Abstract
The accurate segregation of chromosomes requires the kinetochore, a complex protein machine that assembles onto centromeric DNA to mediate attachment of replicated sister chromatids to the mitotic spindle apparatus. This study reveals an important role for the yeast RSC ATP-dependent chromatin-remodeling complex at the kinetochore in chromosome transmission. Mutations in genes encoding two core subunits of RSC, the ATPase Sth1p and the Snf5p homolog Sfh1p, interact genetically with mutations in genes encoding kinetochore proteins and with a mutation in centromeric DNA. RSC also interacts genetically and physically with the histone and histone variant components of centromeric chromatin. Importantly, RSC is localized to centromeric and centromere-proximal chromosomal regions, and its association with these loci is dependent on Sth1p. Both sth1 and sfh1 mutants exhibit altered centromeric and centromere-proximal chromatin structure and increased missegregation of authentic chromosomes. Finally, RSC is not required for centromeric deposition of the histone H3 variant Cse4p, suggesting that RSC plays a role in reconfiguring centromeric and flanking nucleosomes following Cse4p recruitment for proper chromosome transmission.
The segregation of sister chromatids during cell division ensures that each daughter cell receives a complete copy of the genome. The accuracy of chromosome transmission depends on the coordination of events in the chromosome cycle. Sister chromatid cohesion is first established during DNA replication or shortly thereafter and is maintained as chromosomes condense and align themselves on the mitotic spindle until the onset of anaphase. The attachment of chromosomes to the mitotic spindle is mediated by kinetochores, multiprotein complexes that assemble onto centromeric DNA. When all sister chromatids achieve bipolar kinetochore-microtubule orientation, cohesion is dissolved, and sister chromatids segregate to opposite spindle poles (70). Defects in any of these steps can result in aneuploidy, which can lead to unregulated cell growth or death. Cellular surveillance pathways such as the spindle checkpoint closely monitor kinetochore-microtubule interactions to ensure that chromosome alignment, orientation, and segregation occur with fidelity (1, 62).
Centromere function in the budding yeast Saccharomyces cerevisiae is conferred by an unusually compact 125-bp DNA sequence comprised of three conserved elements, CDEI, CDEII, and CDEIII, which are necessary and sufficient to mediate segregation of sister chromatids (12, 29). CDEI and CDEIII are highly conserved palindromic sequences that flank the nonconserved AT-rich CDEII sequence (29). This comparatively simple point yeast centromere is nevertheless capable of assembling a complex kinetochore structure resembling that of higher eukaryotes. Binding of the CBF3 complex to CDEIII is thought to nucleate kinetochore assembly, thereby playing a central role in kinetochore function (32, 41). Binding of the Cbf1p dimer to CDEI induces DNA bending in a manner thought to be important for stabilization of higher-order kinetochore structure (32). In addition to Cbf1p and the CBF3 complex, the centromere-specific histone H3 variant Cse4p, a homologue of the human CENP-A protein, and Mif2p, a homologue of human CENP-C, also interact with centromeric DNA elements (41, 43, 48). The attachment of the kinetochore to spindle microtubules requires outer kinetochore protein complexes. Central kinetochore complexes, such as CTF19, link these outer microtubule-binding complexes to the inner kinetochore (14).
Several studies in the budding yeast S. cerevisiae suggest that chromatin structure is critical for centromere-kinetochore function and chromosome segregation (56, 68). Centromeric chromatin of S. cerevisiae consists of a 160- to 220-bp nuclease-resistant domain demarcated by strong nuclease-hypersensitive sites flanked by highly ordered pericentromeric chromatin (56). The core centromeric chromatin is marked by the presence of the histone H3 variant Cse4p. These specialized H3 histones are present at active centromeres in all eukaryotes examined, from S. cerevisiae to humans (68). Specific mutations in the genes encoding histones H2A, H2B, and H4 and the histone H3 variant as well as in the CDE elements alter the nuclease digestion patterns of both core centromeric and centromere-proximal regions and have been shown to impair mitotic chromosome segregation in S. cerevisiae (25, 43, 51, 54, 55). Genetic evidence from S. cerevisiae suggests that centromeric chromatin includes a histone octamer in which the histone H3 variant replaces histone H3 (63, 68). How this specialized nucleosome is assembled and targeted to the centromere and how it contributes to kinetochore assembly are largely unknown.
Eukaryotic chromatin undergoes dynamic structural changes throughout the cell division cycle. These structural alterations range from the local changes necessary for transcriptional regulation to global changes necessary for chromosome segregation. Several evolutionarily conserved protein complexes capable of modifying histones or altering histone-DNA interactions have been identified. These chromatin-modifying complexes can be grouped into two major classes. The first class of enzymes covalently modifies histones and includes histone acetyltransferases, kinases, histone deacetylases, and histone methyltransferases (66). The second class of remodelers, represented by the SWI/SNF and related ISWI, CHD, Mi-2/NURD, and Ino80 families, has a common subunit in the Snf2p/Swi2p family of ATPases and uses the energy of ATP hydrolysis to disrupt histone-DNA interactions (4, 59, 76). Several of these ATP-dependent remodelers can function in conjunction with histone-modifying complexes to regulate access of transcription factors to nucleosomal DNA (35, 67). Roles for chromatin remodelers in DNA replication, DNA repair, and recombination have also been reported (20). RSC (for remodels the structure of chromatin) is a 15-protein ATP-dependent remodeling complex in the SWI/SNF family that is essential for viability and cell cycle progression (2, 10, 11, 17, 37, 74). Four of the RSC proteins, Sth1p, Sfh1p, Rsc8p, and Rsc6p, are highly similar to the SWI-SNF subunits Snf2p, Snf5p, Swi3p, and Swp73p, respectively. Genetic analysis and genome-wide localization and expression studies suggest roles for RSC in the cellular response to stress, nitrogen and carbohydrate metabolism, mitochondrial function, and the regulation of RNA polymerase III-mediated transcription (2, 13, 15, 17, 45, 47).
Previously, two studies implicated RSC in chromosome and plasmid transmission (73, 77). Here, we report a role for RSC at kinetochores in chromosome segregation. RSC interacts genetically and physically with kinetochore components and is localized to centromeric and centromere-proximal regions. Centromeric chromatin structure is altered in sth1-3ts and sfh1-1ts mutants, and RSC localization is impaired in sth1-3ts mutants. Moreover, sister chromatids are missegregated in both rsc mutants. Interestingly, RSC appears to be dispensable for the centromeric deposition of the kinetochore proteins Cse4p and Mif2p. We propose that the RSC nucleosome remodeler is required for configuring centromeric and flanking chromatin structure that supports proper kinetochore function.
MATERIALS AND METHODS
Strains, plasmids, and genetic methods.
The S. cerevisiae strains used had either the S288c or W303 genetic background and are listed in Table 1. Plasmids used in this study are listed in Table 2. Strain constructions and genetic manipulations were carried out by standard methods (53). All yeast media were prepared as described previously (53). Yeast cultures were grown in rich medium, consisting of yeast extract, peptone, and 2% dextrose (YPD), or selective synthetic complete (SC) medium containing 2% dextrose. Adenine indicator plates for the chromosome segregation assay contained synthetic complete minimal medium supplemented with adenine (6 μg/ml) to enhance coloration. For synchronization of cells in G1 phase, α-factor was added to a final concentration of 15 μg/ml; for S phase, hydroxyurea was added to a final concentration of 0.1 M; and for G2/M phase, nocodazole was added to a final concentration of 15 μg/ml. Temperature shift experiments and fluorescence-activated cell sorting analysis were performed as described previously (11, 17). To determine viability, cells grown at the nonpermissive temperature were briefly sonicated and plated onto YPD medium at a concentration of ≈250 cells/plate at the indicated times.
TABLE 1.
