 15% of children with NF1 [4]. This low-grade pilocytic astrocytoma is rarely fatal, but can lead to visual loss and hypothalamic dysfunction. In contrast to the neurofibroma, the growth of these tumors appear to be limited to the first decade of life, with rare examples of continued growth in adulthood.
Although rare, myeloid malignancies, such as myelodysplastic syndrome (MDS) and juvenile chronic myeloid leukemia (JCML), are overrepresented in children with NF1 [5]. Boys tend to be affected more often than girls. Pheochromocytoma, a tumor of the adrenal gland, is more common in individuals with NF1. Despite its association with NF1, it affects less than 1% of individuals with the disorder.
Children with NF1 also frequently exhibit specific learning disabilities [6]. Approximately 40% to 60% of children with NF1 will manifest below average performance on standardized IQ tests and demonstrate difficulties with traditional learning paradigms. These children are not retarded, but have specific deficits in learning.
Neurofibromatosis 2
Neurofibromatosis type 2 (NF2), also called bilateral acoustic neurofibromatosis, is a dominantly inherited genetic disorder characterized by the development of bilateral vestibular schwannomas, schwannomas of other cranial, spinal, and cutaneous nerves, as well as cranial and spinal meningiomas and ependymomas [1,2,7]. The incidence of NF2 is much lower than NF1, affecting approximately 1/40,000 individuals (vs 1/3,000 for NF1). Bilateral vestibular schwannomas and multiple spinal schwannomas are diagnostic of NF2. These benign Schwann cell neoplasms (World Health Organization [WHO] grade I) develop in the majority of NF2 patients and are the hallmark of the disease. Occasionally, schwannomas may also arise on other sensory nerves, including the fifth (trigeminal) cranial nerve and spinal dorsal roots. Most vestibular schwannomas are located at the internal auditory meatus or within the internal auditory canal and slowly grow toward the cerebellopontine angle to result in increased intracranial pressure, cranial nerve dysfunction, or cerebellar herniation. Malignant progression of schwannomas to MPNSTs is a rare event.
Multiple meningiomas are the second most common tumor in NF2, occurring in nearly half of all affected individuals. These benign neoplasms (WHO grade I), composed of neoplastic arachnoidal cap cells, also grow slowly and elicit neurological signs and symptoms by compression of adjacent structures. Because of their tendency to invade brain tissue, recur after resection, and spread along the leptomeninges to involve multiple regions, some meningiomas represent a therapeutic challenge.
Other CNS tumors include ependymomas and astrocytomas. Occurrence of gliomas is less frequent in NF2 than in NF1 patients and most of these neoplasms are morphologically benign intramedullary spinal tumors. Dysplastic CNS lesions, such as glial microhamartomas, schwannosis, meningioangiomatosis and ependymal ectopias are also encountered in the context of NF2.
Pigmentary abnormalities are uncommonly seen in patients with NF2. About 60% to 80% of NF2 patients develop juvenile posterior subcapsular lens opacities. Therefore, lens changes are considered a valuable, early marker of NF2 in individuals at risk.
NF1 Gene and Function
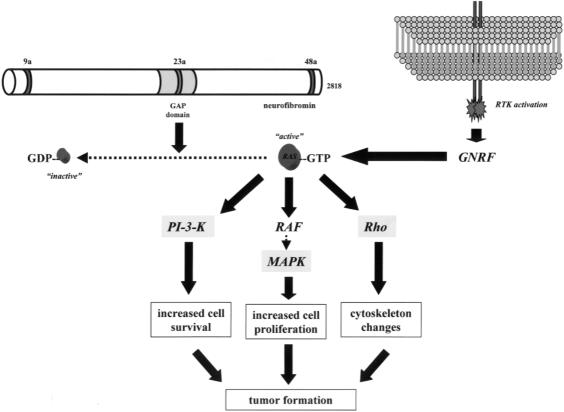
The identification of the NF1 gene ushered in a new era of molecular understanding of the pathogenesis of this disorder. The NF1 gene on chromosome 17q11.2 spans 350 kilobases (kb) of genomic DNA and encodes an 11- to 13-kb mRNA [8–11]. The protein product, neurofibromin, is a large 220- to 250-kDa cytoplasmic protein expressed predominantly in neurons, Schwann cells, oligodendrocytes, astrocytes, leukocytes and the adrenal medulla (Figure 1) [12–14]. Neurofibromin has been shown to associate with the actin and microtubule cytoskeleton, but lacks conventional binding motifs for known protein-protein associations [15–17]. Analysis of the predicted NF1 amino acid sequence revealed a small central region of the protein with sequence similarity to a family of proteins involved in the inactivation of RAS (GTPase-activating protein or GAPs) [18]. RAS is an important intracellular signaling molecule, which mediates cell proliferation in some cells (e.g., astrocytes and leukocytes) and differentiation in other cell types (e.g., neurons and Schwann cells) [19]. The association of the RAS molecule with guanosine phosphates dictates RAS activity: RAS is active when bound to GTP and inactive when associated with GDP (Figure 1). RAS-GAP molecules, like neurofibromin, function as negative regulators of RAS, by accelerating the intrinsic GTPase activity of RAS, thereby converting RAS from an active GTP-bound form to inactive RAS-GDP. Because RAS promotes cell proliferation and transformation in some cells, neurofibromin is believed to function as a tumor suppressor by inactivating RAS mitogenic signaling.
Figure 1.
Neurofibromin, the product of the NF1 gene, encodes a 2818 amino acid protein that contains a central region with RAS GTPase-activating protein (GAP) activity. The GAP domain is denoted by the shaded area. In addition to this functionally important domain, neurofibromin contains three alternatively spliced exons (9a, 23a, and 48a). Neurofibromin, like other GAP molecules, accelerates the conversion of active GTP-bound RAS to the inactive GDP-bound form. Guanosine nucleotide replacing factors (GNRFs) reactivate RAS by exchanging GDP for GTP.
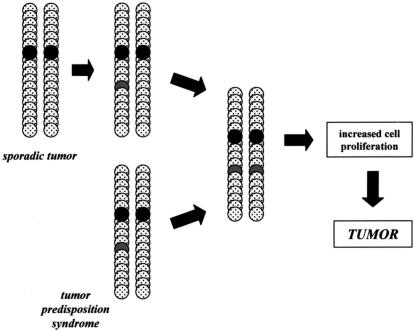
Individuals with NF1 are born with one mutated or nonfunctional NF1 gene. Similar to the retinoblastoma paradigm, tumors in NF1 form only when the one remaining wild-type NF1 allele is inactivated by somatic mutation [20]. This “two-hit” hypothesis has been validated in tumor tissues from individuals with NF1, in which both the somatic and germline NF1 mutations were identified, resulting in no neurofibromin expression (Figure 2). In agreement with the proposed function of neurofibromin as a RAS regulator, loss of neurofibromin in NF1-associated tumors is associated with elevated levels of activated RAS [21–25].
Figure 2.
Knudson “two-hit” hypothesis for tumor suppressor genes. In sporadic cancers, there is sequential inactivation of both copies of a given tumor suppressor gene, whereas in individuals with an inherited cancer syndrome, only one additional genetic “hit” is required. Because individuals with NF1 or NF2 start life with a germline mutation in the NF1 or NF2 genes, respectively, the only genetic event required for tumor formation is inactivation of the one remaining functional NF1 or NF2 gene. Loss of NF1 or NF2 expression results in increased cell proliferation that predisposes to tumor formation.
The NF1 gene has three alternatively spliced exons, 9a, 23a, and 48a, which are differentially expressed in specific tissue types. Exon 9a-containing neurofibromin is restricted to neurons of the forebrain beginning in early postnatal development [26]. Neurofibromin containing exon 48a is only expressed in muscle tissues [27]. It is not known what functions are gained or lost by the insertion of exons 9a or 48a. Lastly, exon 23a inserts a unique 21 amino acids into the GAP domain of neurofibromin and results in a neurofibromin protein with reduced GAP activity [28]. NF1-23a mRNA is found in many tissues during development and in the adult [29].
NF2 Gene and Function
The identification of germline mutations in NF2 patients provided the genetic evidence that this condition is caused by germline alterations in the NF2 gene [30,31]. The majority of germline and somatic mutations in the NF2 gene, resulting in either a premature termination codon, a splicing alteration, or a frameshift, lead to the production of a truncated protein. In-frame deletions and missense mutations have also been found, suggesting that the alteration of particular functional domains may abolish the function of the NF2 protein. In tumors from patients with NF2, mutations in both NF2 alleles have been identified, concomitant with loss of NF2 protein expression. These observations indicate that NF2 is a tumor suppressor gene, although the detailed mechanism by which NF2 mutation leads to transformation of Schwann cells is largely unknown.
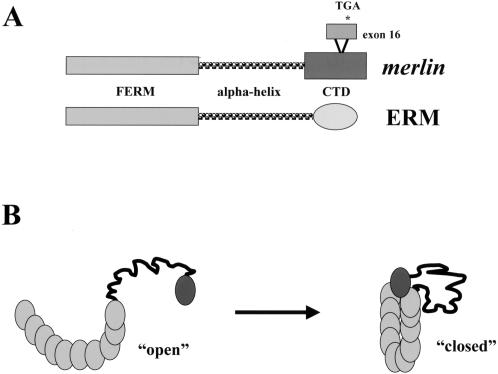
The product of the NF2 gene has been termed merlin [31] or schwannomin [30]. Analysis of the predicted protein sequence indicates that merlin belongs to the band 4.1 family of cytoskeleton-associated proteins and exhibits the greatest similarity to a subset of proteins in this family, notably the ezrin, radixin, and moesin (ERM) proteins [32]. The NF2 gene encodes a 595 amino acid protein with 17 exons (Figure 3A). Alternative splicing yields at least two merlin isoforms, type 1 (lacking exon 16) and type 2 that contains exon 16, but lacks exon 17 sequences, owing to a premature termination signal within exon 16 [33]. These NF2 isoforms are conserved in mouse [34] and rat [35]. There are three predicted regions with potential functional significance: The FERM domain (residues 1–302) and the alpha helical region (residues 303–479) are shared among all ERM family members whereas the carboxyl terminal domain of merlin is unique and lacks a conventional actin binding motif, found in other ERM proteins. Merlin is expressed in Schwann cells, neurons, lens fibers, blood vessels, and leptomeningeal cells in the adult, but exhibits a more widespread tissue expression pattern during embryonic development. The FERM homology domain of merlin appears to be the main determinant that localizes the protein to the plasma membrane [36]. Mutations that lead to an interstitial deletion in this domain have been observed both in the germline of NF2 patients and in sporadic schwannomas, meningiomas, and mesotheliomas. Mutant proteins lacking exon 2 or exons 2 and 3 do not interact with the plasma membrane, and are diffusely distributed in the cytoplasm [36,37].
