Abstract
p300 is a multifunctional transcriptional coactivator that serves as an adapter for several transcription factors including nuclear steroid hormone receptors. p300 possesses an intrinsic histone acetyltransferase (HAT) activity that may be critical for promoting steroid-dependent transcriptional activation. In osteoblastic cells, transcription of the bone-specific osteocalcin (OC) gene is principally regulated by the Runx2/Cbfa1 transcription factor and is stimulated in response to vitamin D3 via the vitamin D3 receptor complex. Therefore, we addressed p300 control of basal and vitamin D3-enhanced activity of the OC promoter. We find that transient overexpression of p300 results in a significant dose-dependent increase of both basal and vitamin D3-stimulated OC gene activity. This stimulatory effect requires intact Runx2/Cbfa1 binding sites and the vitamin D-responsive element. In addition, by coimmunoprecipitation, we show that the endogenous Runx2/Cbfa1 and p300 proteins are components of the same complexes within osteoblastic cells under physiological concentrations. We also demonstrate by chromatin immunoprecipitation assays that p300, Runx2/Cbfa1, and 1α,25-dihydroxyvitamin D3 receptor interact with the OC promoter in intact osteoblastic cells expressing this gene. The effect of p300 on the OC promoter is independent of its intrinsic HAT activity, as a HAT-deficient p300 mutant protein up-regulates expression and cooperates with P/CAF to the same extent as the wild-type p300. On the basis of these results, we propose that p300 interacts with key transcriptional regulators of the OC gene and bridges distal and proximal OC promoter sequences to facilitate responsiveness to vitamin D3.
The rat osteocalcin (OC) gene encodes a 10-kDa bone-specific protein that is induced in osteoblasts with the onset of mineralization at late stages of differentiation (26). Transcription of the OC gene is controlled by modularly distributed basal and hormone-responsive elements, located within two DNase I-hypersensitive sites (distal site, positions −600 to −400; proximal site, positions −170 to −70) that are present only in bone-derived cells expressing this gene (23, 24). Thus, chromatin remodeling of the OC gene promoter accompanies the onset in OC gene expression during osteoblast differentiation (Fig. 1). A key regulatory element that controls OC gene expression is recognized by the 1α,25-dihydroxyvitamin D3 receptor (VDR) complex upon ligand activation. This vitamin D3-responsive element (VDRE) is located in the distal region (Fig. 1) of the OC promoter (positions −465 to −437) and functions as an enhancer to increase OC gene transcription by three- to fivefold (20). Binding of the ligand 1α,25-dihydroxyvitamin D3 (vitamin D3) induces conformational changes in the receptor that enable it to interact with several coactivators, such as NCoA-1/SRC-1 (nuclear receptor coactivator 1/steroid receptor coactivator 1), NCoA-2/GRIP/TIF2 (nuclear receptor coactivator 2/glucocorticoid receptor-interacting protein/transcription intermediary factor 2), CBP (cyclic AMP receptor element binding protein [CREB] binding protein), and p300 (early 1 adenoviral protein [E1A]-associated 300-kDa protein) (31), that are critical for transcriptional activation. In addition, the multisubunit DRIP (for vitamin D receptor-interacting protein) complex binds to the VDR in response to the ligand (28, 29). This interaction occurs through a single motif on the VDR in much the same manner as for the coactivators and also results in transcriptional enhancement (30).
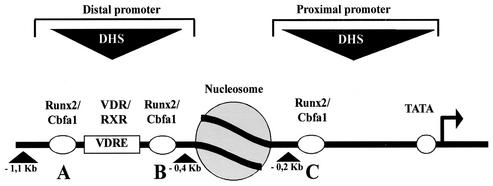
FIG. 1.
Schematic representation of the transcriptionally active OC promoter in an osteoblastic cell. Key regulatory elements of the rat OC gene, such as Runx2/Cbfa1 sites A, B, and C and the VDRE localized within the distal and proximal DNase I-hypersensitive sites (DHS), are shown. A nucleosome positioned between both DHS (shaded circle) and the direction of transcription (black arrow) are shown.
The coactivator p300 and its homologue CBP (cointegrator-associated protein) function as transcriptional adapters interacting with several transcription factors to form multimolecular complexes that regulate transcription of eukaryotic genes (9). Both p300 and CBP have been reported to interact directly with nuclear receptors through a conserved domain in their N termini in a ligand-dependent manner. This interaction is important for p300/CBP involvement in ligand-dependent transactivation of nuclear receptors (5, 12, 18). The importance of p300 in nuclear receptor activity is further demonstrated by the defective transactivation of the retinoic acid receptor (RAR) in p300 null mice (35).
p300 contains a domain with intrinsic histone acetyltransferase (HAT) activity that has been implicated in chromatin structure alterations associated with gene expression (6, 33). In addition, p300 interacts with other proteins containing HAT activity such as P/CAF (p300/CBP-associated factor) and the coactivators SRC-1 and ACTR (activator of RAR), forming large multiprotein complexes with different HAT activities (6, 33). Interestingly, the HAT activity of P/CAF but not of p300 was found to be essential for the transcriptional activation mediated by RAR (15). In contrast, the HAT activity of p300 is essential for its ability to enhance hormone-dependent activation by the thyroid receptor-RAR complex in the context of chromatin (19). Thus, p300 HAT activity may be dependent on the steroid receptor.