S. cerevisiae strains
| Straina | Relevant genotype |
|---|---|
| BLY2 | MATahis3-Δ200 ura3-52 ade2-101 leu2-3,112 trp1-901 URA3::lexAop-lacZ gal4 gal80 |
| BLY46 | MATahis3-11,15 ura3-1 ade2-1 leu2-3,112 trp1-1 can1-100 |
| BLY49 | MATasth1-3ts his3-Δ200 ura3-52 ade2-101 |
| BLY75-1 | MATα sfh1-1::HIS3 his3-11,15 ura3-1 ade2-1 leu2-3,112 trp1-1 can1-100 |
| BLY76 | MATahis3-Δ200 ura3-52 ade2-101 lys2-801 |
| BLY278 | MATaura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 |
| BLY283 | MATahtb1-1 htb2-2 ura3-1 his3-11,15 ade2-1 leu2-3,112 trp1-1 can1-100 (pRS314-HTB1) |
| BLY295 | MATamad1Δ::TRP1 his3-11,15 ura3-1 ade2-1 leu2-3,112 trp1-1 can1-100 |
| BLY296 | MATasfh1-1::HIS3 mad1Δ::TRP1 his3-11,15 ura3-1 ade2-1 leu2-3,112 trp1-1 can1-100 |
| BLY298 | MATaswh3Δ::HIS3 his3-Δ200 ura3-52 lys2-801 pJMH2 (CEN URA3 SWH3-HA) |
| BLY301 | MATaswh3Δ::HIS3 his3-Δ200 ura3-52 lys2-801 leu2-3,112 pJMH1 (CEN URA3 SWH3) |
| BLY309 | MATahtb1-1 htb2-2 ura3-1 his3-11,15 ade2-1 leu2-3,112 trp1-1 can1-100 (pRS314-Flag-HTB1) |
| BLY353 | MATasfh1-1::HIS3 his3-Δ200 ura3-52 ade2-101 lys2-801 leu2-Δ1 trp1-Δ1 |
| BLY360 | MATα sth1-3ts his3-Δ200 ura3-52 ade2-101 leu2-Δ1 lys2-801 CFIII (CEN3L.YPH278) URA3 SUP11 |
| BLY361 | MATα STH1 his3-Δ200 ura3-52 ade2-101 leu2-Δ1 lys2-801 CFIII (CEN3L.YPH278) URA3 SUP11 |
| BLY363 | MATa/MATα his3-Δ200/his3-Δ200 ura3-52/ura3-52 leu2-Δ1/LEU2 lys2-801/lys2-801 ade2-101/ade2-101CFIII (CEN3L.YPH278) URA3 SUP11 |
| BLY394 | MATa/MATα sth1-3ts/sth1-3ts his3-Δ200/his3-Δ200 ura3-52/ura3-52 ade2-101/ade2-101 leu2-Δ1/LEU2 lys2-801/LYS2 CFIII (CEN3L.YPH278) URA3 SUP11 |
| BLY397 | MATaMCD1-6HA ura3 leu2 |
| BLY398 | MATaMCD1 ura3 leu2 |
| BLY413 | MATaura3-52 leu2-3,112 lys2Δ200 HHT hhf1-20 Δ(HHT1-HHF1) Δ(HHT2-HHF2) |
| BLY415 | MATα ura3-52 lys2 his3-Δ200 HHT hhf1-20 Δ(HHT1-HHF1) Δ(HHT2-HHF2) |
| BLY416 | MATα mif2-3 his3-Δ200 ura3 |
| BLY424 | MATaura3-52 ade2-101 trp1-Δ1 lys2-801 his3-Δ200 leu2-Δ1 CFIII(D8B.d) CDEII(+45) URA3 SUP11 CEN6 |
| BLY429 | MATaura3-52 ade2-101 trp1-Δ1 lys2-801 CFIII(D8B.d) CDEI(8-C) URA3 SUP11 CEN6 |
| BLY493 | MATasth1-3ts his3-11,15 ade2-1 can1-100 leu2-3,112::GFP::tetR-LEU2 trp1-1 ura3-1::tetO-URA3 |
| BLY500 | MATα cse4-1 his3 ura3-52 ade2-101 lys2-801 |
| BLY503 | MATα ctf14-42 his3-Δ200 ura3-52 ade2 leu2-Δ1 lys2-801 |
| BLY506 | MATandc10-1 his3-Δ200 ura3 ade2 leu2 lys2 trp1 hom3? bar1:KAM? |
| BLY520 | MATahis3-11,15 ade2-1 can1-100 leu2-3,112::GFP::tetR-LEU2 trp1-1 ura3-1 cen1::URA3-CEN3-tetO |
| BLY526 | MATα sth1-3ts his3-11,15 ade2-1 can1-100 leu2-3,112::GFP::tetR-LEU2 trp1-1 ura3-1 cen1::URA3-CEN3-tetO |
| BLY528 | MATα sth1-3ts ctf14-42 ura3-52 leu2-Δ1 his3-Δ200 lys2-801 ade2-101? |
| BLY531 | MATα sth1-3ts mif2-3 ura3 his3-Δ200 |
| BLY532 | MATα sth1-3ts ndc10-1 his3-Δ200 ura3 ade2-101 hom3? bar1:KAM? |
| BLY536 | MATα sth1-3ts cse4-1 his3 ura3-52 ade2-101 lys2-801 |
| BLY538 | MATα sfh1-1::HIS3 ndc10-1 his3-Δ200 ura3 ade2 leu2 lys2 trp1 hom3? barl:KAM? |
| BLY546 | MATα sth1-3ts ura3-52 ade2-101 his3-Δ200 HHT hhf1-20 Δ(HHT1-HHF1) Δ(HHT2-HHF2) |
| BLY549 | MATα sth1-3ts swh3Δ::HIS3 ura3-52 his3-Δ200 ade2-101 pJMH2 (pRS316-HA-SWH3) |
| BLY552 | MATasfh1-1::HIS3 lys2 ura3-52 trp1-Δ200 HHT hhf1-20 Δ(HHT1-HHF1) Δ(HHT2-HHF2) |
| BLY565 | MATasth1-3ts ura3-52 ade2-101 lys2-801 his3-Δ200 CFIII(D8B.d) CDEI(8-C) URA3 SUP11 CEN6 |
| BLY566 | MATα sth1-3ts ura3-52 ade2-101 leu2-Δ1 lys2-801 his3-Δ200 CFIII(D8B.d) CDEII(+45) URA3 SUP11 CEN6 |
| BLY568 | MATasth1-3ts his3-11,15 ura3-1::GFP-TUB1-URA3 ade2-1 leu2-3, 112 trp1-1 can1-100 |
| BLY570 | MATahis3-11,15 ura3-1::GFP-TUB1-URA3 ade2-1 leu2-3,112 trp1-1 can1-100 |
| BLY572 | MATaura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 pRS315 |
| BLY573 | MATaura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 (Flag-H31) (pR5315) |
| BLY574 | MATaura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2Δ1(pRS424) |
| BLY575 | MATaura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 (Flag-H4) |
| PM1311 | MATaura3-52 ade2-101 his3-Δ200 lys2-801 STH1 CSE4-HA::URA3 |
| PM1312 | MATaura3-52 ade2-101 his3-Δ200 LYS2 sth1-3ts CSE4-3HA::URA3 |
BLY2 is the same as CTY10-5d (obtained from R. Sternglanz); BLY278 and BLY424 are the same as YPH250 and YPH433 respectively (obtained from P. Hieter); BLY397 and BLY398 are the same as 1377A1-4B and 983-8A, respectively (obtained from P. Megee and D. Koshland); and BLY413 is the same as MSY554 (obtained from M. M. Smith).
TABLE 2.
Plasmids
| Plasmid | Descriptiona | Source or reference |
|---|---|---|
| pJMH1 | pRS316-SWH3 (CEN6, URA3) | This study |
| pJMH2 | pRS316-HA-SWH3 (CEN6, URA3) | This study |
| YEp24 | 2μm, URA3, Ampr | 31 |
| pYC5H | SFH1 in YEp24 (2μm, URA3) | 11 |
| pPY3 | CSE4 (2μm, URA3) | 22 |
| pPM47 | NDC10, 2μm, URA3 | 42 |
| pPM290 | ∼11.2-kb KpnI fragment from pRS306- tetOp(224) subcloned into pUC19- based CEN1 replacement vector pPM101 | 42 |
| pACT2 | GAD, 2μm, LEU2 | 38 |
| pJL484 | GAD-CSE4, 2μm, LEU2 | 48 |
| pJL486 | GAD-MIF2, 2μm, LEU2 | 48 |
| pJO196 | GAD-NDC10, 2μm, LEU2 | 48 |
| pGAD-CTF19 | GAD-CTF19, 2μm, LEU2 | 48 |
| pSH2-1 | 2μm, HIS3 | 26 |
| pLexA202-Sth11-1359 | 2μm, HIS3 | 11 |
| pIT331 | GAD-SWH3/RSC8, 2μm, LEU2 | 72 |
| pDJ91 | STH1, 2μm, URA3 | 11 |
| pK3524 | NLS-tetR-GFP LEU2 | 44 |
| pRS306-tetO(224) | URA3 | 44 |
| pAFS125 | pTUB1-GFP | A. W. Murray |
| p307.3 | pGEX-2T (histone H3 residues 21-46) | 28 |
| p272.1 | pGEX-2T (histone H4 15-34) | 28 |
| p284.1 | pGEX-2T (histone H4 1-16) | 28 |
| p107.1 | pGEX-2T (histone H4 1-34) | 28 |
| p289.6 | pGEX-2T (histone H3 1-25) | 28 |
| p184.1 | pGEX-2T (histone H3 1-46) | 28 |
| pFlag-H4 | 1.9-kb EcoRI-BamHI fragment of 2μm-URA3 Flag-H4 (gift of M. A. Osley) cloned into pRS424 | |
| pFlag-H3 | 2.0-kb XhoI-BamHI fragment of pPM416 (pRS314-Flag-H3) into pRS315 | |
| pRS424 | 2μm, TRP1 | 60 |
| pRS315 | CEN6/ARSH4 | 60 |
NLS, nuclear localization signal.
Diploid strain BLY297 (swh3Δ::HIS3/SWH3 ura3/ura3) was transformed with pJMH2 or pJMH1 to uracil prototrophy, and the transformants were sporulated and subjected to tetrad dissection. The recovered swh3Δ spore clones carrying pJMH2 or pJMH1 were named strains BLY298 and BLY301, respectively. Strains BLY295 (mad1Δ) and BLY296 (sfh1-1ts mad1Δ) were constructed by one-step gene disruption with the SmaI-SalI fragment of pUCmad1Δ::TRP1 (73). pJMH2 was created by cloning the EcoRI-SaII fragment of pIT340 (71) into pRS316 (61). pJMH1 differs from pJMH2 in that the hemagglutinin (HA) tag was removed by NotI digestion.
Chromosome segregation assay.
Chromosome missegregation events were analyzed by a visual colony color-sectoring assay, and the rate of artificial chromosome fragment loss was quantified as described previously (30, 36). Mid-logarithmic-phase homozygous wild-type and sth1-3ts cells were briefly sonicated and plated onto adenine indicator plates. The first cell cycle chromosome missegregation events were scored by counting red and white half-sectored colonies for chromosome nondisjunction (2:0) and by counting pink and red half-sectored colonies for chromosome loss events (1:0).
Far-Western assays.
Histones were prepared by acid extraction, separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and blotted to polyvinylidene difluoride membranes as described previously (18). Radiolabeled Sth1p was prepared by translating Sth1p (pDJ91) in vitro in the presence of [35S]methionine in a TNT-coupled reticulocyte lysate system (Promega) (11). Blots were hybridized and washed as described previously (18), and exposed to PhosphorImager screens.
GST pulldown assays.
Glutathione S-transferase (GST) (pGEX-3X) or GST-histone fusion proteins (expressed from plasmids p307.3, p272.1, p284.1, p107.1, p289.6, and p184.1 in Escherichia coli) were prepared and bound to glutathione-Sepharose 4B resin equilibrated in binding buffer (20 mM HEPES [pH 7.5], 50 mM KCl, 2 mM MgCl2, 0.5 mM EDTA, 0.5% [vol/vol] NP-40, 20% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride). Five microliters of the reticulocyte lysate reaction mixture containing labeled Sth1p (as described previously for Far-Western assays) were incubated in binding buffer (with 50 μg of ethidium bromide per ml) with GST or GST-histone-Sepharose beads for 30 min at 25°C and then for 60 min at 4°C. Input levels of GST fusion proteins were normalized by Coomassie blue staining. The beads were then washed three times in binding buffer and resuspended in 1× SDS loading dye.
Immunoprecipitation.
Extracts prepared from strains BLY309, BLY283, BLY573, BLY575, BLY572, and BLY574 expressing Flag-tagged histone H2B, H3, or H4 or control strains expressing untagged histones or containing empty vectors grown in YPD or SC medium selective for the plasmids were used to analyze coimmunoprecipitation of Sth1p with Flag-tagged histones. Extracts were prepared from cells grown to mid-logarithmic phase, and immunoprecipitation carried out in immunoprecipitation buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 0.5% NP-40, 0.5 mg of bovine serum albumin per ml) in the presence of 250 U of DNase I for 2 h at 4°C as described previously (52). For each immunoprecipitation, 30 μl of anti-Flag M2 affinity resin (Sigma) was incubated with 2.5 mg of protein. Proteins released by boiling in SDS sample buffer were separated on an SDS-4 to 20% acrylamide gradient or 15% acrylamide gels and transferred to nitrocellulose for immunoblot analysis with a 1:1,000 dilution of polyclonal anti-Sth1p or a 1:300 dilution of anti-Flag M2 monoclonal antibody.