Figure 3.
Structure of the NF2 gene product, merlin. (A) Merlin is structurally related to the ERM subfamily of Protein 4.1 molecules, including ERM. Merlin contains an amino terminal FERM domain and a central predicted alpha helical domain followed by a unique carboxyl terminal region (CTD) that is not conserved among ERM family members. The conventional ERM actin-binding region is not contained within the CTD of merlin. The NF2 gene has an alternatively spliced exon (exon 16) that encodes 11 unique residues followed by a termination codon, which results in a truncated protein lacking exon 17 sequences. (B) Merlin exists in “open” and “closed” conformations that may be dictated by phosphorylation. In the “open” conformation, merlin is defective as a negative growth regulator, but, in the closed self-associated form is active and binds to CD44 to mediate growth suppression.
Merlin, like other ERM proteins, exists in two conformations, dictated by its ability to form an intramolecular complex. Merlin forms an intramolecular association using residues in the extreme carboxyl terminus (exon 17) to bind to a region at the end of the FERM domain (residues 302–308) [38–40]. In addition to the N-term:C-term intramolecular complex, merlin also forms another intramolecular association within the FERM domain, which is disturbed by mutations within exons 2 and 3 [40,41]. Failure to form either an N-term:C-term or N-term:N-term association leads to an inactive growth suppressor protein (Figure 3B). The crystal structure of the merlin FERM domain has been recently determined and provides physical evidence for the biochemical studies demonstrating that intramolecular associations within the FERM domain are important for merlin function [42].
In addition, there is a good correlation between the ability of the various merlin isoforms to form intramolecular interactions and the growth suppressive activity of these molecules. In particular, merlin isoform I, which is the isoform able to inhibit growth when exogenously expressed in rat schwannoma cells, forms both intramolecular interactions [38,40]. However, merlin isoform II, which lacks the region encoded by exon 17 required for proper folding, does not affect schwannoma cell growth. Similarly, some rare merlin isoforms or naturally occurring missense mutants with variant N-terminal regions likewise exhibit a constitutively “open” conformation and lack growth-suppressive activity [43,44].
Merlin folding is also dictated by the phosphorylation state of merlin: Hyperphosphorylation of merlin leads to protein unfolding and is associated with an “inactive” molecule whereas the “active” merlin growth suppressor is underphosphorylated and associates with specific interactors to modulate cell growth [45]. In keeping with the proposed function of merlin in signal transduction events beginning at the plasma membrane, merlin interacts with the transmembrane protein CD44. Merlin associates with the cytoplasmic tail of CD44 preferentially in its hypophosphorylated “active” form [45]. Under growth-permissive conditions, merlin becomes phosphorylated and inactive and does not associate with CD44. In situations permissive for growth arrest, merlin is hypophosphorylated and can bind to CD44 to abrogate CD44 mitogenic signaling. Recent work demonstrated that merlin is phosphorylated in a Rac1/cdc42-dependent fashion [46–48]. The discovery of the link between a well-known signaling pathway and merlin represents an important step toward understanding the regulation of merlin growth suppression.
One approach to defining how merlin functions involves the identification of molecules that specifically interact with merlin and could potentially transduce its growth inhibitory signal. Several merlin interacting proteins have been identified, including βII-spectrin [49], CD44 [50], sodium-hydrogen exchange regulator factor (NHE-RF) [51], schwannomin-interacting protein-1 (SCHIP-1) [52], hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) [53], actin [43], and β1-integrin [54]. NHE-RF interacts with all ERM family members [55] and is therefore unlikely to be a specific merlin signaling molecule. SCHIP-1 has no known function to date. The interaction between merlin and HRS is intriguing, given the observation that hepatocyte growth factor (HGF) is one of the most potent mitogenic factors for Schwann cells. HRS interacts with merlin better in merlin's “open” conformation and HRS overexpression in rat schwannoma cells has the same effect on cell growth as merlin overexpression [56]. It is not known whether merlin growth suppression requires HRS.
In addition to growth suppression, merlin is involved in actin cytoskeleton-mediated processes, such as motility, spreading, and attachment. Human schwannoma cells deficient in NF2 expression have dramatic alterations in the actin cytoskeleton, which can be partially reversed by attenuation of Rho pathway activity [57] and by wild-type merlin reexpression [58]. Similarly, overexpression of wild-type merlin in rat schwannoma cells results in decreased cell attachment and motility as well as abnormalities in actin cytoskeleton organization during cell spreading [59]. Support for a role of merlin in modulating actin cytoskeleton-mediated processes also derives from experiments on tumors from Nf2+/- mice, which were shown to be highly motile and metastatic [60].
NF Mouse Modeling
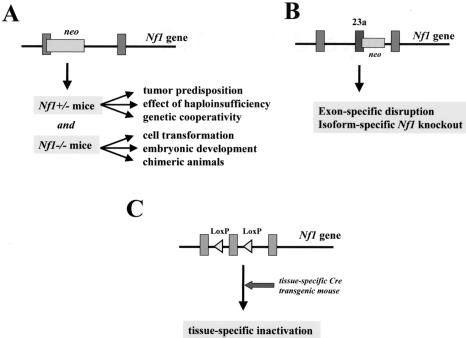
A number of distinct approaches have been taken to develop models for the tumors seen in individuals affected with NF1 (Figure 4 and Table 1). Initial studies focused on the generation of mice with a targeted mutation in the Nf1 gene. Second-generation models included Nf1-/- chimeric mice and Nf1 exon-specific knockout mice. Recently, tissue-specific inactivation of Nf1 has yielded important insights into the function of neurofibromin in specific cell types and the consequence of Nf1 loss on tumorigenesis in NF1. Similar approaches have been taken for the study of NF2 (Figure 5 and Table 2).
Figure 4.
Strategies for Nf1 mouse modeling. (A) Traditional Nf1+/- mice were generated in which one allele of the murine Nf1 gene is inactivated by the insertion of a neomycin (neo) cassette. Nf1 +/- mice can be studied for tumor predisposition, effects of haploinsufficiency, and genetic cooperativity. In addition, Nf1 +/- mice can be intercrossed to generate Nf1-/- mice to analyze the effect of neurofibromin loss on cell transformation and embryonic development. Nf1-/- cells can be used to generate chimeric Nf1 mice. (B) Nf1 mutant mice can also be generated in which specific exons are deleted. In this regard, the effect of ablation of a particular Nf1 alternatively spliced exon can be studied. Alternatively, mice can be generated in which a stop codon is inserted into the alternatively spliced exon to ablate Nf1 expression in tissues that express that particular Nf1 isoform. (C) Tissue-specific Nf1 inactivation can be accomplished by the use of Cre/LoxP technology. Briefly, LoxP recombinatorial sequences are inserted into noncoding regions of the Nf1 gene to produce phenotypically normal Nf1flox mice. Tissue-specific inactivation is mediated by the expression of the bacteriophage Cre recombinase from a tissue-specific promoter (e.g., synapsin-I promoter).
Table 1.
Nf1 Mouse Models.
| Genotype | Phenotype | Reference |
| Nf1-/- | Embryonic lethality Double outlet right ventricle Exencephaly | [61–64] |
| Nf1+/- | Tumor development: pheochromocytoma, leukemia, lymphoma | [62] |
| Astrogliosis; increased astrocyte proliferation | [75,76] | |
| Impaired spatial learning and memory | [80,81] | |
| Neuronal Nf1 conditional knockout | Astrogliosis, growth retardation | [86] |
| Astrocyte Nf1 conditional knockout | Increased astrocyte proliferation | Bajenaru et al., unpublished results Zhu et al., unpublished results |
| Nf1 exon 23a knockout | Impaired spatial learning and contextual discrimination, delayed motor skill acquisition | [85] |
| Nf1-/- chimera | Plexiform neurofibromas, myelodysplasia, neuromotor defects | [82] |
| Nf1+/-: p53+/- | Malignant peripheral nerve sheath tumors | [82,83] |
| Malignant astrocytomas | [84] | |
Figure 5.
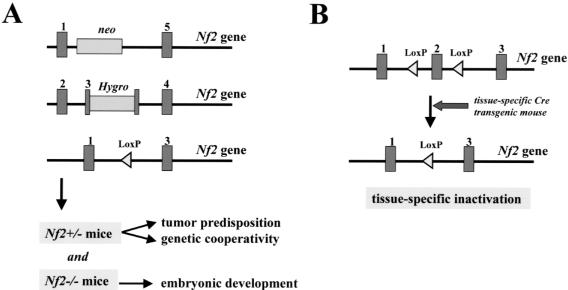
Strategies for Nf2 mouse modeling. (A) Nf2 +/- mice were generated by using three different targeting vectors. In the Nf2 mutant allele generated by McClatchey and associates [88] the 3′ part of exon 2 up to the 5′ part of intron 3 has been replaced by the selection marker leading to a message that has deleted exons 2 to 4. Giovannini and coworkers [87] generated two different mutant Nf2 alleles. One mutant allele, Nf2 KO3, was generated by an insertional mutation in exon 3 leading to a message in which exon 3 is deleted. The other mutant allele, Nf2Δ2, carried an in-frame deletion of exon 2. (B) As for Nf1, tissue-specific Nf2 inactivation can be accomplished by the use of Cre/LoxP technology [91]. LoxP recombinatorial sequences are inserted into noncoding regions flanking exon 2 of the Nf2 gene to produce phenotypically normal Nf2flox mice. Tissue-specific inactivation is mediated by the expression of the bacteriophage Cre recombinase from a tissue-specific promoter (e.g., P0 promoter) or by direct injection of adenoviral Cre.