Bone-specific expression of the OC gene is principally regulated by the Runx2/Cbfa1 (mammalian Runt domain protein 2/core binding factor a1) transcription factor (2, 3, 7, 8). The rat OC promoter contains three recognition sites for Runx2/Cbfa1 interactions, site A (positions −605 to −595), site B (positions −435 to −430), and site C (positions −138 to −130) (Fig. 1). Mutation of sites A and B (which flank the VDRE) abolishes vitamin D3 enhancement of OC promoter transcription, while mutation of all three sites significantly reduces basal OC promoter activity and inhibits chromatin remodeling at the OC gene promoter (11). Genetic inhibition of the Runx2/Cbfa1 gene causes developmental defects in osteogenesis (14), and hereditary mutations in this gene are linked to specific ossification defects as observed in cleidocranial dysplasia (25).
Runx factors (Runx1/Cbfa2) have been shown to interact with p300 in myeloid cells (13), suggesting that members of the Runx/Cbfa family of transcription factors may be targeting p300 complexes to gene regulatory regions. However, the relationship between targeting of p300 and its function as a regulator of both tissue-specific and steroid-dependent transcription has not been established. Here, we have investigated the role of p300 as a regulator of OC gene expression in osteoblastic cells. We report that p300 stimulates basal and vitamin D3-enhanced expression of the OC promoter. Furthermore, we find that p300 interacts with the osteoblast-specific Runx2/Cbfa1 transcription factor and that the Runx2/Cbfa1 binding sites are essential for stimulation of basal and vitamin D3-enhanced OC promoter activity. In addition, p300 requires an intact VDRE to up-regulate vitamin D3-enhanced OC promoter response. Chromatin immunoprecipitation (ChIP) analyses reveal that p300, Runx2/Cbfa1, and VDR interact with the OC gene promoter in intact osteoblastic cells expressing OC. Surprisingly, the stimulatory effect of p300 is independent of its intrinsic HAT activity, as a HAT-less p300 mutant protein up-regulates the OC promoter and cooperates with P/CAF to the same extent as the wild-type p300 does. Together, our results suggest that p300 interacts with key transcriptional regulators of the OC gene and that it may function by connecting factors bound to distal and proximal promoter sequences.
MATERIALS AND METHODS
Expression constructs.
Constructs containing the rat OC gene promoter fused to the luciferase gene (pOC-LUC) were obtained by cloning the EcoRI/HindIII promoter segment (positions −1097 to +23) into the pGL3-basic plasmid (Promega, Madison, Wis.). Similarly, the pmSHE-LUC plasmid containing a mutation in the VDRE of the OC promoter, which prevents the VDR complex from binding (1), and the pmABC-LUC construct containing mutations in Runx2/Cbfa1 sites A, B, and C (11), which block binding of the Runx2/Cbfa1 factor, were cloned into the EcoRI/HindIII sites of the pGL3-basic plasmid. The pCMVβ plasmid coding for enzyme β-galactosidase and driven by the cytomegalovirus (CMV) promoter was purchased from Clontech, Inc. (Palo Alto, Calif.). The CMV-driven constructs coding for the full-length and deletion forms of Runx2/Cbfa1 protein (pcDNA-Runx2/Cbfa1Δ230, pcDNA-Runx2/Cbfa1Δ361, pcDNA-Runx2/Cbfa1Δ391, and pcDNA-Runx2/Cbfa1Δ432) were prepared as described previously (10). The CMV-driven constructs coding for p300 and E1A were kindly provided by Neil Perkins (University of Dundee, Dundee, Scotland). The CMV-driven p300Δ 72 AΤ plasmid coding for a mutated p300 protein which does not display HAT activity was generously provided by Lee Krauss (Cornell University). The pCI-P/CAF plasmid coding for P/CAF was a generous gift of Y. Nakatani (Harvard University). All the constructs were verified by automated DNA sequencing.
Cell cultures, transient-transfection assays, and reporter assays.
ROS 17/2.8 osteoblastic cells were cultured as described previously (21). COS-7 cells were grown in Dulbecco modified Eagle medium with 10% fetal calf serum as described elsewhere (8). Similar transient-transfection protocols were followed for both cell lines (8, 11). Cells plated in six-well plates were transiently transfected with 1 μg of pOC-LUC, pmSHE-LUC, pmAB-LUC, or pmABC-LUC and 0.5 μg of CMV empty vector and cotransfected with the p300, p300Δ 72 AΤ, P/CAF, or Runx2/Cbfa1 expression vector. The total amount of exogenous DNA was maintained at 9 μg/well with salmon sperm DNA (Gibco BRL, Rockville, Md.). Cells were transfected using the Lipofectamine Plus reagent (Gibco BRL) following the manufacturer's recommendations. The pCMVβ plasmid was included as an internal control for transfection efficiency. Cells were harvested 24 to 32 h after transfection and assayed for luciferase and β-galactosidase activities in a luminometer.