Chromatin immunoprecipitation.
In vivo cross-linking and chromatin immunoprecipitation were performed as described previously (21) except that the lysis buffer contained 25 mM Tris-Cl [pH 8.0], 1 mM EDTA, 150 mM NaCl, 1 mM dithiothreitol, 1% Triton X-100, and 0.1% deoxycholate. The lysis and immunoprecipitation buffers used for chromatin immunoprecipitations in Fig. 7 were described previously (41). Sheared chromatin was immunoprecipitated with mouse monoclonal 12CA5 anti-HA (1:1,000), control monoclonal immunoglobulin G2b (the same isotype as the anti-HA antibody) (Roche Molecular Biochemicals), or polyclonal anti-Mif2p (1:250) (41) antibodies. PCR analysis of total and immunoprecipitated chromatin was carried out with primers to amplify target loci. Primer sequences are available upon request. Images were captured with a Bio-Rad Chemi Doc gel documentation system and analyzed with Quantity One quantitation software.
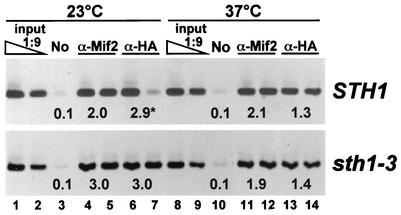
FIG. 7.
Mif2p and Cse4p remain associated with centromeres in sth1-3ts mutant cells. Chromatin was prepared from extracts of wild-type STH1 or mutant sth1-3ts cells expressing HA-tagged Cse4p (PM1311 and PM1312, respectively). Cells grown to mid-logarithmic phase in YPD medium at 23°C were first incubated with α-factor for 3 h to arrest cells in G1 phase (≈85%). Half of each culture was then shifted to 37°C in the continued presence of α-factor for 30 min, after which cells were washed with YPD containing 0.1 mg of pronase per ml at 23°C or 37°C and released into fresh YPD medium containing 0.1 mg of pronase per ml and nocodazole. Cells were incubated for an additional 3.5 h before processing for chromatin immunoprecipitation. Duplicate stained CEN3 PCR products (except for lane 7, upper panel) were imaged and quantified. No, mock immunoprecipitation without antibody.
Chromatin structure analysis.
S. cerevisiae nucleus preparation, micrococcal nuclease (MNase) digestion, and indirect end-labeling analysis of CEN3 chromatin were performed as described previously (21, 51). DNA purified from MNase-treated nuclei was further digested with BamHI, fractionated by agarose gel electrophoresis, and transferred to nitrocellulose membranes. CEN3 DNA 5′ to CDEI was detected with the CEN3 probe as described previously (51). The DraI enzymatic accessibility assay and subsequent Southern blot analysis were performed as described previously (55). DNA was digested with HindIII following purification from DraI-digested chromatin. The 0.9-kb BamHI-HindIII fragment flanking CDEIII was labeled by random priming and used as a probe.
Microscopy.
Strains carrying TetR-GFP (green fluorescent protein) were generated by transforming cells with the EcoRV-linearized plasmid pK3524 to direct integration at LEU2. To introduce TetO (TetR-arrays, approximately 228 bp 3′ of CDEIII of chromosome I and 35 kb from the CEN DNA of chromosome V, TetR-GFP-expressing wild-type and rsc mutant cells were transformed with the EcoRI fragment of pPM290 and the EcoRV-linearized pRS306-tetO(224) plasmid (44), respectively. Correct integrations were verified by Southern blot analysis. Strains carrying GFP-TUB1 were generated by transformation with the StuI-linearized plasmid pAFS125. To visualize TetR-GFP-marked chromosomal loci or GFP-marked tubulin, paraformaldehyde-fixed cells were imaged on a Nikon Microphot 2 microscope with a 100×, 1.4 NA oil immersion lens. Images were acquired with a Spot-RT charge-coupled device-cooled camera with SPOT diagnostic software.
RESULTS
rsc mutants activate the spindle checkpoint and missegregate chromosomes.
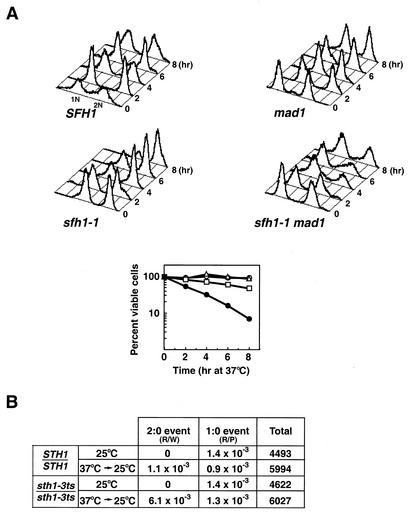
Mutations in the essential genes encoding the core RSC Sth1p ATPase and Sfh1p subunits sth1-3ts, nps1-105 (a different sth1 allele), and sfh1-1ts were previously shown to cause conditional cell cycle arrest at G2/M, suggesting that RSC function is required for cell cycle progression (11, 17, 73). Initial analysis of several classes of genes encoding proteins required for mitotic cell division, including cyclins (CLN3, CLB2), kinetochore components (CSE4, NDC10), and the mitotic segregation apparatus (TUB1, TUB3) revealed no obvious defects in the transcription of these genes in the sth1-3ts or sfh1-1ts mutant (13, 17; J.-M. Hsu and B. C. Laurent, unpublished data). We tested whether the preanaphase checkpoints that respond to damaged DNA or defective kinetochore-microtubule interactions were activated in these mutants. We found that two key components of the DNA damage checkpoint, RAD9 and MEC1, were not required for arrest of sth1-3ts or sfh1-1ts cells (17; I. Nasir and B. C. Laurent, unpublished). In contrast, both the sth1-3ts and sfh1-1ts mutations activated the MAD1 spindle checkpoint (Fig. 1A; only sfh1-1ts is shown). Flow cytometry analysis of DNA content revealed that inactivation of the spindle checkpoint by the mad1Δ mutation permitted a significant percentage of the sfh1-1ts mad1Δ cells to complete mitosis (Fig. 1A, upper panel). Furthermore, fewer than 7% of the sfh1-1ts mad1Δ double mutants were viable, while the corresponding single-mutant viability remained high 8 h following a shift to 37°C (Fig. 1A, lower panel), indicating that the spindle checkpoint is required to arrest sfh1-1ts at G2/M and to maintain its viability. These results are in agreement with previous results showing that nps1-105, a different allele of sth1, and rsc3-3 similarly engaged the MAD1-dependent checkpoint to arrest cells in G2/M (2, 73).
FIG. 1.
rsc mutants exhibit phenotypes characteristic of mutants defective in chromosome segregation. (A) sfh1-1ts activates the MAD1-dependent spindle checkpoint pathway. Upper panel: Flow cytometric analysis of DNA contents in SFH1 (BLY46), mad1 (BLY295), sfh1-1ts (BLY75-1), and sfh1-1ts mad1 (BLY296) strains at the times indicated. Cells grown to mid-logarithmic phase at 25°C were shifted to 37°C, and aliquots were removed for analysis at the times indicated. The number of cells was plotted versus the relative intensity of emitted light. Lower panel: At the times indicated, the SFH1 (open circles), mad1 (open triangles), sfh1-1ts (open squares), and sfh1-1ts mad1 (solid circles) cells described above were removed from 37°C and plated onto YPD medium at 25°C. Viability was determined as the number of cells capable of forming colonies at 25°C. Values reported are the averages for two independent determinations with <15% error. (B) Homozygous sth1-3ts diploid cells exhibit increased 2:0 missegregation of a chromosome fragment. STH1/STH1 (BLY363) and sth1-3ts/sth1-3ts (BLY394) cells carrying a single copy of CFIII were incubated at 37°C for 4 h before shifting back to 25°C. Red sectors indicate loss of chromosomes, and white sectors indicate gain of chromosomes. Half-sectored colonies were scored as R/W (half red/half white) or R/P (half red/half pink) after 2 to 4 days of incubation. The rates of CFIII nondisjunction (2:0 segregation) and loss (1:0 segregation) were calculated by dividing the number of half red/half white and half red/half pink cells, respectively, by the total number of pink parental cells indicated. The data from two independent experiments were combined.
Activation of the spindle checkpoint by the rsc mutations indicates a potential role for RSC in centromere-kinetochore function, as this checkpoint monitors the integrity of kinetochores and microtubules, kinetochore-microtubule interactions, and the tension resulting from bipolar attachment (1, 14). Consistent with this idea, we found that both sth1-3ts and sfh1-1ts mutants were sensitive to the microtubule-depolymerizing agents benomyl and thiabendazole (TBZ), which perturb the chromosome segregation machinery (data not shown; see also Fig. 2B) (13). We also found that the homozygous sth1-3ts diploids missegregated a linear reporter chromosome fragment by using a colony color-sectoring assay. The sth1-3ts mutants exhibited sixfold higher rates of 2:0 chromosome nondisjunction than isogenic wild-type strains (6.1 × 10−3 versus 1.1 × 10−3) and no significant change in rates of 1:0 chromosome loss (1.3 × 10−3 versus 0.9 × 10−3) (Fig. 1B). Thus, chromosome destabilization in sth1-3ts mutants occurs predominantly by sister chromatid nondisjunction, consistent with another sth1 allele (73). These results suggest that cells require RSC for the faithful transmission of chromosomes during mitosis.
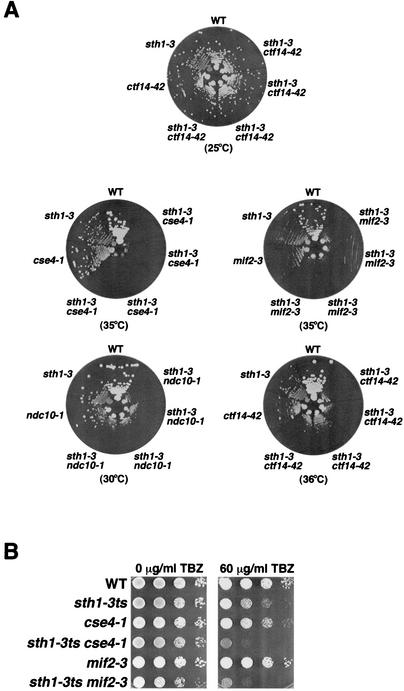
FIG. 2.