Table 2.
Nf2 Mouse Models.
| Genotype | Phenotype | Reference |
| Nf2-/- | Embryonic lethality | [88] |
| Nf2+/- | Tumor development: osteogenic tumors, fibrosarcoma, liver tumors | [60,92] |
| P0-Sch-Δ(39–121) transgenic mice | Schwann cell hyperplasia, Schwann cell tumors (schwannoma, malignant peripheral nerve sheath tumors) | [87] |
| Schwann cell Nf2 conditional knockout | Schwann cell hyperplasia, Schwann cell tumors (schwannoma, malignant peripheral nerve sheath tumors) | [92] |
| Leptomeningeal cell Nf2 conditional knockout | Meningioma | [107] |
| Schwann cell Nf2 conditional knockout; Nf1+/- | Schwann cell hyperplasia, Schwann cell tumors (schwannoma, neurofibroma, malignant peripheral nerve sheath tumors) | Niwa-Kawakita et al., unpublished results, 2002 |
Traditional Nf1 Knockout Mice
Traditional gene-targeted studies initially focused on the development of mice in which the murine Nf1 gene was disrupted. Mice heterozygous for a mutation in the Nf1 gene (Nf1+/-) are viable and fertile, whereas those homozygous for an Nf1 mutation die during embryonic development. The embryonic lethality resulting from homozygous inactivation of the Nf1 gene reflects the critical role of neurofibromin during tissue development and organogenesis. Nf1-/- embryos die of heart failure and edema secondary to a developmental cardiac vessel defect in which the aortic and pulmonary outflow vessels remain fused (double outlet right ventricle) [61,62]. In addition, some of these mice exhibit neural tube closure defects (exencephaly) and endocardial cushion abnormalities [63,64]. The death of Nf1-/- embryos between E12.5 and 13.5 precludes an analysis of the role of the Nf1 gene in postnatal tissues and cell types affected in adults and children with NF1.
Studies on Nf1-deficient cell types have demonstrated that loss of neurofibromin is associated with increased cell proliferation or survival, increased RAS pathway activation, and tumorigenesis. Nf1-/- Schwann cells exhibit activation of RAS and but do not initially hyperproliferate in vitro [65]. After serum removal, individual clones of hyperproliferating cells emerge that lose expression of myelin P0 and display angiogenic and invasive properties [66]. Neurofibromin-deficient fibroblasts also exhibit abnormal cell proliferation and cannot form perineurium in vitro [67]. In hematopoietic progenitor cells, neurofibromin loss is associated with increased cell proliferation in response to multiple mitogenic cytokines, increased RAS pathway signaling, and chronic myeloid leukemia [22,68,69]. This hyperactivation of RAS is mediated in part through Rac2, which provides cross talk between phosphoinositide 3-kinase (PI-3K) and the Raf-MAPK pathways to modulate cell fate [70]. Lastly, in neurons, activation of RAS is associated with increased cell survival and not proliferation. Sympathetic ganglion neurons deficient in Nf1 expression display neurotrophin-independent survival [71] that is associated with hyperactivation of RAS and its downstream effector, PI-3K [72,73].
Nf1+/- Mice
Analysis of Nf1+/- mice has yielded some important insights into NF1 pathophysiology. Nf1+/- animals are prone to tumor development. By 15 to 18 months of age, Nf1+/- mice die with leukemias and pheochromocytomas [61,62]. Loss of the wild-type Nf1 gene can be demonstrated in these tumors, fulfilling the two-hit hypothesis proposed by Alfred Knudson for retinoblastoma. In addition, Nf1+/- mice are more susceptible to carcinogen-induced pigmentation and papilloma formation [74].
Nf1+/- mice also demonstrate subtle cell growth abnormalities. Mice heterozygous for a targeted mutation in the Nf1 gene demonstrate 50% more brain astrocytes with no effects on other glia populations [75]. In contrast, mice heterozygous for a targeted mutation in another RAS GAP molecule, p120-Gap, do not manifest increased astrocyte numbers. Nf1+/- astrocytes have a cell-autonomous growth advantage in vitro associated with increased activation of the downstream RAS effectors, AKT and MAPK [76]. Genetic cooperativity between Nf1 and the p53 and retinoblastoma cell cycle regulators can be seen in mice doubly heterozygous for targeted mutations in Nf1 and p53 as well as Nf1 and RB. Nf1+/- astrocytes also display abnormalities in cytoskeleton-associated processes, including motility, attachment and actin cytoskeleton organization during cell spreading, supporting the notion that some of the nontumor phenotypes in NF1 may result from additional alterations in cell physiology, not strictly due to increased cell proliferation [77]. Similarly, Nf1+/- fibroblasts proliferate faster in vitro than wild-type fibroblasts, display abnormalities in collagen deposition, and exhibit abnormal wound healing in vivo [78]. As observed with fibroblasts and astrocytes, Nf1+/- melanocytes and mast cells demonstrate increased cell proliferation and RAS pathway activation [79].
Nf1+/- mice exhibit spatial learning deficits reminiscent of the specific learning disabilities in children with NF1 [80]. The mechanism for these learning deficits appears to be related to increased GABA-mediated inhibition and specific defects in long-term potentiation (LTP) [81]. Decreasing RAS function in Nf1+/- mice reverses both the inhibition and LTP deficits, suggesting that brain dysfunction in NF1 might be a direct consequence of abnormal RAS activation.
Nf1+/- mice develop malignant tumors when bred with mice heterozygous for a mutation in another tumor suppressor gene, p53. Nf1+/-;p53+/- mice are prone to the development of high-grade MPNSTs that are histologically similar to their human counterparts [82,83]. The spectrum of tumors in these Nf1+/-;p53+/- mice can be modulated by the genetic background of the mouse [84].
Exon-Specific Nf1 Knockout Mice
Mice in which exon 23a has been selectively ablated are viable, fertile, and do not succumb to tumors, as reported for Nf1+/- mice. Interestingly, loss of exon 23a results in mice with significant abnormalities in spatial learning and memory, similar to Nf1+/- mice [85]. This phenotype is unexpected given the lack of Nf1 mRNA containing exon 23a in cultured neurons [29], but clearly argues that exon 23a is important for neurofibromin's function in cognition, memory and learning.
Exon 9a knockout mice have recently been generated by Alcino Silva and his colleagues (Y. Elgersma and A. Silva, personal communication). Mice in which Nf1 is inactivated in exon 9a-expressing neurons, produced by the insertion of a termination codon in exon 9a, demonstrate deficits in long-term potentiation and spatial learning, as well as a slight increase in astrocyte number. In contrast, ablation of only exon 9a-containing Nf1 results in mice with normal learning, but a similar increase in astrocyte number. These results suggest that whereas loss of neurofibromin in neurons results in learning deficits, exon 9a itself does not appear to be essential for learning and memory in mice.
Nf1 Chimeric Mice
A limitation to the conventional Nf1 knockout strategy is the inability to study the growth advantage and tumorigenic properties of a small population of cells deficient in neurofibromin expression in the context of the whole mouse. Two approaches have been taken to circumvent this problem. The first involves the generation of chimeric mice in which the developing embryo is composed of a small number of Nf1-/- cells. The chimeric approach allows one to more accurately model the human condition, because tumors arising in individuals with NF1 involve small numbers of cells or clones in which somatic inactivation of NF1 results in loss of neurofibromin expression. Chimeric mice composed in part of Nf1-/- cells develop neurofibromas, reminiscent of plexiform neurofibromas seen in patients with NF1 [82]. Similarly, adoptive transfer of Nf1-/- myeloid cells into an irradiated recipient normal host induces myeloproliferative disease through hypersensitivity to granulocyte-macrophage colony-stimulating factor (GM-CSF) [68].
Conditional Nf1 Knockout Mice
A second approach involves the generation of tissue-specific conditional knockout mice. Recently, mice have been generated by Luis Parada and colleagues in which the Nf1 gene is conditionally disrupted using Cre-LoxP technology. In this approach, specific exons of the Nf1 gene are flanked by LoxP recombinatorial sequences placed in non-coding introns. These floxNf1 mice are phenotypically indistinguishable from wild-type mice because the insertion of these LoxP sequences does not disrupt Nf1 gene expression. Inactivation of the Nf1 gene occurs in the presence of the bacteriophage Cre recombinase enzyme, which can be expressed in vitro by adenoviral delivery or in vivo using tissue-specific promoters in transgenic mice. Several Nf1 conditional knockout mice have been generated including those with neuron-specific, astrocyte-specific, and Schwann cell-specific Nf1 inactivation. Ablation of Nf1 in neurons using the synapsin-I promoter results in viable, growth-retarded pups that exhibit massive brain astrogliosis and abnormalities in cortical development [86]. The astrogliosis in this model appears to be reactive in response to abnormal neuronal function and is not related to increased astrocyte proliferation. In contrast, astrocyte-specific Nf1 inactivation appears to be insufficient for astrocytoma formation, but does confer a significant growth advantage for astrocytes in vitro and in vivo (Bajenaru, Zhu, Parada, and Gutmann, unpublished observations). It is not clear whether the failure to generate astrocytomas reflects additional genetic events or other cooperating factors necessary for brain tumor formation in NF1.