Immunoprecipitations.
Nuclear extracts from ROS 17/2.8 osteoblastic cells cultured in the presence or absence of 10−8 M vitamin D3 were prepared by a modified version of the Dignam method (10). Nuclear extracts (200 μg) were resuspended in 4.6 volumes of buffer D (20 mM HEPES [pH 7.9], 20% glycerol, and a complete proteinase cocktail [Roche, Indianapolis, Ind.]) with 0.1% Nonidet NP-40. The samples were then precleared with 10 μl of protein A/G Plus agarose beads (Santa Cruz Biotechnology, Santa Cruz, Calif.) for 1 h at 4°C. The beads were collected by centrifugation at 1,000 × g for 10 min at 4°C. One microgram of anti-p300 monoclonal antibody (BD Pharmingen, San Diego, Calif.) or mouse purified immunoglobulin G (IgG) (Santa Cruz Biotechnology) was added to the precleared nuclear extract, followed by an incubation for 1 h at 4°C with agitation. To collect the immunocomplexes, 10 μl of protein A/G Plus agarose beads were added and further incubated at 4°C with agitation. The beads were washed three times with washing buffer (20 mM HEPES [pH 7.9], 75 mM KCl, 2.5 mM MgCl2, 0.1% NP-40, and a proteinase inhibitor mixture) and then collected by centrifugation at 3.000 × g for 10 min at 4°C. The beads were resuspended in sodium dodecyl sulfate (SDS)-polyacrylamide gel loading buffer, incubated at 80°C for 5 min, and analyzed by Western blotting.
GST pull-down assays.
The pGEX-Runx2/Cbfa1Δ361 construct coding for the fusion protein GST-Runx2/Cbfa1Δ361 was obtained by digesting the plasmid pcDNA3.1/Runx2/Cbfa1Δ361 (10) with the restriction enzymes BamHI and XhoI and then ligating the insert into the pGEX-4XT1 vector (Pharmacia Biotech, Uppsala, Sweden). The fusion proteins GST-Runx2/Cbfa1 and GST-Runx2/Cbfa1Δ361 were obtained by expression in Escherichia coli BL-21 as previously reported (27). For the pull-down assays, 20 μl of glutathione-Sepharose beads was incubated with 4 μg of GST-Runx2/Cbfa1, GST-Runx2/Cbfa1Δ361, or glutathione S-transferase (GST) alone in a final volume of 500 μl of elution buffer (20 mM Tris-HCl [pH 8.0], 70 mM KCl, 1 mM dithiothreitol [DTT], 1 mM EDTA, 0.5% NP-40, and 0.05 mM phenylmethylsulfonyl fluoride [PMSF]) for 45 min at 4°C with gentle agitation. The beads were collected by centrifugation at 9,000 × g for 30 s and then incubated with 150 μg of nuclear extracts isolated from ROS 17/2.8 cells in 480 μl of BP buffer supplemented with 0.5% nonfat milk for 2 h at 4°C with agitation. The complexes bound to the beads were recovered by centrifugation at 9,000 × g for 30 s, resuspended in 15 μl of loading buffer, incubated at 95°C for 5 min, and fractionated by SDS-polyacrylamide gel electrophoresis (PAGE) in a 6% polyacrylamide gel. The presence of p300 associated with the GST-Runx2/Cbfa1 proteins was confirmed by Western blot analysis using an anti-p300 polyclonal antibody (Santa Cruz Biotechnology). Similarly, the presence of equivalent concentrations of the GST-Runx2/Cbfa1 proteins in the pull-down assay was determined by Western blotting, using an anti-Runx2/Cbfa1 antibody (Oncogene Research, Boston, Mass.).
Western blot analysis.
Transfected or untransfected cells cultured on 100-mm-diameter dishes were lysed on the plate by adding 300 μl of SDS lysis buffer (2% SDS, 10 mM DTT, 10% glycerol, 2 M urea, 1 mM PMSF, 10 mM Tris-HCl [pH 6.8], 0.002% bromophenol blue, and a proteinase inhibitor mixture). Proteins were fractionated in SDS-polyacrylamide gels with different amounts of polyacrylamide (8% polyacrylamide to detect p300 and 10% polyacrylamide to detect Runx2/Cbfa1, E1A, and P/CAF) and transferred to a nitrocellulose membrane (Bio-Rad, Hercules, Calif.). The membrane was blocked with 1.5% low-fat milk in phosphate-buffered saline (PBS) for 1 h at room temperature and then washed four times for 10 min each time with a solution of PBS and 0.05% Tween 20. The proteins were revealed using the chemiluminescence reagent plus system (Perkin-Elmer Life Sciences, Boston, Mass.). Antibodies were purchased from Oncogene Research (anti-Runx/Cbfa1), BD Pharmingen (anti-E1A), Santa Cruz Biotechnology (anti-p300), and Upstate Biotechnology (Lake Placid, N.Y.) (anti-P/CAF).