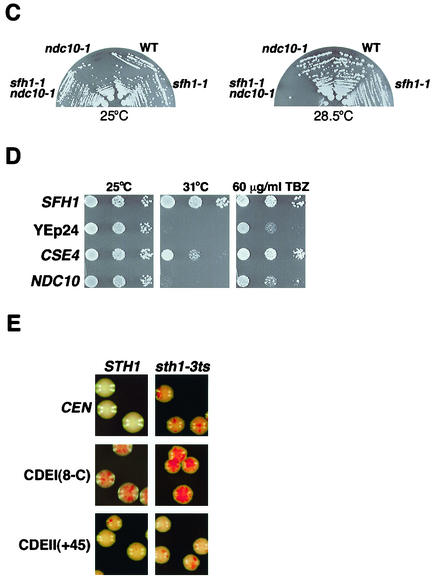
RSC interacts genetically with components of the kinetochore. (A) sth1-3ts interacts with mutations in kinetochore components. The genotypes refer to the following strains: wild type (WT, BLY76), sth1-3 (BLY49), ctf14-42 (BLY503), sth1-3 ctf14-42 (BLY528), cse4-1 (BLY500), sth1-3 cse4-1 (BLY536), mif2-3 (BLY416), sth1-3 mif2-3 (BLY531), ndc10-1 (BLY506), and sth1-3 ndc10-1 (BLY532). Strains were grown on YPD plates at the indicated temperatures for 3 to 4 days. Four independently isolated double mutant strains were tested on each plate. (B) sth1-3ts mutations in combination with either cse4-1 or mif2-3 mutations cause enhanced hypersensitivity to TBZ. Tenfold serial dilutions of mid-logarithmic-phase cells were spotted onto YPD plates containing 0 or 60 μg of TBZ per ml. Growth of wild-type (BLY76), sth1-3ts (BLY49), cse4-1 (BLY500), sth1-3ts cse4-1 (BLY536), mif2-3 (BLY416), and sth1-3ts mif2-3 (BLY531) cells was compared at 25°C after 3 to 4 days. (C) The sfh1-1ts mutation interacts synthetically with the ndc10-1 mutation. Wild-type (BLY278), sfh1-1ts (BLY353), ndc10-1 (BLY506), and sfh1-1 ndc10-1 (BLY538) strains were grown on YPD plates at the temperatures indicated for 3 to 4 days. (D) Increased dosage of CSE4 partially suppresses the temperature sensitivity and TBZ sensitivity of sfh1-1ts mutants. Equal numbers of tenfold serially diluted mid-logarithmic-phase sfh1-1ts cells (BLY353) carrying YEp24 or YEp24-derived high-copy plasmids expressing SFH1 (pYC5H), CSE4 (pPY3), or NDC10 (pPM47) were spotted onto SC plates lacking uracil and incubated at 25°C or 31°C or onto SC plates lacking uracil plates containing 60 μg of TBZ per ml and incubated at 25°C. Cell growth was compared after 3 to 4 days. (E) sth1-3ts mutations cause synergistic chromosome instability when combined with a CDEI but not a CDEII mutation. The stabilities of chromosome fragments containing either a wild-type CEN or mutant CEN sequence CDEI(8-C) or CDEII(+45) in STH1 (BLY361, BLY429, and BLY424, respectively) and sth1-3ts (BLY360, BLY565, and BLY566, respectively) cells were determined by plating cells onto adenine indicator plates. Cells were grown at 34°C and photographed after 3 to 4 days. Red sectors indicate missegregation of the chromosome fragment.
sth1-3ts and sfh1-1ts interact genetically with mutations in both kinetochore components and a centromeric DNA element.
Proper chromosome segregation requires centromeric (CEN) DNA and the kinetochore, a protein complex that assembles onto CEN DNA to mediate the attachment of chromosomes to the spindle microtubules. The engagement of the spindle checkpoint and the chromosome segregation defects in rsc mutants suggest a role for RSC in kinetochore function. To test whether RSC interacts with kinetochores, we constructed double mutants between the sth1-3ts or sfh1-1ts mutation and mutations in one of the kinetochore genes CSE4, MIF2, NDC10/CTF14, and CTF13. The sth1-3ts cse4-1 and sth1-3ts mif2-3 mutants exhibited conditional synthetic lethality at 35°C, and the sth1-3ts ndc10-1 and sth1-3ts ctf14-42 double mutants grew dramatically more slowly than either of the single mutants at semipermissive temperatures (Fig. 2A). All mutant strains grew as well as wild-type cells at the permissive temperature (Fig. 2A; only ctf14-42 is shown). We also found that sth1-3ts combined with either the cse4-1 or mif2-3 mutation caused enhanced hypersensitivity to TBZ (Fig. 2B). This suggests that RSC exhibits functionally distinct as well as overlapping roles with kinetochore proteins in microtubule-dependent processes.
Like sth1-3ts, the sfh1-1ts mutation interacted synthetically with the ndc10-1 mutation (Fig. 2C), shown previously to prevent formation of the CBF3-CEN DNA complex in vitro and in vivo (48, 65). In addition, our failure to recover sfh1-1ts ctf13-30 or sfh1-1ts cft14-42 double mutants from at least 40 tetrads examined from each strain suggests that these ctf mutations are lethal in combination with sfh1-1ts.
As rsc mutants interacted synthetically with kinetochore mutants, we next tested whether overexpression of kinetochore proteins could suppress the temperature-sensitive (Ts−) phenotypes of rsc mutants. We found that high-copy CSE4 but not NDC10, MIF2, or CBF1 partially suppressed the Ts− phenotype of sfh1-1ts at 31°C (Fig. 2D; only the CSE4 and NDC10 data are shown). High-copy CSE4 also partially suppressed the TBZ sensitivity of sfh1-1ts cells (Fig. 2D). Interestingly, a strain bearing a deletion of the homologous SNF5 gene, which encodes a core component of the Snf-Swi complex, was also sensitive to TBZ, although high-copy CSE4 was unable to suppress the TBZ sensitivity (data not shown), suggesting that the interaction with centromeric components of the kinetochore is specific to RSC components.
Mutations in genes encoding kinetochore proteins have been shown to enhance chromosome missegregation when combined with mutations in centromere DNA elements (CDEs) (3, 34, 42). For example, chromosomes are lost at unusually high rates in mutants carrying cse4 mutations plus CDEI or CDEII mutations (34). To determine if RSC interacts with centromeric DNA sequences, we tested whether combining sth1-3ts and the CDEI(8-C), CDEII(+45), or CDEII(Δ31) mutation caused synthetic impairment of chromosome segregation. At the semipermissive temperature (34°C), the sth1-3ts mutation caused elevated missegregation of a chromosome fragment containing wild-type CEN sequences, as described previously above (Fig. 1B), and this missegregation was greatly exacerbated when the chromosome fragment carried the CDEI(8-C) mutation (Fig. 2E). However, neither the sth1-3ts CDEII(+45) nor the sth1-3ts CDEII(Δ31) double mutant showed higher missegregation rates than the single mutants [Fig. 2E; only CDEII(+45) is shown]. This analysis suggests that RSC interacts with centromeric DNA element CDEI but not with CDEII. Taken together, the genetic interactions between rsc and both kinetochore and centromere DNA element mutations strongly imply an important role for RSC in centromere-kinetochore function.
rsc interacts with a histone H4 mutation that disrupts centromeric structure and function.
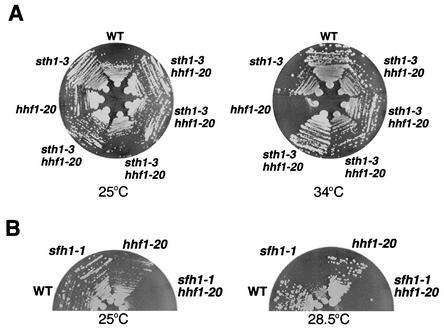
Centromeric chromatin is distinguished by the presence of a histone H3 variant. In humans, equimolar amounts of histones H2A, H2B, and H4 and the histone H3 variant CENP-A assemble to form a specialized nucleosome in vitro (68). Considering the specific genetic interactions between RSC and the Cse4p component of centromeric chromatin and the ability of RSC to remodel nucleosome structure in vitro, we reasoned that rsc would interact with hhf1-20, a histone H4 mutation that disrupts centromeric chromatin structure and missegregates chromosomes (22, 43, 64). The hhf1-20 single mutant was unable to grow at 34°C, but its growth was restored in the sth1-3ts mutant background. Thus, the sth1-3ts mutation suppressed the temperature sensitivity of hhf1-20 (Fig. 3A). In contrast, the sfh1-1ts mutation showed synthetic sickness when combined with the hhf1-20 mutation, even at the permissive temperature (Fig. 3B). These data suggest that Sth1p and Sfh1p interact differently with centromeric histones and are consistent with the differential cross-linking of these proteins to nucleosomes (57). One possibility is that individual RSC subunits contribute differently to the distinct chromatin-remodeling activities of RSC (e.g., histone octamer transfer and nucleosome sliding) (39).
FIG. 3.
sth1-3ts and sfh1-1ts mutations interact with a histone H4 mutation, hhf1-20. (A) sth1-3ts suppresses the temperature-sensitive phenotype of hhf1-20 mutants. Growth of sth1-3ts hhf1-20 double mutants (BLY546) was compared to that of hhf1-20 (BLY415), sth1-3ts (BLY49), and wild-type (WT, BLY76) cells at 25°C and 34°C. Plates were incubated for 2 to 3 days. Four independently isolated double mutant strains were tested. (B) The sfh1-1ts mutation combined with hhf1-20 results in synthetic growth defects. The growth of wild-type (BLY278), sfh1-1ts (BLY353), hhf1-20 (BLY415), and sfh1-1ts hhf1-20 (BLY552) cells on YPD was compared at the indicated temperatures after 3 days.
Sth1p interacts directly with the histone H3 variant Cse4p and histones H3, H4, and H2B.
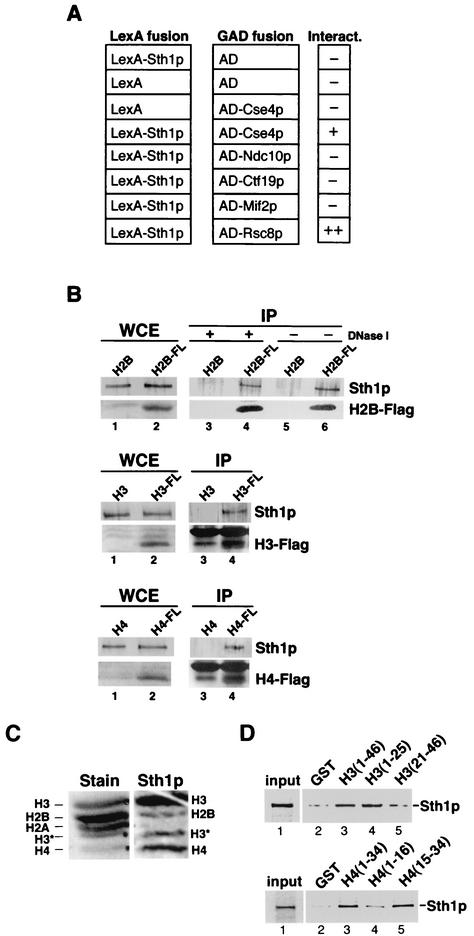
A prediction from the genetic interactions observed in combining rsc and kinetochore component mutations is that RSC interacts physically with these proteins. We investigated the ability of Sth1p to interact with the kinetochore components Ndc10p, Ctf19p, Mif2p, and Cse4p in the yeast two-hybrid assay. Sth1p fused to the LexA DNA-binding domain interacted with Cse4p fused to the Gal4p activation domain but not with the other kinetochore components when β-galactosidase activity was analyzed in the reporter strain (Fig. 4A). The interaction was slightly weaker than that between Sth1p and a second conserved RSC component, Rsc8p (71). Thus, Sth1p interacts specifically with a histone component of centromeric chromatin.