Traditional Nf2 Knockout Mice
Two different groups have reported the development of Nf2 knockout mice [87,88]. In the two studies, similar genetargeting approaches were used, with the primary differences relating to how much of the endogenous Nf2 gene was deleted during the targeting process (Figure 5). In our approach to model more closely human hereditary (NF2-related) and sporadic schwannomas in the mouse, we have initially generated two mouse lines carrying different Nf2 mutant alleles [87]. The first mutant allele, Nf2KO3, was generated by an insertional mutation in exon 3. This Nf2KO3 allele differed from the mutant allele described by McClatchey and associates [88], here called “Nf2KO2–3.” In the latter, the 3′ part of exon 2 up to the 5′ part of intron 3 has been replaced by the selection marker leading to a message that lacks exons 2 to 3. The second mutant allele, Nf2Δ2, carried an in-frame deletion of exon 2. Both the Nf2KO3 and Nf2Δ2 alleles mimicked two different, naturally occurring, human mutant NF2 alleles and allowed us to compare the phenotypic effects of these two human NF2 mutations in mice. Because the three different targeting approaches resulted in germline Nf2 homozygous mutant mice, which were not viable, it is likely that all have Nf2 null alleles. Nf2 null mice die early during embryonic development of a failure to initiate gastrulation as a result of an absence of organized extraembryonic ectoderm [88]. The underlying defect in these Nf2-/- embryos does not appear to be related to cell proliferation abnormalities in the embryo itself, but rather a failure to produce the extraembryonic structures required to generate a mesoderm-inducing signal from the embryo proper. The inability of the developing Nf2-deficient embryo to generate or respond to important differentiation cues suggests a role for the Nf2 protein in cell-cell signaling events critical for extraembryonic formation.
Heterozygosity for any of the three mutant alleles leads to a high incidence of bone tumors showing loss of the wild-type Nf2 allele. However, differences were observed with respect to the grade of malignancy. Metastases of osteosarcoma were found less frequently in Nf2KO3/+ and Nf2Δ2/+ mice in comparison to the mice described by McClatchey and associates [60] (29% vs. 95%, P<0.0001, in case of Nf2KO3/+ mice). The different time windows of the histologic analyses may account for this difference, because mice were followed up to 24 months in our study and up to 30 months in the study of McClatchey and colleagues. The finding of osteomas in Nf2KO3/+ and Nf2Δ2/+ mice contrasts with the absence of benign bone tumors described by McClatchey [60]. This difference might be explained by the use of different genetic backgrounds. Regardless, our data indicate that Nf2 mutant alleles lacking only exon 2 or exon 3 do not permit normal embryonic development and lack tumor-suppressor properties. Thus, both exon 2 and exon 3 carry sequences that are essential for Nf2 function and regardless of the mutation type, all three hemizygous mice do not show clinical features of human NF2. In particular they demonstrate a tumor spectrum that differs significantly from that observed in NF2 patients. Therefore, these animals do not accurately recapitulate the cognate human genetic disease.
Schwannomas and Schwann Cell Hyperplasia Are Induced by Overexpression of a Mutant Nf2
When transiently expressed in various cell types, mutant proteins corresponding to naturally occurring NF2 mutations demonstrate distinct subcellular localizations [36,89,90]. C-terminal deletion mutants of various lengths remain located at the cell membrane. In contrast, mutants with an intact C-terminal domain but with a deleted or altered N-terminal domain are mislocalized mainly to the perinuclear cytoplasmic region. Such mislocalization was observed for a mutant protein modeled from naturally occurring mutations where exons 2–3 are deleted without a frameshift, Sch-Δ(39–121) [36].
To develop a system by which to identify functional domains of the NF2 protein that may play a role in merlin growth suppression, we have generated transgenic mice expressing various Nf2 mutants under the control of the Schwann cell-specific P0 promoter [91], including a mutant schwannomin modeled from a naturally occurring mutation, Sch-Δ(39–121) and a mutant schwannomin prototypic for C-terminal deletion mutants, Sch-ΔCter. Mice expressing Sch-Δ(39–121) showed a high prevalence of Schwann cell-derived tumors and Schwann cell hyperplasia, whereas those expressing Sch-ΔCter were normal [87]. These results indicate that a subset of mutant NF2 alleles observed in patients may encode products with dominant-negative properties when overexpressed in specific cell lineages. However, the endogenous expression level of a mutant Nf2 allele may not be sufficient to elicit this “potential” oncogenic effect. Indeed, proteins generated from endogenous mutant alleles in Nf2KO3/+ and Nf2Δ2/+ mice are hardly detectable and recent data demonstrate that mutant products of endogenous alleles are efficiently degraded by the ubiquitin-dependent proteasome pathway (Gautreau et al., submitted for publication, 2002).
Conditional Nf2 Knockout Mice
To examine the consequences of Nf2-deficiency in selected cell types of adult mice, we have generated a conditional Nf2 allele (Nf2flox2) [92]. Upon successive matings with transgenic P0-Cre mice, the Nf2 conditional mutant mice recapitulate several features observed in NF2 patients, specifically Schwann cell hyperplasia, Schwann cell tumors, cataracts, and cerebral calcifications (Figure 6). All these manifestations have been exclusively observed in mice with both alleles of the Nf2 gene inactivated in Schwann cells and in a subset of neural crest cells. Meningioma, the second tumor hallmark of human NF2, was not observed in these mice, suggesting that this tumor originates from cells that do not express the P0 protein. To determine whether Nf2 disruption is also sufficient for meningioma formation, we inactivated Nf2 in homozygous conditional knockout mice by adenoviral Cre delivery. Mice with arachnoidal cell Cre-mediated excision of Nf2 exon 2 developed a range of meningioma subtypes histologically similar to the human tumors [107].
Figure 6.
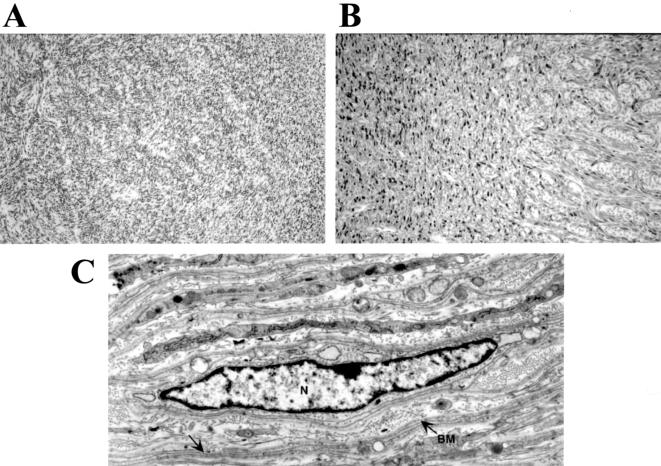
Schwannomas and Schwann cell hyperplasia in P0CreC;Nf2flox2/flox2 mice. (A) Schwannoma in the uterus of a P0CreC;Nf2flox2/flox2 mouse. The tumor appeared as a rather uniform Schwann cell growth extending from the corpus uteri to the uterine horns in combination with (B) Schwann cell hyperplasia in the peripheral part of the myometrium showing strong S-100 protein immunoreactivity. (C) Ultrastructural examination of the tumor demonstrated Schwann cells (Schwann cell nucleus denoted by N) with long, thin cytoplasmic processes (arrow), which was variably associated with a basement membrane (BM).
Conditional Nf2 knockout mice also provide a powerful tool for investigating genetic interactions and tumorigenic cooperativity. In an attempt to assess the cooperativity of the Nf2 gene with other cancer-associated genes, we and others have crossed Nf2 mutant (germline or conditional) mice to other strains of knockout mice. In such crosses, if the offspring containing both parental oncogenic alleles develop tumors faster or of a different type than either tumor-prone parent, then genetic cooperativity is operative. The identification of cooperating tumor suppressor genes, which synergize with Nf2 loss, can provide important mechanistic insights into Nf2-associated tumorigenesis pathways. Table 2 describes the results from some representative crosses performed to date. Remarkably, some crosses result in accelerated tumorigenesis in the bitransgenic offspring compared to either parent. For example, mice carrying two mutant Nf2 (conditional; P0) and one Nf1 null allele(s) not only show accelerated tumorigenesis, but also exhibit novel tumor types (e.g., neurofibroma) not seen in the parental mutant mice.
Future Approaches
The failure of Nf1+/- and Nf2+/- knockout mice to develop tumor spectra similar to the human conditions reflects the major limitation of first-generation knockout modeling strategies. The generation of mouse models that accurately mimic the human disease requires additional sophisticated approaches, such as tissue-specific and conditional mutations. However, the failure of astrocyte-specific knockout mice to develop astrocytomas indicates that even the newest generation of mouse model has a number of limitations (Figure 7).
Figure 7.
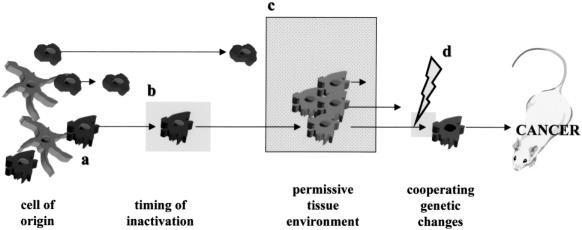
Development of future mouse models for NF1-associated tumors. The formation of tumors or cancer requires a number of events, both genetic and environment, that culminate in tumorigenesis. First, Nf1 inactivation must occur in the correct cell of origin at the proper developmental time. In some cell types, the timing of inactivation might be critical. Second, in the brain, some regions might be permissive for tumor development. This permissiveness may reflect genetic factors, regional differences in cellular composition, and brain environment. Third, cooperating genetic changes may influence tumor expansion. These genetic changes might be subtle genetic alterations, not previously thought to be conventional initiating events for tumorigenesis, or modifying loci that promote tumor formation.