ChIP assays.
ChIP studies were performed as described earlier (32), with modifications. All the steps were done at 4°C. Ros 17/2.8 cell cultures (100-mm-diameter plates) were incubated for 5 min with 1% formaldehyde and gentle agitation. The cross-linking was stopped by the addition of 0.125 M glycine for 5 min. The cells were then washed with 10 ml of PBS, scraped off in the same volume of PBS, and collected by centrifugation at 1,000 × g for 5 min. The cell pellet was resuspended in 3 ml of lysis buffer (85 mM Tris-HCl [pH 8.0], 85 mM KCl, 0.5% NP-40, and a cocktail of proteinase inhibitors) and incubated for 10 min on ice. The cell extract was then collected by centrifugation at 1,000 × g for 5 min, resuspended in 1.5 ml of nucleus lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA, 1% SDS, and a cocktail of proteinase inhibitors), and incubated for 10 min on ice.
To reduce the length of the chromatin fragments to approximately 500 bp (confirmed by electrophoretic analysis), the extract was sonicated with a Misonix sonicator (model 3000), using three 15-s pulses at 30% power. After centrifugation at 16,000 × g, the supernatant was collected, frozen in liquid nitrogen, and kept at −80°C. An aliquot was kept for protein measurements. Cross-linked extracts (150 μg) were resuspended in buffer E (20 mM HEPES [pH 7.9], 0.5 mM DTT, 20% glycerol, and a cocktail of proteinase inhibitors) to a final volume of 500 μl. The samples were precleared by incubation with 50 μl of protein A-agarose beads preblocked with bovine serum albumin (Santa Cruz Biotechnology) for 15 min at 4°C with agitation. After centrifugation at 1,000 × g for 5 min, the supernatant was collected and immunoprecipitated with either an anti-p300 monoclonal antibody (BD Pharmingen), an anti-VDR monoclonal antibody (Oncogene Research), or an anti-Runx2/Cbfa1 polyclonal antibody (Oncogene Research). The immune complexes were recovered with the addition of 50 μl of protein A-agarose beads and subsequent incubation for 15 min at 4°C with agitation. The complexes were washed twice with dialysis buffer (2 mM EDTA and 50 mM Tris-HCl [pH 8.0]) and four times with 1 ml of immunoprecipitation buffer (100 mM Tris-HCl [pH 8.0], 500 mM LiCl, 1% NP-40, and 1% deoxycholic acid), incubating the solution at each washing for 5 min at 4°C. The protein-DNA complexes were then eluted by two incubations with 150 μl of elution buffer (50 mM NaHCO3 and 1% SDS) and brief agitation. After centrifugation at 1,000 × g for 5 min, the supernatant was collected and incubated with 10 μg of RNase A per ml at 67°C to reverse the cross-linking. The proteins were then digested with 200 μg of proteinase K per ml for 1 h at 42°C. The DNA was recovered by phenol-chloroform extraction and ethanol precipitation using tRNA (5 μg/ml) as a precipitation carrier. The PCR conditions and the sequences of the primers used were reported previously (32). Primers used were as follows (only the starting positions are indicated): for p300, forward (position −459) and reverse (position −28); for Runx2/Cbfa1, forward (position −459) and reverse (position −118); and for VDR, forward (position −773) and reverse (position −433).
RESULTS
p300 stimulates basal and vitamin D3-enhanced OC promoter activity.
Transcription of the rat OC gene is regulated by the coordinated activity of basal tissue-specific and steroid hormone enhancer elements that are modularly distributed across distal and proximal promoter sequences (20). Functional and physical interactions between transcription factors bound to these two regions of the OC promoter are necessary to respond to the physiological mediators that either up- or down-regulate OC expression in osteoblastic cells. Establishment of these interactions can be facilitated by a defined three-dimensional chromatin organization of the promoter (20) and by proteins that can interact simultaneously and specifically with several promoter-bound regulatory transcription factors. p300 is a large coactivator protein that functions as a transcriptional adapter interacting with various types of transcription factors, including the nuclear steroid receptors upon ligand stimulation (6, 33).