FIG. 4.
Sth1p interacts physically with the histone H3 variant Cse4p and histones H3, H4, and H2B. (A) Sth1p interacts with Cse4p in the yeast two-hybrid assay. β-Galactosidase activity was assayed in the lexAop-lacZ (BLY2) reporter strain expressing pLexA or pLexA202-Sth11-1359 and Gal4p activation domain (GAD) or GAD fusion proteins expressed from pACT2, pJL484, pJL486, pJO196, pGAD-CTF19, or pIT331. β-Galactosidase activity in permeabilized cells was determined as described previously (11). No significant interactions were detected in cells expressing the LexA DNA-binding partner alone and GAD fusion proteins or in cells expressing the GAD activating partner alone and LexA202-Sth11-1359. The interaction between LexA202-Sth11-1359 and GAD-Cse4p was confirmed in β-galactosidase liquid assays (data not shown). AD, activation domain. (B) Sth1p interacts with Flag-tagged histones H2B, H3, and H4 in vivo. Anti-Flag M2 monoclonal antibody affinity resin was incubated with whole-cell extracts (WCE) prepared from cells carrying plasmids expressing Flag-H2B (BLY309), H2B (BLY283), Flag-H3 (BLY573), or Flag-H4 (BLY575) and cells carrying pRS315 (BLY572) and pRS425 (BLY574) as controls for Flag-H3 and Flag-H4, respectively. Immune complexes were separated on SDS-4 to 20% polyacrylamide gradient or SDS-15% acrylamide gels and immunoblotted with anti-Sth1p (1:1,000) polyclonal or anti-Flag M2 (1:300) monoclonal antibodies. Lanes 1 and 2, 1/83 of input lysate for both anti-Sth1p and anti-Flag immunoblots; lanes 3, 4, 5, and 6, entire immunoprecipitation pellets for both anti-Sth1p and anti-Flag Western blots. Immunoprecipitations (IPs) were performed in the presence of 250 U of DNase I except for lanes 5 and 6, from which DNase I was omitted. Nonspecific proteins comigrated with immunoprecipitated Flag-H3 and Flag-H4 proteins (compare lanes 3 and 4). (C) Sth1p interacts directly with histones H3, H4, and H2B by Far-Western analysis. Samples of acid-soluble enriched histones were separated by SDS-21% PAGE, blotted to polyvinylidene difluoride membranes, and probed with radiolabeled Sth1p (Sth1p). A parallel lane of transferred histones was stained with Ponceau S (Stain). H3* is a spontaneous breakdown product of histone H3. (D) Sth1p interacts directly with the N termini of histones H3 and H4. [35S]methionine-labeled Sth1p was incubated with GST (lanes 2) or GST fusion proteins containing the N termini of histone H3 (amino acids 1 to 46) or H4 (amino acids 1 to 34) (lanes 3), H3 (1 to 25) or H4 (1 to 16) (lanes 4), or H3 (21 to 46) or H4 (15 to 34) (lanes 5) (28) immobilized on glutathione-Sepharose beads. We analyzed 25% of the input fractions and 100% of the bound and unbound fractions by SDS-8% PAGE, and labeled Sth1p was visualized by PhosphorImager analysis. Similar results were observed in at least four independent experiments.
We tested further whether Sth1p interacts with histones in vivo by immunoprecipitation. We found that Sth1p specifically coprecipitated with Flag-H2B when anti-Flag antibodies were used. Sth1p also interacted with Flag-H2B in immunoprecipitations with anti-Sth1p antibody (data not shown). In the presence of DNase I, Sth1p still associated with Flag-H2B, although at a slightly reduced level (Fig. 4B, compare lanes 4 and 6). Similarly, Sth1p also immunoprecipitated with Flag-tagged histones H3 and H4 in the presence of DNase I (bottom two panels, compare lanes 3 and 4), suggesting that these interactions occurred through protein-protein interactions. However, these experiments do not distinguish whether these interactions occur directly through Sth1p or through another protein(s).
We further investigated the interactions between Sth1p and histones by Far-Western and GST pulldown analyses. In vitro-translated Sth1p bound specifically and strongly to histones H3 and H4 and to histone H2B but not detectably to histone H2A (Fig. 4C). The lack of a significant signal with H2A suggests that the Sth1p interactions with H3, H2B, and H4 are specific and not due simply to electrostatic interactions. In addition, we showed that Sth1p bound to the N-terminal tails of histones H3 (amino acids 1 to 46) and H4 (amino acids 1 to 34) (Fig. 4D, upper and lower panels, lane 3) in GST pulldown assays. Interestingly, Sth1p interacted specifically with amino acids 1 to 25 of histone H3 (Fig. 4D, upper panel, compare lanes 4 and 5) and residues 15 to 34 of histone H4 (Fig. 4D, lower panel, compare lanes 4 and 5). These selective interactions with specific regions of histone tails may have important implications for the mechanism of RSC remodeling.
RSC localizes to centromeric and flanking chromatin in vivo.
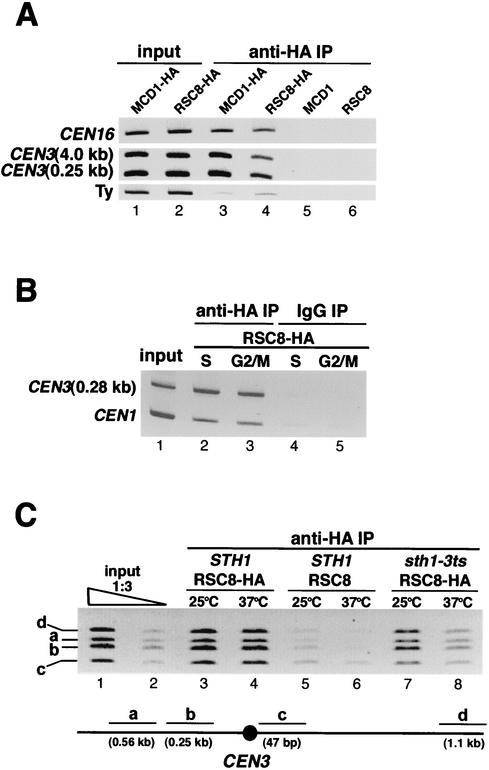
The genetic and physical interactions between RSC and kinetochore components are consistent with a functional role for RSC at or near the centromere. Thus, we investigated the association of RSC with centromere and centromere-proximal DNAs in vivo by monitoring the presence of functional hemagglutinin (HA)-tagged Rsc8p in the chromatin immunoprecipitation assay. Like Mcd1p, a subunit of cohesin, Rsc8p-HA protein interacts with centromeric DNA in vivo. In addition to its presence at the centromere of chromosome XVI (CEN16), Rsc8p-HA was also present at centromere-proximal regions at distances 2.5 kb and 4.0 kb 3′ to CDEIII of CEN3 but not at a Ty element ≈30 kb 5′ to CDEI of CEN3 (Fig. 5A). As cohesin has been shown to localize to centromeric and centromere-flanking sequences (7, 69), we sought to determine the localization pattern for RSC in this region. We found that RSC was associated with the contiguous 4.0-kb regions on either side of CEN3 (data not shown). Nearly identical localization patterns for RSC were observed whether Sth1p-HA or Sfh1p-HA strains were used (data not shown). To determine if the presence of RSC at centromeric chromatin is cell cycle dependent, we synchronized cells in the S and G2/M phases of the cell cycle. RSC was associated with the centromere of chromosome I (CEN1) and the centromere-proximal region 0.28 kb 3′ to CDEIII of CEN3 in both cell cycle stages (Fig. 5B).
FIG. 5.
Rsc8p associates with centromeric and flanking chromosomal regions in vivo. (A) Rsc8p is localized to centromeric and flanking regions. Chromatin prepared from extracts of cells expressing Mcd1p-HA (BLY397) or Rsc8p-HA (BLY298), and untagged Mcd1p (BLY398) or Rsc8p (BLY301) grown to mid-logarithmic phase in YPD was immunoprecipitated (IP) with anti-HA antibodies. Coimmunoprecipitated DNAs were amplified by PCR with primers specific for the indicated chromosomal loci. (B) Rsc8p localizes to CEN3-flanking and CEN1 regions in S- and G2/M-phase-synchronized cells. Chromatin was prepared from cells expressing HA-tagged Rsc8p (BLY298) synchronized in S or G2/M phase by treatment with hydroxyurea or nocodazole, respectively, for 3 h at 25°C. (C) Association of Rsc8p with CEN DNA and flanking regions is defective in sth1-3ts mutants. Chromatin was prepared from mid-logarithmic-phase RSC8-HA (BLY298), RSC8 (BLY301), and sth1-3ts RSC8-HA (BLY549) cells grown at 25°C or 37°C for 8 h. A schematic map showing the positions of the PCR products relative to CEN3 is shown.
We next examined whether the association of RSC with chromosomal DNA is dependent on functional Sth1p by comparing the enrichment of CEN and flanking DNAs in sth1-3ts cells grown at the permissive and nonpermissive temperatures. The levels of immunoprecipitated DNAs in sth1-3ts cells grown at 37°C were greatly reduced compared to those in mutant strains grown at 25°C and wild-type cells at either 25°C or 37°C (Fig. 5C, compare lane 8 with lane 7 and lane 7 with lanes 3 and 4). The reduced association was not due to decreased Sth1p levels or DNA inputs, as both the levels of Sth1p that coimmunoprecipitated with Rsc8p-HA and the initial DNA inputs for chromatin immunoprecipitation assays were comparable in the mutant and wild-type strains at both temperatures (data not shown). We infer that Sth1p is required for recruitment or stable association of RSC with chromosomal loci. Interestingly, Sth1p is one of three RSC components that is cross-linked to nucleosomal DNA prior to remodeling (57). These results suggest that RSC function is required at both centromeric and flanking chromosomal regions.