First, NF1-associated astrocytomas typically occur in specific brain regions (optic nerve, chiasm, hypothalamus, and brainstem). It is possible that not all regions of the brain are permissive for NF1-associated brain tumor formation. In this regard, pilocytic astrocytomas in NF1 typically occur within the optic pathway, whereas in the general population, the most frequent brain location for a pilocytic astrocytoma is the cerebellum. Astrocytes from different regions of the brain have been reported to have differing responses to agents that elevate cyclic AMP [93], regional specificity in gap junction coupling [94], differences in glutamate and serotonin uptake [95], and distinctive growth patterns [96]. Future studies aimed at inactivating Nf1 specifically in cerebellar or optic nerve astrocytes might be useful in determining the importance of regional differences in astrocyte biology or brain environment.
Second, the cell of origin of the pilocytic astrocytoma may not be a type 1 fibrillary astrocyte, based on recent studies demonstrating gene expression similarities between pilocytic astrocytomas and oligodendrocyte-astrocyte precursors [103]. Pilocytic astrocytomas represent a unique grade of glial neoplasm that does not progress to a fibrillary astrocytoma. Several studies have highlighted these inherent differences, both with regard to genetic changes and glial cell protein expression patterns [97–101]. For instance, pilocytic astrocytomas express the unique PEN5 epitope shared with cells and tumors of an oligodendrocyte lineage [102]. This marker is not expressed in fibrillary astrocytes or astrocytomas. Recent studies from our laboratory have demonstrated expression of oligodendrocyte-associated transcripts, such as PLP, PMP-22, MBP, and oligodendrocyte myelin glycoprotein (OMgP) in pilocytic astrocytomas, suggesting that these tumors have at least some gene expression features of oligodendrocyte-type 2 astrocyte (O2A) cells [103]. In this regard, it may be necessary to inactivate Nf1 in the correct progenitor cell, perhaps an O2A lineage cell, to generate NF1-associated astrocytomas in mice.
Third, NF1-associated astrocytomas do not develop in the context of a genetically normal (Nf1+/+) brain environment. In contrast, these tumors develop in a brain environment in which the rest of the brain is Nf1+/-. In the neurofibroma, it has been suggested that Nf1+/- mast cells and fibroblasts play a role in tumorigenesis. Another environmental effect is the contribution of modifying genes to tumor development. In the case of glioblastoma, Reilly et al. observed genetic background-specific effects on the spectrum of tumors in Nf1+/-;p53+/- mice [84]. Further work will be required to formally address this genetic environment effect as well as the mechanisms that underlie the modifier gene effect on tumor spectrum.
Lastly, it is possible that NF1-associated astrocytomas only form when other cooperating genetic alterations are present. It is unlikely that these additional genetic events are the common alterations seen in high-grade astrocytomas, like p53 loss [101], despite the observation that high-grade glioblastomas form in mice doubly heterozygous for mutations in both Nf1 and p53 [84]. Work is ongoing in many laboratories using gene and protein expression profiling technologies to identify subtle cooperating factors that promote or inhibit tumorigenesis. The characterization of these additional proteins may provide additional targets for rational drug design for the effective management of tumors in NF1.
In the case of NF2 modeling, the refinement of current targeting approaches has allowed the generation of relevant models of NF2-related tumor development and provides powerful tools for investigating meningioma progression and for the preclinical evaluation of potential therapeutic interventions. In particular, the availability of arachnoid cell-specific Nf2 conditional mutant mice will greatly facilitate the elucidation of the critical genetic factors that influence meningioma development and progression.
Recently, inactivation of Protein 4.1B (or DAL-1), a second protein 4.1-related tumor suppressor, was identified as an early event in meningioma tumorigenesis [104,105]. In contrast to NF2, Protein 4.1B loss occurs in meningiomas and not in schwannomas [104], and overexpression of Protein 4.1B suppresses meningioma cell line proliferation, but has no effect on schwannoma cell line growth in vitro [106]. Further work will need to be performed to determine the relationship of merlin loss to Protein 4.1B expression as well as the contribution of Protein 4.1B to meningioma pathogenesis in the mouse. The generation of a preclinical mouse model for meningiomas through the targeted disruption of the Protein 4.1B and Nf2 genes could facilitate the assessment of potential targeted therapies for this common brain tumor in NF2.
Acknowledgements
We thank E. Robanus-Maandag, M. van der Valk and J. Woodruff for histology pictures.
Abbreviations
- ERM
ezrin-radixin-moesin
- GAP
GTPase-activating protein
- MPNST
malignant peripheral nerve sheath tumor
- NF1
neurofibromatosis type 1
- NF2
neurofibromatosis type 2
- WHO
World Health Organization
Footnotes
D. H. G. is supported by grants from the American Cancer Society and the National Institutes of Health. M. G. is supported by Grants from the US Army Medical Research and Materiel Command, Ligue Nationale Française contre le Cancer, Association pour la Recherche sur le Cancer, and the Association Neurofibromatoses et Recklinghausen.
References
- 1.Friedman JM, Gutmann DH, MacCollin M, Riccardi VM. Neurofibromatosis: phenotype, natural history and pathogenesis. Baltimore, MD: Johns Hopkins Press; 1999. [Google Scholar]
- 2.Gutmann DH, Aylsworth A, Carey JC, Korf B, Marks J, Pyeritz RE, Rubinstein A, Viskochil D. The diagnostic evaluation and multidisciplinary management of neurofibromatosis 1 and neurofibromatosis 2. JAMA. 1997;278:51–57. [PubMed] [Google Scholar]
- 3.Korf BR. Plexiform neurofibroma. Am J Med Genet. 1999;89:31–37. doi: 10.1002/(sici)1096-8628(19990326)89:1<31::aid-ajmg7>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 4.Listernick R, Louis DN, Packer PJ, Gutmann DH. Optic pathway gliomas in children with neurofibromatosis 1: consensus statement from the NF1 optic pathway glioma task force. Ann Neurol. 1997;41:143–149. doi: 10.1002/ana.410410204. [DOI] [PubMed] [Google Scholar]
- 5.Side L, Taylor B, Cayouette M, Conner E, Thompson P, Luce M, Shannon K. Homozygous inactivation of the NF1 gene in bone marrow cells from children with neurofibromatosis type 1 and malignant myeloid disorders. N Engl J Med. 1997;336:1713–1720. doi: 10.1056/NEJM199706123362404. [DOI] [PubMed] [Google Scholar]
- 6.North KK, Riccardi V, Samango-Sprouse C, Ferner R, Moore B, Legius E, Ratner N, Denckla MB. Cognitive function and academic performance in neurofibromatosis 1: consensus statement from the NF1 cognitive disorders task force. Neurology. 1997;48:1121–1127. doi: 10.1212/wnl.48.4.1121. [DOI] [PubMed] [Google Scholar]
- 7.McKusick VA. Mendelian inheritance in man: a catalogue of human genes and genetic disorders. Baltimore: The John Hopkins University Press; 1994. http://www.ncbi.nlm.nih.gov/omim. [Google Scholar]
- 8.Cawthon RM, Weiss M, Xu G, Viskochil D, Culver M, Stevens J, Robertson M, Dunn D, Gesteland R, OConnell P, White R. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell. 1990;62:193–201. doi: 10.1016/0092-8674(90)90253-b. [DOI] [PubMed] [Google Scholar]
- 9.Viskochil D, Buchberg AM, Xu G, Cawthon RM, Stevens J, Wolff RK, Culver M, Carey JC, Copeland NG, Jenkins NA, White R, O'Connell P. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990;62:187–192. doi: 10.1016/0092-8674(90)90252-a. [DOI] [PubMed] [Google Scholar]
- 10.Wallace MR, Marchuk DA, Andersen LB, Letcher R, Odeh HM, Saulino AM, Fountain JW, Brereton A, Nicholson J, Mitchell AL, Brownstein BH, Collins FS. Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science. 1990;249:181–186. doi: 10.1126/science.2134734. [DOI] [PubMed] [Google Scholar]
- 11.Marchuk DA, Saulino AM, Tavakkol R, Swaroop M, Wallace MR, Andersen LB, Mitchell AL, Gutmann DH, Boguski M, Collins FS. cDNA cloning of the type 1 neurofibromatosis gene: complete sequence of the NF1 gene product. Genomics. 1991;11:931–940. doi: 10.1016/0888-7543(91)90017-9. [DOI] [PubMed] [Google Scholar]