We analyzed whether p300 plays a role in regulating rat OC gene transcription by assessing the effect of forced expression of p300 on the activity of a full-length (1.1-kb) OC promoter-luciferase reporter gene construct in the osteoblastic ROS 17/2.8 cell line. Similar to mineralized normal diploid osteoblasts, ROS 17/2.8 cells express high levels of OC and respond to vitamin D3 with a three- to fivefold increase in OC gene transcription (23). The results indicate that p300 significantly enhances OC promoter activity (three- to fourfold) in a dose-dependent manner (Fig. 2A). We confirmed by Western blot analysis that increasing levels of p300 are obtained in the transiently transfected cells (Fig. 2B). Transient overexpression of p300 in ROS 17/2.8 cells also resulted in higher levels of OC mRNA, measured by reverse transcription-PCR (data not shown). Interestingly, the effect of p300 on OC promoter activity was significantly higher when the cells were treated with vitamin D3 (two- to threefold [Fig. 2A]), suggesting that p300 may be functionally coupled to the liganded VDR complex that interacts with the OC promoter (4, 22, 27).
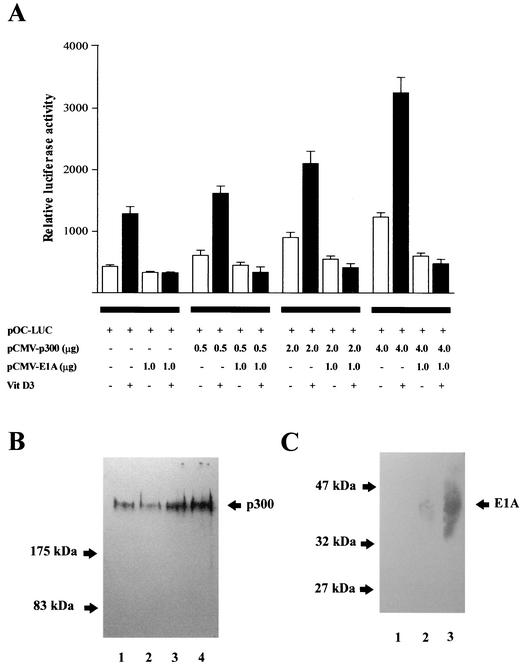
FIG. 2.
The p300 coactivator up-regulates OC gene expression. (A) ROS 17/2.8 cells were transiently cotransfected with increasing amounts of p300 expression plasmid (pCMV-p300) and 1.0 μg of 1.1-kb pOC-LUC reporter construct. The presence (+) or absence (−) and the amount of different plasmids and vitamin (Vit) D3 are shown below the bars. Cells were cultured either under control conditions (white bars) or in the presence of 10−8 M vitamin D3 (black bars) for 24 h and harvested, and luciferase activities were determined. The data were normalized to values for pCMV-β-galactosidase activity as an internal control. In some transfections, 1 μg of E1A expression vector (pCMV-E1A) was included. Pooled data from at least three independent experiments are presented. Each bar represents the mean ± standard error of the mean (n = 9; P < 0.05). (B) ROS 17/2.8 cells plated on 100-mm-diameter dishes were transiently transfected in parallel with 1.0 μg of empty vector or with increasing concentrations of the p300 expression construct. Proteins (30 μg) were resolved on SDS-8% polyacrylamide gels, transferred to nitrocellulose membranes, and the presence of p300 was revealed by Western blotting. Lane 1, 1.0 μg of empty vector; lanes 2 to 4, 0.5 μg (lane 2), 2.0 μg (lane 3), and 4.0 μg (lane 4) of p300 vector. (C) As in panel B, ROS 17/2.8 cells were transfected with empty vector or with increasing concentrations of the pE1A expression construct, and proteins were revealed by Western blotting. Lane 1, 1.0 μg of empty vector; lanes 2 and 3, 0.5 μg (lane 2) and 2.0 μg (lane 3) of E1A vector. The positions of the molecular mass markers are shown at the left of the blots.
The ability of p300 to form large molecular complexes can be abolished by the viral oncoprotein E1A, which inhibits the p300 coactivator functions. Thus, it has been shown that E1A blocks the association of p300 with P/CAF (34) and with coactivators SRC-1 and P/CIP (p300/CBP cointegrator-associated protein) (17) which recruit p300 to steroid receptor complexes in a ligand-dependent manner. We found that the stimulatory effect of p300 on both basal and vitamin D3-enhanced OC promoter activity is inhibited when the viral oncoprotein E1A is also coexpressed (Fig. 2A). Furthermore, higher concentrations of E1A result in complete loss of OC promoter activity (data not shown). As a control, E1A overexpression was confirmed by Western blot analysis (Fig. 2C).
Thus, our results indicate that p300 stimulates OC promoter activity likely by interacting directly with transcription factors that modulate OC expression in osteoblastic cells.
p300 requires specific regulatory elements to enhance OC promoter activity.
The Runx2/Cbfa1 transcription factor and the VDR complex are the principal regulators of bone-specific OC gene expression (2, 3, 7, 8). We assessed the relationship between p300 and these OC transcriptional modulators by evaluating the effect of forced expression of p300 on rat OC promoter-luciferase reporter constructs in which the binding sites for Runx2/Cbfa1 or for the VDR complex (VDRE) have been mutated. These mutations were previously shown to prevent binding of these transcription factors and to inhibit their stimulatory effect on OC promoter activity (1, 11).