Centromeric chromatin structure is altered in rsc mutants.
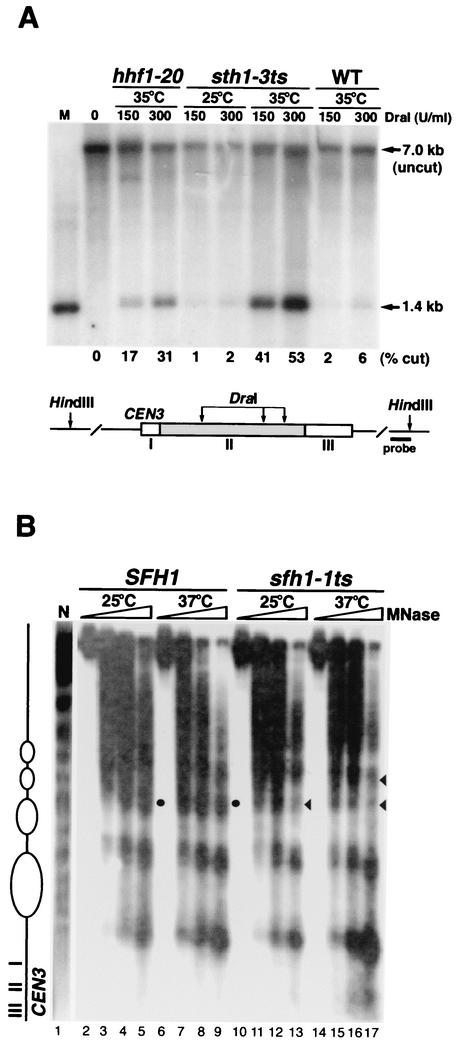
To test whether RSC function is required for the proper configuration of centromeric chromatin, we analyzed the centromeric and adjacent chromatin structure of CEN3 in rsc mutants with both the endonuclease accessibility assay and indirect end-labeling analysis of MNase-digested chromatin. The three DraI restriction sites in the CDEII region of CEN3 are normally protected from endonuclease digestion, presumably due to the presence of the centromere-kinetochore protein complex. Mutations in CEN DNA or kinetochore components as well as depletion of histones H2B or H4 were previously shown to increase the accessibility of CDEII to DraI endonuclease digestion (54, 55).
The DraI sites within CDEII of CEN3 remained inaccessible in wild-type cells grown at 35°C and in sth1-3ts mutants grown at the permissive temperature (25°C). However, upon shifting mutants to 35°C, DraI accessibility increased dramatically in sth1-3ts mutants (Fig. 6A). This increase in accessibility resembled that in hhf1-20 mutants, whose mutation was previously shown to cause centromeric chromatin structural alterations (43).
FIG. 6.
Centromeric and flanking chromatin structure is altered in rsc mutants. (A) DraI accessibility to CDEII of CEN3 is increased in sth1-3ts mutants. Nuclei isolated from sth1-3ts (BLY49) grown at 25°C or hhf1-20 (BLY413), sth1-3ts (BLY49), and wild-type (WT, BLY76) cells grown at 35°C for 8 h were first digested with DraI. Southern blot analysis of the purified DNA subsequently digested with HindIII is shown. M, ≈1.4-kb DraI-HindIII genomic fragment as a marker. The 7.0-kb band is the intact HindIII fragment uncut by DraI. The fraction of CDEII chromatin digested by DraI is shown as a percentage. (B) Pericentromeric chromatin structure is altered in the sfh1-1ts mutant. Indirect end labeling of microccocal nuclease-digested chromatin prepared from SFH1 (BLY278) and sfh1-1ts (BLY353) cells grown at 25°C or 37°C for 8 h. CDEI-proximal pericentromeric chromatin structure is shown. The arrowheads indicate regions in the mutant with altered digestion patterns; solid circles indicate the corresponding regions in wild-type cells. N, naked DNA control.
We next examined the centromeric chromatin structure around CEN3 in sfh1-1ts mutants by indirect end labeling of MNase-digested chromatin. Both SFH1 and sfh1-1ts cells showed ordered nucleosome arrays flanking the CEN3 nuclease-resistant centromeric core (8, 51). At both the permissive (25°C) and nonpermissive (37°C) temperatures, the MNase digestion patterns of chromatin in wild-type SFH1 cells were comparable. However, the pattern was altered in sfh1-1ts mutants, and this change was most evident at 37°C. The levels of MNase digestion between nucleosomes were diminished at two positions in the region 5′ to CDEI (Fig. 6B, compare lanes 17 and 13 with lanes 9 and 5). Interestingly, we did not observe any significant differences in the MNase digestion pattern between the wild-type and the sfh1-1ts mutant 3′ to CDEIII (data not shown). Taken together, these results indicate that the core centromeric chromatin and flanking chromatin structures are significantly altered in rsc mutants.
Kinetochore components remain associated with the centromere in sth1-3ts mutants.
One explanation for the rsc mutant defects in centromeric chromatin structure and chromosome segregation is that kinetochore proteins or proteins necessary for centromeric cohesion are not deposited properly at centromeres. To test this, we assessed binding of Mif2p and Cse4p-3HA to CEN3 DNA by chromatin immunoprecipitation analysis of chromatin prepared from wild-type and sth1-3ts cells grown at the permissive (23°C) and nonpermissive (37°C) temperatures by chromatin immunoprecipitation analysis. Comparable levels of CEN3 DNAs were immunoprecipitated in each case from sth1-3ts cells at 23°C and 37°C (Fig. 7; lower panel, compare lanes 4 through 7 to lanes 11 through 14, respectively), and these levels were similar to those in wild-type cells grown under the same conditions (compare upper and lower panels, lanes 4 through 7 and lanes 11 through 14, respectively). These results suggest that RSC is not required for localization of these proteins to the kinetochore.
rsc mutants are defective in sister chromatid segregation.
The structural alterations of centromeric chromatin and missegregation of a chromosome fragment in rsc mutants are consistent with defects in several aspects of chromosome segregation, including kinetochore assembly, sister kinetochore biorientation, sister chromatid cohesion, and the dynamic oscillatory movements of chromatids prior to the onset of anaphase. To more directly assess chromosome behavior in rsc mutants, we followed sister chromatid segregation with GFP-TetR-marked chromosomes tagged either 206 bp from CEN1 or 35 kb from CEN5 on the left arm of chromosome V.
Sister chromatids marked at CEN1 exhibited obvious segregation defects in sth1-3ts mutant cells shifted to 37°C for 8 h. Among large-budded cells with two GFP dots within one cell body, 36% were accompanied by bilobed or binucleated DNA masses within the same cell body, and 7% exhibited an uncoupling of bulk DNA segregation from sister chromatid segregation (Fig. 8A, columns b and c). Even in the remaining 54% of cells in which two sister chromatids separated within a single DNA mass, the nucleus was frequently positioned away from the mother bud neck (Fig. 8A, column d). Examination of mitotic spindles in sth1-3ts mutants by labeling microtubules with GFP-Tub1p revealed that while the majority of large-budded cells with a single nucleus contained characteristically short spindles, a subpopulation of mutants with either single or separated DNA masses contained aberrant spindles. In cells with a single DNA mass, the spindle was frequently elongated and incorrectly positioned compared to wild-type cells in similar cell cycle stages (Fig. 8B, compare column b with column a). In cells containing two separated DNA masses within the same cell body, the spindles were entirely contained within the mother cell. The middle portions of these spindles were often not visible (Fig. 8B, column c). Thus, the chromosome segregation defects in rsc mutants could result from defects in both kinetochore and microtubule functions.
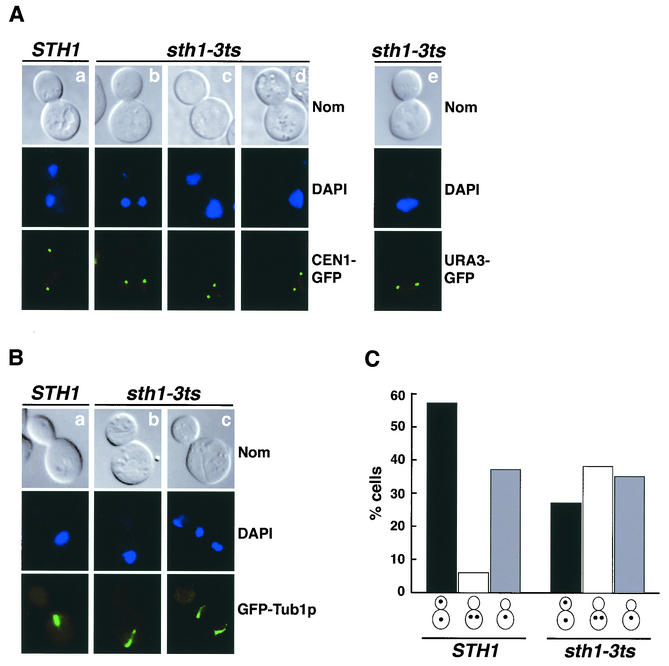
FIG. 8.
rsc mutants missegregate authentic chromosomes. (A) Sister chromatid segregation is defective in sth1-3ts mutants. STH1 (BLY520) and sth1-3ts (BLY526) cells marked at CEN1 were grown to mid-logarithmic phase at 25°C, diluted, and shifted to 37°C for 8 h. Representative mutant cells with separated sister chromatids in one cell body are shown (columns b to d). Mutant sth1-3ts cells (BLY493) marked at URA3 (35 kb from CEN5) with separated sister chromatids are shown in column e. Nuclei were visualized by 4,6-diamidino-2-phenylindole (DAPI) staining. Nom, Nomarski images. (B) Spindle morphology is aberrant in sth1-3ts mutants. Mid-logarithmic-phase wild-type (BLY570) and sth1-3ts (BLY568) cells expressing the GFP-Tub1p fusion protein were grown at 37°C for 8 h. Microtubule structures were analyzed by fluorescence microscopy. (C) Sister chromatid segregation is delayed in sth1-3ts cells. Wild-type (BLY520) and sth1-3ts (BLY526) strains marked at CEN1 were arrested in G1 with α-factor, released at 37°C, and scored for sister chromatid separation and segregation. The percentages of cells with segregated, separated, and unseparated sister chromatids 100 min after α-factor release are plotted.
Interestingly, when chromosome arms were marked 35 kb from CEN5, we observed sister chromatid separation within single nuclear DNA masses in arrested mutants (Fig. 8A, column e). This was unexpected because chromosome arms in wild-type cells do not normally undergo transient separation prior to the onset of segregation. Similar separation defects were also observed in sfh1-1ts cells with chromosomes tagged 35 kb from CEN5 (data not shown).