- 12.Dasto MM, Scrable H, Norlund M, Sturbaum AK, Nissen LM, Ratner N. The protein product of the neurofibromatosis type 1 gene is expressed at highest abundance in neurons, Schwann cells and oligodendrocytes. Neuron. 1992;8:415–428. doi: 10.1016/0896-6273(92)90270-n. [DOI] [PubMed] [Google Scholar]
- 13.DeClue JE, Cohen BD, Lowy DR. Identification and characterization of the neurofibromatosis type 1 gene product. Proc Natl Acad Sci USA. 1991;88:9914–9918. doi: 10.1073/pnas.88.22.9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutmann DH, Wood DL, Collins FS. Identification of the neurofibromatosis type 1 gene product. Proc Natl Acad Sci USA. 1991;88:9658–9662. doi: 10.1073/pnas.88.21.9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bollag G, McCormick F, Clark R. Characterization of full-length neurofibromin: tubulin inhibits Ras GAP activity. EMBO J. 1993;12:1923–1927. doi: 10.1002/j.1460-2075.1993.tb05841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregory PE, Gutmann DH, Boguski M, Mitchell AM, Parks S, Jacks T, Wood DL, Jove R, Collins FS. The neurofibromatosis type 1 gene product, neurofibromin, associates with microtubules. Somatic Cell Mol Genet. 1993;19:265–274. doi: 10.1007/BF01233074. [DOI] [PubMed] [Google Scholar]
- 17.Li C, Cheng Y, Gutmann DH, Mangoura D. Differential localization of the neurofibromatosis 1 (NF1) gene product, neurofibromin, with the F-actin or microtubule cytoskeleton during differentiation of telencephalic neurons. Dev Brain Res. 2001;130:231–248. doi: 10.1016/s0165-3806(01)00190-0. [DOI] [PubMed] [Google Scholar]
- 18.Xu G, O'Connell P, Viskochil D, Cawthon R, Robertson R, Culver M, Dunn D, Stevens J, Gesteland R, White R, Weiss R. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990;62:599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- 19.Vojtek AB, Der CJ. Increasing complexity of the Ras signaling pathway. J Biol Chem. 1998;273:19925–19928. doi: 10.1074/jbc.273.32.19925. [DOI] [PubMed] [Google Scholar]
- 20.Knudson AG. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basu TN, Gutmann DH, Fletcher JA, Glover TW, Collins FS, Downward J. Aberrant regulation of ras proteins in tumour cells from type 1 neurofibromatosis patients. Nature. 1992;356:713–715. doi: 10.1038/356713a0. [DOI] [PubMed] [Google Scholar]
- 22.Bollag G, Clapp DW, Shih S, Adler F, Zhang YY, Thompson P, Lange BJ, Freedman MH, McCormick F, Jacks T, Shannon K. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat Genet. 1996;12:144–148. doi: 10.1038/ng0296-144. [DOI] [PubMed] [Google Scholar]
- 23.DeClue JE, Papageorge AG, Fletcher JA, Diehl SR, Ratner N, Vass WC, Lowy DR. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Recklinghausen (type 1) neurofibromatosis. Cell. 1992;69:265–273. doi: 10.1016/0092-8674(92)90407-4. [DOI] [PubMed] [Google Scholar]
- 24.Lau N, Feldkamp MM, Roncari L, Loehr AH, Shannon P, Gutmann DH, Guha A. Loss of neurofibromin is associated with activation of ras/MAPK and PI3-K/akt signaling in a neurofibromatosis 1 astrocytoma. J Neuropathol Exp Neurol. 2000;59:759–767. doi: 10.1093/jnen/59.9.759. [DOI] [PubMed] [Google Scholar]
- 25.Sherman LS, Atit R, Rosenbaum T, Cox AD, Ratner N. Single cell Ras-GTP analysis reveals altered ras activity in a single population of neurofibroma Schwann cells but not fibroblasts. J Biol Chem. 2000;275:30740–30745. doi: 10.1074/jbc.M001702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutmann DH, Zhang Y, Hirbe A. Developmental regulation of a neuron-specific neurofibromatosis 1 (NF1) isoform. Ann Neurol. 1999;46:777–782. doi: 10.1002/1531-8249(199911)46:5<777::aid-ana15>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 27.Gutmann DH, Geist RT, Rose K, Wright DE. Expression of two new protein isoforms of the neurofibromatosis type 1 gene product, neurofibromin, in muscle tissues. Dev Dyn. 1995;202:302–311. doi: 10.1002/aja.1002020309. [DOI] [PubMed] [Google Scholar]
- 28.Andersen LB, Ballester R, Marchuk DA, Chang E, Gutmann DH, Saulino AM, Camonis J, Wigler M, Collins FS. A conserved alternative splice in the von Recklinghausen neurofibromatosis (NF1) gene produces two neurofibromin isoforms, both with GAP activity. Mol Cell Biol. 1993;13:487–495. doi: 10.1128/mcb.13.1.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutmann DH, Geist RT, Wright DE, Snider WD. Expression of the neurofibromatosis 1 (NF1) isoforms in developing and adult rat tissues. Cell Growth Differ. 1995;6:315–322. [PubMed] [Google Scholar]
- 30.Rouleau GA, Mérel P, Lutchman M, Sanson M, Zucman J, Marineau C, Hoang-Xuan K, Demczuk S, Desmaze C, Plougastel B, Pulst SM, Lenoir G, Bijlsma E, Fashold R, Dumanski J, de Jong P, Parry D, Eldrige R, Aurias A, Delattre O, Thomas G. Alteration in a new gene encoding a putative membrane-organizing protein causes neurofibromatosis type 2. Nature. 1993;363:515–521. doi: 10.1038/363515a0. [DOI] [PubMed] [Google Scholar]
- 31.Trofatter JA, MacCollin MM, Rutter JL, Murrell JR, Duyao MP, Parry DM, Eldridge R, Kley N, Menon AG, Pulaski K, Haase VH, Ambrose CM, Munroe D, Bove C, Haines JL, Martuza RL, MacDonald ME, Seizinger BR, Short MP, Buckler AJ, Gusella JF. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell. 1993;72:791–800. doi: 10.1016/0092-8674(93)90406-g. [DOI] [PubMed] [Google Scholar]
- 32.Bretscher A, Chambers D, Nguyen R, Reczek D. ERM-Merlin and EBP50 protein families in plasma membrane organization and function. Annu Rev Cell Dev Biol. 2000;16:113–143. doi: 10.1146/annurev.cellbio.16.1.113. [DOI] [PubMed] [Google Scholar]
- 33.Arakawa H, Hayashi H, Nagase N, Ogawa M, Nakamura Y. Alternative splicing of the NF2 gene and its mutation analysis of breast and colorectal cancers. Hum Mol Genet. 1994;3:565–568. doi: 10.1093/hmg/3.4.565. [DOI] [PubMed] [Google Scholar]
- 34.Huynh DP, Nechiporuk T, Pulst SM. Alternative transcripts in the mouse neurofibromatosis type 2 (NF2) gene are conserved and code for schwannomins with distinct C-terminal domains. Hum Mol Genet. 1994;3:1075–1107. doi: 10.1093/hmg/3.7.1075. [DOI] [PubMed] [Google Scholar]
- 35.Gutmann DH, Wright DE, Geist RT, Snider WD. Expression of the neurofibromatosis 2 (NF2) gene isoforms during rat embryonic development. Hum Mol Genet. 1995;4:471–478. doi: 10.1093/hmg/4.3.471. [DOI] [PubMed] [Google Scholar]
- 36.Deguen B, Merel P, Goutebroze L, Giovannini M, Reggio H, Arpin M, Thomas G. Impaired interaction of naturally occurring mutant NF2 protein with actin-based cytoskeleton and membrane. Hum Mol Genet. 1998;7:217–226. doi: 10.1093/hmg/7.2.217. [DOI] [PubMed] [Google Scholar]
- 37.Koga H, Araki N, Takeshima H, Nishi T, Hirota T, Kimura Y, Nakao M, Saya H. Impairment of cell adhesion by expression of the mutant neurofibromatosis type 2 (NF2) genes which lack exons in the ERM-homology domain. Oncogene. 1998;17:801–810. doi: 10.1038/sj.onc.1202010. [DOI] [PubMed] [Google Scholar]
- 38.Sherman L, Xu H-M, Geist RT, Saporo-Irwin S, Howells N, Ponta H, Herrlich P, Gutmann DH. Interdomain binding mediates tumor growth suppression by the NF2 gene product. Oncogene. 1997;15:2505–2509. doi: 10.1038/sj.onc.1201418. [DOI] [PubMed] [Google Scholar]
- 39.Gutmann DH, Geist RT, Xu H, Kim JS, Saporito-Irwin S. Defects in neurofibromatosis 2 protein function can arise at multiple levels. Hum Mol Genet. 1998;7:335–345. doi: 10.1093/hmg/7.3.335. [DOI] [PubMed] [Google Scholar]
- 40.Gutmann DH, Haipek CA, Hoang Lu K. Neurofibromatosis 2 tumor suppressor protein, merlin, forms two functionally important intramolecular associations. J Neurosci Res. 1999;58:706–716. [PubMed] [Google Scholar]
- 41.Brault E, Gautreau A, Lamarine M, Callebaut I, Thomas G, Goutebroze L. Normal membrane localization and actin association of the NF2 tumor suppressor protein are dependent on folding of its N-terminal domain. J Cell Sci. 2001;114:1901–1912. doi: 10.1242/jcs.114.10.1901. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu T, Seto A, Maita N, Hamada K, Tsukita S, Hakoshima T. Structural basis for neurofibromatosis type 2: crystal structure of the merlin FERM domain. J Biol Chem. 2001;276:52. doi: 10.1074/jbc.M109979200. [DOI] [PubMed] [Google Scholar]
- 43.Xu HM, Gutmann DH. Merlin differentially associates with the microtubule and actin cytoskeleton. J Neuroscience Res. 1998;51:403–415. doi: 10.1002/(SICI)1097-4547(19980201)51:3<403::AID-JNR13>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 44.Gutmann DH, Hirbe AC, Haipek CA. Functional analysis of neurofibromatosis 2 (NF2) missense mutations. Hum Mol Genet. 2001;10:1519–1529. doi: 10.1093/hmg/10.14.1519. [DOI] [PubMed] [Google Scholar]