We found that the OC promoter construct containing all three Runx2/Cbfa1 sites (sites A, B, and C) mutated does not respond to increased expression of p300 (Fig. 3A). Moreover, the addition of vitamin D3 does not change the inability of this promoter to respond to p300, suggesting that binding of the Runx2/Cbfa1 transcription factor is a requirement for the stimulatory effect of p300. Interestingly, mutation of the VDRE does not prevent p300 from enhancing basal OC promoter activity (Fig. 3B) but completely blocks the stimulatory effect of p300 on the vitamin D3-dependent increase of this promoter activity (Fig. 3B). In agreement with a previous report (10), we found that the absence of functional Runx2/Cbfa1 sites A and B inhibits the vitamin D3-dependent enhancement of the OC promoter (Fig. 3C). Similarly, we observed that mutation of these two sites prevents p300 from further stimulating basal OC promoter activity in ROS 17/2.8 cells treated with vitamin D3 (Fig. 3C).
FIG. 3.
p300-dependent up-regulation of the OC promoter requires functional Runx2/Cbfa1 and VDRE sequences. Constructs (1.0 μg each) carrying wild-type (pOC-LUC), mutated Runx2/Cbfa1 sites A, B, and C (pmABC-OC-LUC) (A), mutated VDRE (pmSHE-OC-LUC) (B), or mutated Runx2/Cbfa1 sites A and B (pmAB-OC-LUC) in the context of the full-length OC promoter (1.1 kb) were cotransfected with p300 expression plasmid (3.0 μg) into ROS 17/2.8 cells. The presence (+) or absence (−) of different plasmids and vitamin (Vit) D3 are shown below the bars. Cells were cultured for 24 h in the absence (white bars) or presence (black bars) of 10−8 M vitamin D3. Luciferase reporter activities were then determined and normalized to β-galactosidase values. Pooled data from at least three independent experiments are presented. Each bar represents the mean ± standard error of the mean (n = 9; P < 0.05)
Taken together, our results indicate that p300 requires the Runx2/Cbfa1 sites to stimulate OC promoter expression. Our findings also indicate that the VDRE and Runx2/Cbfa1 sites A and B are necessary for p300 to up-regulate the vitamin D3-dependent enhancement of this promoter.
p300 interacts with the bone-specific Runx2/Cbfa1 transcription factor in osteoblastic cells.
Our results suggest that the transcription factor Runx2/Cbfa1 is important in the recruitment of p300 to the OC promoter and its subsequent stimulatory effect on OC promoter activity. Additionally, it has been shown that p300 interacts with Runx1/Cbfa2, a Runx family member present principally in myeloid cells and highly homologous with the osteoblast-specific Runx2/Cbfa1 (13). Hence, by combining coimmunoprecipitation and Western blot analyses, we assessed whether p300 interacts with Runx2/Cbfa1 in the osteoblastic ROS 17/2.8 cells.
As shown in Fig. 4A, we immunoprecipitated p300 from nuclear extracts of ROS 17/2.8 cells (lanes 1 and 3). This immunoprecipitation was specific, as p300 is not detected when a nonspecific mouse IgG fraction is used in the reaction (Fig. 4A, lane 2). More interestingly, the bone-specific transcription factor Runx2/Cbfa1 is present in the immunoprecipitated material (Fig. 4B, lanes 3 and 4), strongly indicating that in osteoblastic cells expressing OC, p300 and Runx2/Cbfa1 are components of the same nuclear protein complex. For a control, we also show that when the nonspecific mouse IgG is incubated with the ROS 17/2.8 nuclear extracts, Runx2/Cbfa1 is not found in the immunoprecipitated protein fraction (Fig. 4B, lanes 1 and 2). We did not detect significant differences in the immunoprecipitated Runx2/Cbfa1 levels between nuclear extracts obtained from ROS 17/2.8 cells cultured either in the presence or absence of vitamin D3 (Fig. 4B, compare lanes 3 and 4). Thus, the p300-Runx2/Cbfa1 interaction is not altered by the presence of the ligand-activated vitamin D3 receptor.
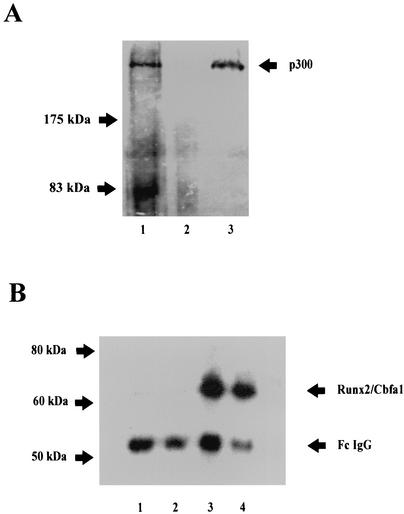
FIG. 4.