To further explore the chromosome missegregation phenotype in rsc mutants, wild-type and sth1-3ts mutant cells marked at CEN1 were arrested in G1 with α-factor and then released to study sister chromatid separation and segregation in synchronous cells. Wild-type and sth1-3ts mutant cells showed similar budding kinetics following release from α-factor. At 60 min following release, both wild-type and mutant cells had similar percentages of budded cells with single GFP dots. As cells progressed through the cell cycle, the population of wild-type cells with segregated sister chromatids increased. In contrast, in the sth1-3ts mutants, the percentage of cells with segregated chromatids remained low. Instead, the population of cells with two separated but not segregated sister chromatids increased. For example, 100 min following release, 57% of the wild-type cells and only 27% of sth1-3ts mutant cells had segregated their sister chromatids (Fig. 8C, black bars). While the percentages of wild-type and mutant cells with single GFP-CEN1 fluorescent dots was comparable (36% versus 35%; gray bars), there was a significant increase in sth1-3ts mutants with separated sister chromatids in one cell body (38% versus 6%; open bars). Accumulation of separated but not segregated sister chromatids in sth1-3ts cells might result from activation of the spindle checkpoint and is consistent with our previous genetic and structural analyses.
DISCUSSION
In this study, we present several lines of evidence that RSC function is required at the kinetochore during chromosome segregation. First, the sth1 and sfh1 mutations interact genetically with mutations in kinetochore components, centromeric histones, and centromeric DNA elements. Second, RSC is localized to centromeric and centromere-flanking chromatin and interacts genetically and physically with histone and histone variant components of chromatin. Finally, centromeric nucleosome structure is altered and TetR-GFP-tagged chromosomes are missegregated in both sth1 and sfh1 mutants. These data suggest a model in which the RSC ATP-dependent remodeling complex is required to restructure centromeric and centromere-flanking nucleosome structure for the accurate transmission of chromosomes.
Both genetic and biochemical data support direct interactions between RSC and Cse4-containing nucleosomes at kinetochores. Processes that may require such interactions include the recruitment, assembly, and maintenance of Cse4-containing nucleosomes. The ability of Cse4p to associate with centromeric DNA in sth1-3ts mutants suggests that the recruitment of Cse4p to kinetochores does not require RSC. However, the alterations in centromeric chromatin structure in rsc mutants suggest that RSC could function at kinetochores in the postrecruitment assembly or maintenance of centromeric chromatin. One model is that RSC functions early in centromeric nucleosome assembly, possibly with factors such as CAF-1/Hir1 (58), during which Cse4p is postulated to compete with histone H3 for binding to histone H4 (22, 43). We found that high-copy Cse4p could partially suppress both the Ts− and TBZ sensitivity phenotypes of sfh1-1ts mutants. Presumably, compromised RSC function would result in a shifted balance between Cse4/H4 and H3/H4 assembly in sfh1-1ts mutants. High-dosage CSE4 would then suppress the sfh1-1ts mutation by driving assembly of Cse4p/H4. Alternatively, RSC could promote the proper positioning of Cse4p-containing nucleosomes on centromeric DNA. During S phase, centromeres of all chromosomes are clustered near spindle pole bodies (33). Recruitment of Cse4p could result in high local concentrations of Cse4p that spread to immediate adjacent regions, similar to what has been observed in experiments in which human CENP-A was overexpressed (75). Thus, the transfer of these ectopic Cse4p-containing nucleosomes catalyzed by RSC would ensure their proper association with centromeric DNA. RSC could catalyze the sliding of Cse4p-containing octamers to finely adjust their positions relative to CDEs, a role that would be especially important in the G2/M phase, when microtubules attach to kinetochores and pull the sister chromatids apart. Proper positioning of the Cse4p-containing nucleosomes relative to the centromeric DNA sequences provides important structural support for the overall configuration and function of kinetochores.
Although our chromosome tagging experiments revealed that kinetochore-microtubule interactions are largely intact in rsc mutants, these rsc mutations were found to activate the MAD1-dependent spindle checkpoint. Thus, subtle alterations of centromeric chromatin could nevertheless impact microtubule-kinetochore function in chromosome segregation. Recent studies have identified several protein complexes that function at the kinetochore. Assembly of the kinetochore involves the initial binding of CBF3 to CDEIII, followed by additional protein-protein and protein-CEN DNA interactions (14). Centromeric DNA conformation is also important for facilitating protein-protein interactions at the kinetochore (48). Interestingly, the sth1-3ts and mif2 mutations show similar genetic interactions with CDEI but not CDEII mutations. Thus, RSC could facilitate the bending of centromeric DNA for kinetochore assembly, as has been proposed for Mif2p (9, 41, 42, 48). The centromeric chromatin structural changes in rsc mutants could reflect altered positions of Cse4-containing nucleosomes relative to centromeric DNA or instead alterations in the overall structural configuration of the kinetochore.
RSC also localizes to centromere-proximal regions, suggesting additional roles for RSC in maintaining the structure of this region that is necessary to support kinetochore function. Indeed, centromere-proximal chromatin structure is perturbed in both sfh1-1ts and sth1/nps1-105 mutants (73; this study). Interestingly, Rsc2p is required to maintain the correct nucleosome structure of the STB plasmid locus for 2μm plasmid partitioning (77), a process that likely shares host factors necessary for chromosome segregation (40). Centromere-proximal regions up to 10 to 13 kb on either side of the CDEs undergo dynamic structural changes before kinetochores establish stable bipolar interactions with the mitotic spindle (23, 27, 49). These changes in centromere-proximal higher-order chromatin structure may reflect the localized condensation and decondensation of chromosomes. We envision that RSC is required to remodel flanking nucleosomes throughout these oscillatory chromosome movements to facilitate kinetochore-microtubule interactions. At the same time, RSC, cohesin, and other factors may be coordinately regulated to limit these changes to a range of 10 to 13 kb surrounding the centromere. In both rsc mutants, sister chromatid separation occurred when chromosomes were marked 35 kb from CEN5, beyond the 10- to 13-kb limit for transient sister separation (49). Although these data support a role for RSC in remodeling centromere-proximal chromatin for kinetochore function, we cannot rule out the possibility that chromosome condensation and/or cohesion is defective in rsc mutants, as both local chromatin changes and the reorganization of chromatin by condensin/cohesin could contribute to reversible centromeric chromatin changes (46).
In this study, we have shown that RSC associates with centromeric and centromere-flanking chromatin and functions in chromosome segregation. Recent genome-wide chromatin immunoprecipitation studies have localized RSC to several other chromosomal loci (15, 47). Together, these results imply distinct functions for RSC in chromosome segregation and other cellular processes such as transcription. The dynamic structural changes at centromeric and centromere-flanking regions are likely to involve multiple chromatin-modifying activities. Indeed, in Schizosaccharomyces pombe and Drosophila melanogaster, histone deacetylase and methyltransferase activities are required for heterochromatic centromere structure and function (5, 6, 16, 19, 50). Like RSC, the related human SWI/SNF-B complex is localized to kinetochores (78). Therefore, at least two classes of chromatin-remodeling complexes may function coordinately at the kinetochore. Interestingly, another human ATP-dependent chromatin-remodeling complex was recently shown to load cohesin onto chromosomes (24). Thus, it appears likely that these remodeling activities function in conjunction with cohesins, condensins, and other factors that regulate local centromeric and centromere-proximal chromatin structure for chromosome segregation. Further investigation is necessary to elucidate the mechanisms by which ATP-dependent remodelers contribute to chromosome transmission.
Acknowledgments
We thank K. Bloom, D. Burke, M. Carlson, M. Grunstein, P. Hieter, D. Koshland, J. Lechner, P. Megee, A. Murray, M. A. Osley, I. Pinto, E. Tsuchiya, F. Winston, and E. Yeh for generous gifts of plasmids and/or strains, N. Azizian for contributions to Fig. 4, M. Ruiz-Noriega for contributions to Fig. 7, and M. Braunstein for assistance with CSE4 suppression experiments. Laurent laboratory members F. Geng and B. Chai are thanked for helpful comments on the manuscript.
This work was supported by Public Health Service grant GM56700 from the NIH.