- 45.Morrison H, Sherman LS, Legg J, Banine F, Isacke C, Haipek CA, Gutmann DH, Ponta H, Herrlich P. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 2001;15:968–980. doi: 10.1101/gad.189601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaw RJ, Paez JG, Curto M, Yaktine A, Pruitt WM, Saotome I, O'Bryan JP, Gupta V, Ratner N, Der CJ, Jacks T, McClatchey AI. The Nf2 tumor suppressor, merlin, functions in Rac-dependent signaling. Dev Cell. 2001;1:63–72. doi: 10.1016/s1534-5807(01)00009-0. [DOI] [PubMed] [Google Scholar]
- 47.Kissil JL, Johnson KC, Eckman MS, Jacks T. Merlin phosphorylation by p21-activated kinase 2 and effects of phosphorylation on merlin localization. J Biol Chem. 2002;277:10394–10399. doi: 10.1074/jbc.M200083200. [DOI] [PubMed] [Google Scholar]
- 48.Xiao GH, Beeser A, Chernoff J, Testa JR. p21-Activated kinase links Rac/Cdc42 signaling to merlin. J Biol Chem. 2002;277:883–886. doi: 10.1074/jbc.C100553200. [DOI] [PubMed] [Google Scholar]
- 49.Scoles DR, Huynh DP, Morcos PA, Coulsell ER, Robinson NGG, Tamanoi F, Pulst SM. Neurofibromatosis 2 tumour suppressor schwannomin interacts with βII-spectrin. Nat Genet. 1998;18:354–359. doi: 10.1038/ng0498-354. [DOI] [PubMed] [Google Scholar]
- 50.Sainio M, Zhao F, Heiska L, Turunen O, den Bakker M, Zwarthoff E, Lutchman M, Rouleau GA, Jääskeläinen J, Vaheri A, Carpén O. Neurofibromatosis 2 tumor suppressor protein colocalizes with ezrin and CD44 and associates with actin-containing cytoskeleton. J Cell Sci. 1997;110:2249–2260. doi: 10.1242/jcs.110.18.2249. [DOI] [PubMed] [Google Scholar]
- 51.Murthy A, Gonzalez-Agosti C, Cordero E, Pinney D, Candia C, Solomon F, Gusella J, Ramesh V. NHE-RF, a regulatory cofactor for Na+-H+ exchange, is a common interactor for merlin and ERM (MERM) proteins. J Biol Chem. 1998;273:1273–1276. doi: 10.1074/jbc.273.3.1273. [DOI] [PubMed] [Google Scholar]
- 52.Goutebroze L, Der Sarkissian H, Brault E, Thomas G. Assignment of the schwannomin-interacting protein 1 (SCHIP1) gene to human chromosome band 3q25 by in situ hybridization and with somatic cell hybrids. Cytogenet Cell Genet. 2001;94:96–97. doi: 10.1159/000048795. [DOI] [PubMed] [Google Scholar]
- 53.Scoles DR, Huynh DP, Chen MS, Burke SP, Gutmann DH, Pulst SM. The neurofibromatosis 2 tumor suppressor protein interacts with hepatocyte growth factor-regulated tyrosine kinase substrate. Hum Mol Genet. 2000;9:1567–1574. doi: 10.1093/hmg/9.11.1567. [DOI] [PubMed] [Google Scholar]
- 54.Obremski VJ, Hall AM, Fernandez-Valle C. Merlin, the neurofibromatosis type 2 gene product, and beta1 integrin associate in isolated and differentiating Schwann cells. J Neurobiol. 1998;37:487–501. [PubMed] [Google Scholar]
- 55.Reczek D, Berryman M, Bretscher A. Identification of EBP50: a PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J Cell Biol. 1997;139:169–179. doi: 10.1083/jcb.139.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gutmann DH, Haipek CA, Burke SP, Sun CX, Scoles DR, Pulst SM. The NF2 interactor, hepatocyte growth factor-regulated tyrosine kinase substrate (HRS), associates with merlin in the “open” conformation and suppresses cell growth and motility. Hum Mol Genet. 2001;10:825–834. doi: 10.1093/hmg/10.8.825. [DOI] [PubMed] [Google Scholar]
- 57.Pelton PD, Sherman L, Rizvi TA, Marchionni MA, Wood P, Friedman RA, Ratner N. Ruffling membrane, stress fiber, cell spreading and proliferation abnormalities in human Schwannoma cells. Oncogene. 1998;17:2195–2209. doi: 10.1038/sj.onc.1202141. [DOI] [PubMed] [Google Scholar]
- 58.Bashour AM, Meng JJ, Ip W, MacCollin M, Ratner N. The neurofibromatosis type 2 gene product, merlin, reverses the F-actin cytoskeletal defects in primary human schwannoma cells. Mol Cell Biol. 2002;22:1150–1157. doi: 10.1128/MCB.22.4.1150-1157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gutmann DH, Sherman L, Seftor L, Haipek C, Hoang Lu K, Hendrix M. Increased expression of the NF2 tumor suppressor gene product, merlin, impairs cell motility, adhesion and spreading. Hum Mol Genet. 1999;8:267–275. doi: 10.1093/hmg/8.2.267. [DOI] [PubMed] [Google Scholar]
- 60.McClatchey AI, Saotome I, Mercer K, Crowley D, Gusella JF, Bronson RT, Jacks T. Mice heterozygous for a mutation at the Nf2 tumor suppressor locus develop a range of highly metastatic tumors. Genes Dev. 1998;12:1121–1133. doi: 10.1101/gad.12.8.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brannan CI, Perkins AS, Vogel KS, Ratner N, Nordlund ML, Reid SW, Buchberg AM, Jenkins NA, Parada LF, Copeland NG. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- 62.Jacks T, Shih TS, Schmitt EM, Bronson RT, Bernards A, Weinberg RA. Tumor predisposition in mice heterozygous for a targeted mutation in NF1. Nat Genet. 1994;7,:353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- 63.Lakkis MM, Epstein JA. Neurofibromin modulation of ras activity is required for normal endocardial-mesenchymal transformation in the developing heart. Development. 1998;125:4359–4367. doi: 10.1242/dev.125.22.4359. [DOI] [PubMed] [Google Scholar]
- 64.Lakkis MM, Golden JA, O'Shea S, Epstein JA. Neurofibromin deficiency in mice causes exencephaly and is a modifier for Splotch neural tube defects. Dev Biol. 1999;212:80–92. doi: 10.1006/dbio.1999.9327. [DOI] [PubMed] [Google Scholar]
- 65.Kim HA, Rosenbaum T, Marchionni MA, Ratner N, DeClue JE. Schwann cells from neurofibromin-deficient mice exhibit activation of p21-ras, inhibition of cell proliferation and morphological changes. Oncogene. 1995;11:325–335. [PubMed] [Google Scholar]
- 66.Kim HA, Ling B, Ratner N. Nf1-deficient mouse Schwann cells are angiogenic and invasive and can be induced to hyperproliferate: reversion of some phenotypes by an inhibitor of farsenyl protein transferase. Mol Cell Biol. 1997;17:862–872. doi: 10.1128/mcb.17.2.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosenbaum T, Boissy YL, Kombrinck K, Brannan CI, Jenkins NA, Copeland NG, Ratner N. Neurofibromin-deficient fibroblasts fail to form perineurium in vitro. Development. 121:3583–3592. doi: 10.1242/dev.121.11.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Largaespada DA, Brannan CI, Jenkins NA, Copeland NG. Nf1 deficiency causes Ras-mediated granulocyte/macrophage colony stimulating factor hypersensitivity and chronic myeloid leukaemia. Nat Genet. 1996;12:137–143. doi: 10.1038/ng0296-137. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Y-Y, Vik TA, Ryder JW, Srour EF, Jacks T, Shannon K, Clapp DW. Nf1 regulates hematopoietic progenitor cell growth and ras signaling in response to multiple cytokines. J Exp Med. 1998;187:1893–1902. doi: 10.1084/jem.187.11.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ingram DA, Hiatt K, King AJ, Fisher L, Shivakumar R, Derstine C, Wenning MJ, Diaz B, Travers JB, Hood A, Marshall M, Williams DA, Clapp DW. Hyperactivation of p21-ras and the hematopoietic-specific Rho GTPase, Rac2, cooperate to alter the proliferation of neurofibromin-deficient mast cells in vivo and in vitro. J Exp Med. 2001;194:57–69. doi: 10.1084/jem.194.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vogel KS, Brannan CI, Jenkins NA, Copeland NG, Parada LF. Loss of neurofibromin results is neurotrophin-independent survival of embryonic sensory and sympathetic neurons. Cell. 1995;82:733–742. doi: 10.1016/0092-8674(95)90470-0. [DOI] [PubMed] [Google Scholar]
- 72.Klesse LJ, Parada LF. P21 ras and phosphatidylinositol-3 kinase are required for survival of wild-type and NF1 mutant sensory neurons. J Neuroscience. 1998;18:10420–10428. doi: 10.1523/JNEUROSCI.18-24-10420.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vogel KS, El-Afandi M, Parada LF. Neurofibromin negatively regulates neurotrophin signaling through p21-ras in embryonic sensory neurons. Mol Cell Neurosci. 2000;15:398–407. doi: 10.1006/mcne.2000.0836. [DOI] [PubMed] [Google Scholar]
- 74.Atit RP, Mitchell K, Nguyen L, Warshawsky D, Ratner N. The neurofibromatosis type 1 (Nf1) tumor suppressor is a modifier of carcinoegn-induced pigmentation and papilloma formation in C57Bl/6 mice. J Invest Dermatol. 2000;114:1093–1100. doi: 10.1046/j.1523-1747.2000.00994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gutmann DH, Loehr A, Zhang Y, Kim J, Henkemeyer M, Cashen A. Haploinsufficiency for the neurofibromatosis 1 (NF1) tumor suppressor results in increased astrocyte proliferation. Oncogene. 1999;18:4450–4459. doi: 10.1038/sj.onc.1202829. [DOI] [PubMed] [Google Scholar]