Runx2/Cbfa1 and p300 are components of the same nuclear protein complexes in osteoblastic cells expressing OC. Interaction between Runx2/Cbfa1 and p300 is shown by immunoprecipitation. Nuclear extracts from ROS 17/2.8 cells were immunoprecipitated with an anti-p300 monoclonal antibody or with a nonspecific mouse IgG fraction as described in Materials and Methods. Immunoprecipitated complexes were resolved by SDS-PAGE with 8% (A) and 10% (B) polyacrylamide gels, followed by Western blotting using either anti-p300 (A) or anti-Runx2/Cbfa1 (B) antibodies. (A) Lane 1, input; lane 2, nonspecific mouse IgG fraction; lane 3, immunoprecipitated p300. (B) Samples immunoprecipitated from ROS 17/2.8 cells cultured in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of 10−8 M vitamin D3. Samples were immunoprecipitated with the nonspecific mouse IgG fraction (lanes 1 and 2) or with the p300 antibody (lanes 3 and 4). The positions of molecular mass markers are shown to the left of the blots.
We next analyzed whether a specific domain of the Runx2/Cbfa1 protein is required to functionally interact with p300 within the context of the OC promoter. Full-length p300 and a series of Runx2/Cbfa1 deletion mutants (Fig. 5) were coexpressed in COS-7 cells and analyzed for their ability to cooperatively stimulate the OC promoter activity. As COS-7 cells do not express Runx2/Cbfa1 factors, they constitute a good system to evaluate Runx2/Cbfa1-dependent gene expression. As shown in Fig. 5, OC promoter activity is significantly increased when most of the Runx2/Cbfa1 isoforms are coexpressed. This Runx2/Cbfa1-dependent activity was maximal when either the full-length Runx2/Cbfa1 or a Runx2/Cbfa1 isoform that lacks 82 amino acids from the C terminus (Runx2/Cbfa1Δ432) were evaluated. In agreement with our studies in ROS 17/2.8 cells, coexpression of p300 and Runx2/Cbfa1 resulted in marked enhancement of the OC promoter activity (Fig. 5, black bars), further indicating that both factors cooperate to stimulate this promoter. The cooperative effect of p300 and Runx2/Cbfa1 was highest when either full-length Runx2/Cbfa1 or the Runx2/Cbfa1Δ432 mutant were analyzed, indicating that the last 82 amino acids of the C-terminal region in Runx2/Cbfa1 (residues 433 to 514) do not contribute to the interaction with p300.
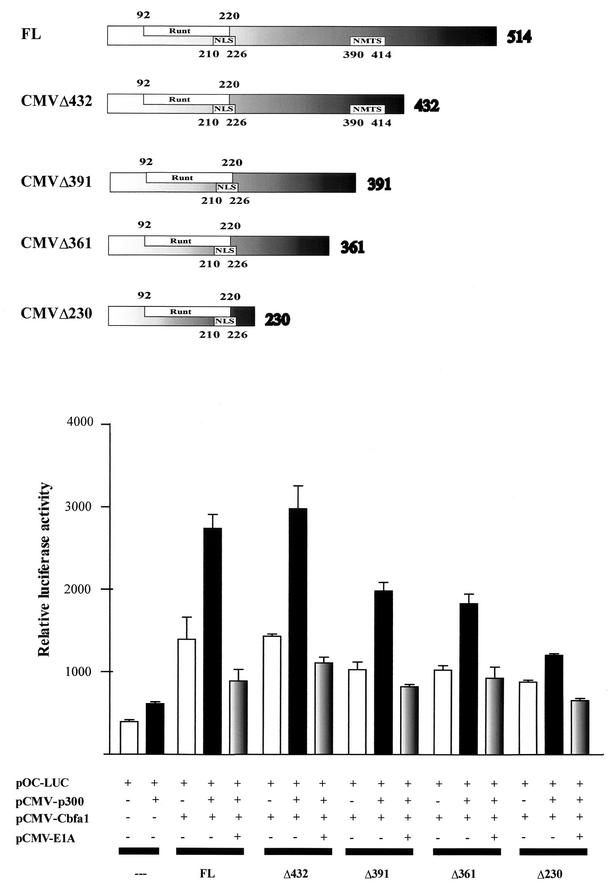
FIG. 5.
Functional interaction of Runx2/Cbfa1 and p300 requires the C-terminal region of Runx2/Cbfa1. CMV-driven constructs coding for full-length Runx2/Cbfa1 (FL) and deletion mutants of Runx2/Cbfa1 are shown schematically at the top of the figure. The Runt homology domain, the nuclear localization signal (NLS), and the nuclear matrix targeting signal (NMTS) are indicated. The functional cooperation between Runx2/Cbfa1 and p300 is demonstrated in COS-7 cells which were transiently cotransfected with different combinations of plasmids. The presence (+) or absence (−) of different plasmids is shown under the bars. The cells were transiently cotransfected with the following plasmids: CMV-empty vector (0.5 μg) (---); plasmids (0.5 μg each) coding for full-length Runx2/Cbfa1 (FL) and deletion mutant Runx2/Cbfa1 proteins (Δ432, Δ391, Δ361, and Δ230); CMV-p300 expression vector (1.0 μg); and pOC-LUC reporter construct (1.0 μg). Cells were cultured for 24 h and harvested, and luciferase activity was determined. The data were normalized to values for pCMV-β-galactosidase activity as an internal control. In some transfections, 1.0 μg of E1A expression vector (pCMV-E1A) was included.