REFERENCES
- 1.Amon, A. 1999. The spindle checkpoint. Curr. Opin. Genet. Dev. 9:69-75. [DOI] [PubMed] [Google Scholar]
- 2.Angus-Hill, M. L., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst, and B. R. Cairns. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7:741-751. [DOI] [PubMed] [Google Scholar]
- 3.Baker, R. E., K. Harris, and K. Zhang. 1998. Mutations synthetically lethal with cep1 target S. cerevisiae kinetochore components. Genetics 149:73-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. [DOI] [PubMed] [Google Scholar]
- 5.Berger, S. L. 2001. The histone modification circus. Science 292:64-65. [PubMed] [Google Scholar]
- 6.Bjerling, P., R. A. Silverstein, G. Thon, A. Caudy, S. Grewal, and K. Ekwall. 2002. Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol. Cell. Biol. 22:2170-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blat, Y., and N. Kleckner. 1999. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98:249-259. [DOI] [PubMed] [Google Scholar]
- 8.Bloom, K. S., and J. Carbon. 1982. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell 29:305-317. [DOI] [PubMed] [Google Scholar]
- 9.Brown, M. T., L. Goetsch, and L. H. Hartwell. 1993. MIF2 is required for mitotic spindle integrity during anaphase spindle elongation in Saccharomyces cerevisiae. J. Cell Biol. 123:387-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairns, B. R., Y. Lorch, Y. Li, M. Zhang, L. Lacomis, H. Erdjument-Bromage, P. Tempst, J. Du, B. Laurent, and R. D. Kornberg. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87:1249-1260. [DOI] [PubMed] [Google Scholar]
- 11.Cao, Y., B. R. Cairns, R. D. Kornberg, and B. C. Laurent. 1997. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol. Cell. Biol. 17:3323-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carbon, J. 1984. Yeast centromeres: structure and function. Cell 37:351-353. [DOI] [PubMed] [Google Scholar]
- 13.Chai, B., J.-M. Hsu, J. Du, and B. C. Laurent. 2002. Yeast RSC function is required for organization of the cellular cytoskeleton via an alternative PKC1 pathway. Genetics 161:575-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheeseman, I. M., D. G. Drubin, and G. Barnes. 2002. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157:199-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damelin, M., I. Simon, T. I. Moy, B. Wilson, S. Komili, P. Tempst, F. P. Roth, R. A. Young, B. R. Cairns, and P. A. Silver. 2002. The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol. Cell 9:563-573. [DOI] [PubMed] [Google Scholar]
- 16.Dillon, N., and R. Festenstein. 2002. Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet. 18:252-258. [DOI] [PubMed] [Google Scholar]
- 17.Du, J., I. Nasir, B. K. Benton, M. P. Kladde, and B. C. Laurent. 1998. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics 150:987-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edmondson, D., M. Smith, and S. Roth. 1996. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 10:1247-1259. [DOI] [PubMed] [Google Scholar]
- 19.Ekwall, K., T. Olsson, B. M. Turner, G. Cranston, and R. C. Allshire. 1997. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91:1021-1032. [DOI] [PubMed] [Google Scholar]
- 20.Fyodorov, D. V., and J. T. Kadonaga. 2001. The many faces of chromatin remodeling: SWItching beyond transcription. Cell 106:523-525. [DOI] [PubMed] [Google Scholar]
- 21.Geng, F., Y. Cao, and B. C. Laurent. 2001. Essential roles of Snf5p in Snf-Swi chromatin remodeling in vivo. Mol. Cell. Biol. 21:4311-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glowczewski, L., P. Yang, T. Kalashnikova, M. S. Santisteban, and M. M. Smith. 2000. Histone-histone interactions and centromere function. Mol. Cell. Biol. 20:5700-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goshima, G., and M. Yanagida. 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100:619-633. [DOI] [PubMed] [Google Scholar]
- 24.Hakimi, M. A., D. A. Bochar, J. A. Schmiesing, Y. Dong, O. G. Barak, D. W. Speicher, K. Yokomori, and R. Shiekhattar. 2002. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature 418:994-998. [DOI] [PubMed] [Google Scholar]
- 25.Han, M., M. Chang, U. J. Kim, and M. Grunstein. 1987. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell 48:589-597. [DOI] [PubMed] [Google Scholar]
- 26.Hanes, S. D., and R. Brent. 1989. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell 57:1275-1283. [DOI] [PubMed] [Google Scholar]
- 27.He, X., S. Asthana, and P. K. Sorger. 2000. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 101:763-775. [DOI] [PubMed] [Google Scholar]
- 28.Hecht, A., T. Laroche, S. Strahl-Bolsinger, S. Gasser, and M. Grunstein. 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80:583-592. [DOI] [PubMed] [Google Scholar]
- 29.Hegemann, J. H., and U. N. Fleig. 1993. The centromere of budding yeast. Bioessays 15:451-460. [DOI] [PubMed] [Google Scholar]
- 30.Hieter, P., C. Mann, M. Snyder, and R. W. Davis. 1985. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell 40:381-392. [DOI] [PubMed] [Google Scholar]
- 31.Hurley, J. L., and J. E. Donelson. 1980. Nucleotide sequence of the yeast plasmid. Nature 286:860-865. [DOI] [PubMed] [Google Scholar]
- 32.Hyman, A. A., and P. K. Sorger. 1995. Structure and function of kinetochores in budding yeast. Annu. Rev. Cell. Dev. Biol. 11:471-495. [DOI] [PubMed] [Google Scholar]
- 33.Jin, Q. W., J. Fuchs, and J. Loidl. 2000. Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 113:1903-1912. [DOI] [PubMed] [Google Scholar]
- 34.Keith, K. C., and M. Fitzgerald-Hayes. 2000. CSE4 genetically interacts with the Saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III: implications for the path of the centromere DNA around a Cse4p variant nucleosome. Genetics 156:973-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 36.Koshland, D., and P. Hieter. 1987. Visual assay for chromosome ploidy. Methods Enzymol. 155:351-372. [DOI] [PubMed] [Google Scholar]
- 37.Laurent, B. C., X. Yang, and M. Carlson. 1992. An essential Saccharomyces cerevisiae gene homologous to SNF2 encodes a helicase-related protein in a new family. Mol. Cell. Biol. 12:1893-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, L., S. J. Elledge, C. A. Peterson, E. S. Bales, and R. J. Legerski. 1994. Specific association between the human DNA repair proteins XPA and ERCC1. Proc. Natl. Acad. Sci. USA 91:5012-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lorch, Y., M. Zhang, and R. Kornberg. 2001. RSC unravels the nucleosome. Mol. Cell 7:89-95. [DOI] [PubMed] [Google Scholar]
- 40.Mehta, S., X. M. Yang, C. S. Chan, M. J. Dobson, M. Jayaram, and S. Velmurugan. 2002. The 2 micron plasmid purloins the yeast cohesin complex: a mechanism for coupling plasmid partitioning and chromosome segregation? J. Cell Biol. 158:625-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meluh, P. B., and D. Koshland. 1997. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11:3401-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meluh, P. B., and D. Koshland. 1995. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell 6:793-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meluh, P. B., P. Yang, L. Glowczewski, D. Koshland, and M. M. Smith. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94:607-613. [DOI] [PubMed] [Google Scholar]
- 44.Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35-45. [DOI] [PubMed] [Google Scholar]
- 45.Moreira, J. M., and S. Holmberg. 1999. Transcriptional repression of the yeast CHA1 gene requires the chromatin-remodeling complex RSC. EMBO J. 18:2836-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nasmyth, K. 2002. Segregating sister genomes: the molecular biology of chromosome separation. Science 297:559-565. [DOI] [PubMed] [Google Scholar]
- 47.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2002. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 16:806-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ortiz, J., O. Stemmann, S. Rank, and J. Lechner. 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13:1140-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pearson, C. G., P. S. Maddox, E. D. Salmon, and K. Bloom. 2001. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 152:1255-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pidoux, A. L., and R. C. Allshire. 2000. Centromeres: getting a grip of chromosomes. Curr. Opin. Cell Biol. 12:308-319. [DOI] [PubMed] [Google Scholar]
- 51.Pinto, I., and F. Winston. 2000. Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. EMBO J. 19:1598-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Recht, J., and M. A. Osley. 1999. Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi/Snf in yeast. EMBO J. 18:101-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 54.Saunders, M., M. Fitzgerald-Hayes, and K. Bloom. 1988. Chromatin structure of altered yeast centromeres. Proc. Natl. Acad. Sci. USA 85:175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saunders, M. J., E. Yeh, M. Grunstein, and K. Bloom. 1990. Nucleosome depletion alters the chromatin structure of Saccharomyces cerevisiae centromeres. Mol. Cell. Biol. 10:5721-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schulman, I., and K. S. Bloom. 1991. Centromeres: an integrated protein/DNA complex required for chromosome movement. Annu. Rev. Cell Biol. 7:311-336. [DOI] [PubMed] [Google Scholar]
- 57.Sengupta, S. M., M. VanKanegan, J. Persinger, C. Logie, B. R. Cairns, C. L. Peterson, and B. Bartholomew. 2001. The interactions of yeast SWI/SNF and RSC with the nucleosome before and after chromatin remodeling. J. Biol. Chem. 276:12636-12644. [DOI] [PubMed] [Google Scholar]
- 58.Sharp, J. A., A. A. Franco, M. A. Osley, and P. D. Kaufman. 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16:85-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen, X., G. Mizuguchi, A. Hamiche, and C. Wu. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406:541-544. [DOI] [PubMed] [Google Scholar]
- 60.Sikorski, R. S., and J. D. Boeke. 1991. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 199:302-318. [DOI] [PubMed] [Google Scholar]
- 61.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skibbens, R. V., and P. Hieter. 1998. Kinetochores and the checkpoint mechanism that monitors for defects in the chromosome segregation machinery. Annu. Rev. Genet. 32:307-337. [DOI] [PubMed] [Google Scholar]
- 63.Smith, M. M. 2002. Centromeres and variant histones: what, where, when, and why? Curr. Opin. Cell Biol. 14:279-285. [DOI] [PubMed] [Google Scholar]
- 64.Smith, M. M., P. Yang, M. S. Santisteban, P. W. Boone, A. T. Goldstein, and P. C. Megee. 1996. A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol. Cell. Biol. 16:1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sorger, P. K., K. F. Doheny, P. Hieter, K. M. Kopski, T. C. Huffaker, and A. A. Hyman. 1995. Two genes required for the binding of an essential Saccharomyces cerevisiae kinetochore complex to DNA. Proc. Natl. Acad. Sci. USA 92:12026-12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 67.Sudarsanam, P., and F. Winston. 2001. The Swi/Snf family: nucleosome-remodeling complexes and transcriptional control. Trends Genet. 16:345-351. [DOI] [PubMed] [Google Scholar]
- 68.Sullivan, K. F. 2001. A solid foundation: functional specialization of centromeric chromatin. Curr. Opin. Genet. Dev. 11:182-188. [DOI] [PubMed] [Google Scholar]
- 69.Tanaka, T., M. P. Cosma, K. Wirth, and K. Nasmyth. 1999. Identification of cohesin association sites at centromeres and along chromosome arms. Cell 98:847-858. [DOI] [PubMed] [Google Scholar]
- 70.Tanaka, T. U. 2002. Bi-orienting chromosomes on the mitotic spindle. Curr. Opin. Cell Biol. 14:365-371. [DOI] [PubMed] [Google Scholar]
- 71.Treich, I., and M. Carlson. 1997. Interaction of a Swi3 homolog with Sth1 provides evidence for a Swi/Snf-related complex with an essential function in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:1768-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Treich, I., L. Ho, and M. Carlson. 1998. Direct interaction between Rsc6 and Rsc8/Swh3, two proteins that are conserved in SWI/SNF-related complexes. Nucleic Acids Res. 26:3739-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsuchiya, E., T. Hosotani, and T. Miyakawa. 1998. A mutation in NPS1/STH1, an essential gene encoding a component of a novel chromatin-remodeling complex RSC, alters the chromatin structure of Saccharomyces cerevisiae centromeres. Nucleic Acids Res. 26:3286-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsuchiya, E., M. Uno, A. Kiguchi, K. Masuoka, Y. Kanemori, S. Okabe, and T. Mikayawa. 1992. The Saccharomyces cerevisiae NPS1 gene, a novel CDC gene which encodes a 160 kDa nuclear protein involved in G2 phase control. EMBO J. 11:4017-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Hooser, A. A., Ouspenski, I. I., H. C. Gregson, D. A. Starr, T. J. Yen, M. L. Goldberg, K. Yokomori, W. C. Earnshaw, K. F. Sullivan, and B. R. Brinkley. 2001. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114:3529-3542. [DOI] [PubMed] [Google Scholar]
- 76.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wong, M. C., S. R. Scott-Drew, M. J. Hayes, P. J. Howard, and J. A. Murray. 2002. RSC2, encoding a component of the RSC nucleosome remodeling complex, is essential for 2μm plasmid maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:4218-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xue, Y., J. C. Canman, C. S. Lee, Z. Nie, D. Yang, G. T. Moreno, M. K. Young, E. D. Salmon, and W. Wang. 2000. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. USA 97:13015-13020. [DOI] [PMC free article] [PubMed] [Google Scholar]