- 76.Bajenaru ML, Donahoe J, Corral T, Keilly KM, Brophy S, Pellicer A, Gutmann DH. Neurofibromatosis 1 (NF1) heterozygosity results in a cell-autonomous growth advantage for astrocytes. Glia. 2001;33:314–323. doi: 10.1002/1098-1136(20010315)33:4<314::aid-glia1030>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 77.Gutmann DH, Wu YL, Hedrick NM, Zhu Y, Guha A, Parada LF. Heterozygosity for the neurofibromatosis 1 (NF1) tumor suppressor results in abnormalities in cell attachment, spreading and motility in astrocytes. Hum Mol Genet. 2001;10:3009–3016. doi: 10.1093/hmg/10.26.3009. [DOI] [PubMed] [Google Scholar]
- 78.Atit RP, Crowe MJ, Greenhalgh DG, Wenstrup RJ, Ratner N. The Nf1 tumor suppressor regulates mouse skin wound healing, fibroblast proliferation, and collagen deposited by fibroblasts. J Invest Dermatol. 1999;112:835–842. doi: 10.1046/j.1523-1747.1999.00609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ingram DA, Yang F-C, Travers JB, Wenning MJ, Hiatt K, New S, Hood A, Shannon K, Williams DA, Clapp DW. Genetic and biochemical evidence that haploinsufficiency of the Nf1 tumor suppressor gene modulates melanocyte and mast cell fates in vivo. J Exp Med. 2000;191:181–187. doi: 10.1084/jem.191.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Silva AJ, Frankland PW, Marowitz Z, Friedman E, Lazlo G, Cioffi D, Jacks T, Bourtchuladze R. A mouse model for the learning and memory deficits associated with neurofibromatosis type 1. Nat Genet. 1997;15:281–284. doi: 10.1038/ng0397-281. [DOI] [PubMed] [Google Scholar]
- 81.Costa RM, Federov MB, Kogan JH, Murphy GG, Stern J, Ohno M, Kucherlapati R, Jacks T, Silva AJ. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 2002;415:526–530. doi: 10.1038/nature711. [DOI] [PubMed] [Google Scholar]
- 82.Cichowski K, Shih TS, Schmitt E, Santiago S, Reilly K, McLaughlin ME, Bronson RT, Jacks T. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999;286:2172–2176. doi: 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 83.Vogel KS, Kleese LJ, Velasco-Miguel S, Meyers K, Rushing EJ, Parada LF. Mouse tumor model for neurofibromatosis type 1. Science. 1999;286:2176–2179. doi: 10.1126/science.286.5447.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reilly KM, Loisel DA, Bronson RT, McLaughlin ME, Jacks T. Nf1;Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects. Nat Genet. 2000;26:109–113. doi: 10.1038/79075. [DOI] [PubMed] [Google Scholar]
- 85.Costa RM, Yang T, Huynh DP, Pulst SM, Viskochil DH, Silva AJ, Brannan CI. Learning deficits, but normal development and tumor predisposition, in mice lacking exon 23a of Nf1. Nat Genet. 2001;27:399–405. doi: 10.1038/86898. [DOI] [PubMed] [Google Scholar]
- 86.Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, Rushing EJ, Marth JD, Parada LF. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15:859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Giovannini M, Robanus-Maandag E, Niwa-Kawakita M, van der Valk M, Woodruff JM, Goutebroze L, Merel P, Berns A, Thomas G. Schwann cell hyperplasia and tumors in transgenic mice expressing a naturally occurring mutant NF2 protein. Genes Dev. 1999;13:978–986. doi: 10.1101/gad.13.8.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McClatchey AI, Saotome I, Ramesh V, Gusella JF, Jacks T. The NF2 tumor suppressor gene product is essential for extraembryonic development immediately prior to gastrulation. Genes Dev. 1997;11:1253–1265. doi: 10.1101/gad.11.10.1253. [DOI] [PubMed] [Google Scholar]
- 89.Shaw RJ, McClatchey AI, Jacks T. Localization and functional domains of the neurofibromatosis type II tumor suppressor, merlin. Cell Growth Differ. 1998;9:287–296. [PubMed] [Google Scholar]
- 90.Xu L, Gonzalez-Agosti C, Beauchamp R, Pinney D, Sterner C, Ramesh V. Analysis of molecular domains of epitope-tagged merlin isoforms in Cos-7 cells and primary rat Schwann cells. Exp Cell Res. 1998;238:231–240. doi: 10.1006/excr.1997.3843. [DOI] [PubMed] [Google Scholar]
- 91.Messing A, Behringer RR, Hammang JP, Palmiter RD, Brinster RL, Lemke G. P0 promoter directs expression of reporter and toxin genes to Schwann cells of transgenic mice. Neuron. 1992;8:507–520. doi: 10.1016/0896-6273(92)90279-m. [DOI] [PubMed] [Google Scholar]
- 92.Giovannini M, Robanus-Maandag E, van der Valk M, Niwa-Kawakita M, Abramowski V, Goutebroze L, Woodruff JM, Berns A, Thomas G. Conditional biallelic Nf2 mutation in the mouse promotes manifestations of human neurofibromatosis type 2. Genes Dev. 2000;14:1617–1630. [PMC free article] [PubMed] [Google Scholar]
- 93.Cholewinski AJ, Wilkin GP. Astrocytes from forebrain, cerebellum, and spinal cord differ in their responses to vasoactive intestinal peptide. J Neurochem. 1988;51:1626–1633. doi: 10.1111/j.1471-4159.1988.tb01133.x. [DOI] [PubMed] [Google Scholar]
- 94.Lee SH, Kim WT, Cornell-Bell AH, Sontheimer H. Astrocytes exhibit regional specificity in gap-junction coupling. Glia. 1994;11:315–325. doi: 10.1002/glia.440110404. [DOI] [PubMed] [Google Scholar]
- 95.Amundson RH, Goderlie SK, Kimelberg HK. Uptake of 3H-serotonin and 3H-glutamate by primary astrocyte cultures: II. Differences in cultures prepared from different brain regions. Glia. 1992;6:9–18. doi: 10.1002/glia.440060103. [DOI] [PubMed] [Google Scholar]
- 96.Escalona-Zapara J, Diez-Nau MD. Distinctive growth patterns between cerebral and cerebellar astrocytomas — a tissue culture study. Histopathology. 1981;5:639–650. doi: 10.1111/j.1365-2559.1981.tb01830.x. [DOI] [PubMed] [Google Scholar]
- 97.White FV, Anthony DC, Yunis EJ, Tarbell NJ, Scott RM, Schofield DE. Nonrandom chromosomal gains in pilocytic astrocytomas of childhood. Hum Pathol. 1995;26:979–986. doi: 10.1016/0046-8177(95)90087-x. [DOI] [PubMed] [Google Scholar]
- 98.Zattara-Cannoni H, Gambarelli D, Lena G, Dufour H, Choux M, Grisoli F, Vagner-Capodano AM. Are juvenile pilocytic astrocytomas benign tumors? A cytogenetic study in 24 cases. Cancer Genet Cytogenet. 1998;104:157–160. doi: 10.1016/s0165-4608(97)00455-x. [DOI] [PubMed] [Google Scholar]
- 99.Sanoudou D, Tingby O, Ferguson-Smith MA, Collins VP, Coleman N. Analysis of pilocytic astrocytomas by comparative genomic hybridization. Br J Cancer. 2000;82:1218–1222. doi: 10.1054/bjoc.1999.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cheng Y, Pang JCS, Ng H-K, Ding M, Zhang SF, Zheng J, Liu DG, Poon WS. Pilocytic astrocytomas do not show most of the genetic changes commonly seen in diffuse astrocytomas. Histopathology. 2000;37:437–444. doi: 10.1046/j.1365-2559.2000.01005.x. [DOI] [PubMed] [Google Scholar]
- 101.Li J, Perry A, James CD, Gutmann DH. Cancer-related gene expression in neurofibromatosis 1 (NF1)-associated pilocytic astrocytomas. Neurology. 2001;56:885–890. doi: 10.1212/wnl.56.7.885. [DOI] [PubMed] [Google Scholar]
- 102.Figarella-Branger D, Daniel L, Andre P, Guia S, Renaud W, Monti G, Vivier E, Rougon G. The PEN5 epitope identifies an oligodendrocyte precursor cell population and pilocytic astrocytomas. Am J Pathol. 1999;155:1261–1269. doi: 10.1016/S0002-9440(10)65228-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gutmann DH, Hedrick NM, Li J, Nagarajan R, Perry A, Watson MA. Comparative gene expression profile analysis of neurofibromatosis 1 (NF1)-associated and sporadic pilocytic astrocytomas. Cancer Res In press. 2002 [PubMed] [Google Scholar]
- 104.Gutmann DH, Donahoe J, Perry A, Lemke N, Gorse K, Kittiniyom K, Rempel SA, Gutierrez JA, Newsham IF. Loss of DAL-1, a protein 4.1-related tumor suppressor, is an important early event in the pathogenesis of meningiomas. Hum Mol Genet. 2000;9:1495–1500. doi: 10.1093/hmg/9.10.1495. [DOI] [PubMed] [Google Scholar]
- 105.Perry A, Cai DX, Scheithauer BW, Swanson PE, Lohse CM, Newsham IF, Weaver A, Gutmann DH. Merlin, DAL-1, and progesterone receptor expression in clinicopathologic subsets of meningioma: a correlative immunohistochemical study of 175 cases. J Neuropathol Exp Neurol. 2000;59:872–879. doi: 10.1093/jnen/59.10.872. [DOI] [PubMed] [Google Scholar]
- 106.Gutmann DH, Hirbe AC, Huang ZY, Haipek CA. The protein 4.1 tumor suppressor, DAL-1, impairs cell motility, but regulates proliferation in a cell-type-specific fashion. Neurobiol Dis. 2001;8:266–278. doi: 10.1006/nbdi.2000.0376. [DOI] [PubMed] [Google Scholar]
- 107.Kalamarides M, Niwa-Kawakita M, Leblois H, Abramowski V, Perricaudet M, Janin A, Thomas G, Gutmann DH, Giovanni M. Nf2 gene inactivation in arachnoidal cells is rate-limiting for meningioma development in the mouse. Genes Dev. doi: 10.1101/gad.226302. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]