In contrast, we did not find cooperation between p300 and a short isoform of Runx2/Cbfa1 (Runx2/Cbfa1Δ230) which lacks the C-terminal residues 231 to 514. This result indicates that the region that interacts with p300 is located C terminal of the Runt homology DNA binding domain of Runx2/Cbfa1. Interestingly, two other Runx2/Cbfa1 deletion mutants which lack residues 392 to 514 and 362 to 514 (Runx2/Cbfa1Δ391 and Runx2/Cbfa1Δ361, respectively) (Fig. 5) show reduced but significant ability to cooperate with p300 and stimulate the OC promoter activity (Fig. 5). As we found earlier for ROS 17/2.8 cells, coexpression of E1A inhibited the cooperative effect of p300 and Runx2/Cbfa1 (Fig. 5).
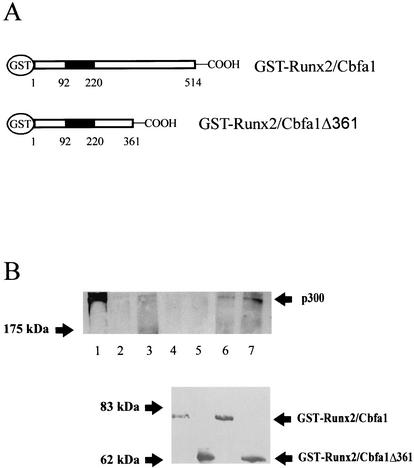
The ability of Runx2/Cbfa1 to interact with p300 was also confirmed by GST pull-down experiments. The overexpression studies in COS-7 cells indicated that the Runx2/Cbfa1Δ361 mutant is able to functionally interact with p300 (Fig. 5). Hence, recombinant full-length GST-Runx2/Cbfa1 and GST-Runx2/Cbfa1Δ361 proteins were produced in E. coli (Fig. 6A) and analyzed for their ability to bind the p300 protein present in nuclear extracts from ROS 17/2.8 cells. As shown in Fig. 6B, both Runx2/Cbfa1 protein isoforms interact with p300 (Fig. 6B, lanes 6 and 7), indicating that there is a p300 binding domain within the first 361 amino acid residues of Runx2/Cbfa1. The interactions were specific, as p300 was not precipitated by GST alone (Fig. 6B, lane 2).
FIG. 6.
Runx2/Cbfa1 and p300 interact in vitro. The genes coding for full-length Runx2/Cbfa1 and Runx2/Cbfa1Δ361 were cloned in the pGEX-4XT1 vector and expressed as a GST fusion protein in bacteria. The abilities of these two proteins to interact with p300 in vitro were evaluated by GST pull-down experiments (see Materials and Methods). (A) Schematic illustration of the GST-Runx2/Cbfa1 and GST-Runx2/Cbfa1Δ361 proteins. The Runt homology DNA binding domain (black box) and amino acid positions are shown. (B) Glutathione-Sepharose beads (20 μl) were preincubated with 4 μg (each) of GST (lane 3), GST-Runx2/Cbfa1 (lanes 4 and 6), and GST-Runx2/Cbfa1Δ361 (lanes 5 and 7) protein for 45 min at 4°C. Nuclear extracts from ROS 17/2.8 cells (150 μg) were added to some samples (lanes 2, 3, 6, and 7) and incubated for 2 h at 4°C. The samples were then fractionated by SDS-PAGE (6% polyacrylamide gel), and the presence of bound p300 was detected by Western blot analysis. Lane 1, input; lane 2, glutathione-Sepharose plus nuclear extracts; lane 3, GST plus nuclear extracts; lane 4, GST-Runx2/Cbfa1; lane 5, GST-Runx2/Cbfa1Δ361; lane 6, GST-Runx2/Cbfa1 plus nuclear extracts; lane 7, GST-Runx2/Cbfa1Δ361 plus nuclear extracts. The bottom blot shows the results of a Western blot analysis done with an anti-Runx2/Cbfa1 polyclonal antibody to confirm that equivalent amounts of GST-Runx2/Cbfa1 and GST-Runx2/Cbfa1Δ361 are bound to the glutathione-Sepharose beads in the samples in lanes 4 to 7. The positions of the molecular mass markers are shown to the left of the blots.
Taken together, these results indicate that p300 can be recruited to the OC promoter by forming a complex with the osteoblast-specific Runx2/Cbfa1 factor. Our results also indicate that the interaction between p300 and Runx2/Cbfa1 requires the presence of a domain located C-terminal of the Runt homology DNA binding domain of Runx2/Cbfa1.
p300 interacts with the OC promoter in intact osteoblastic cells.
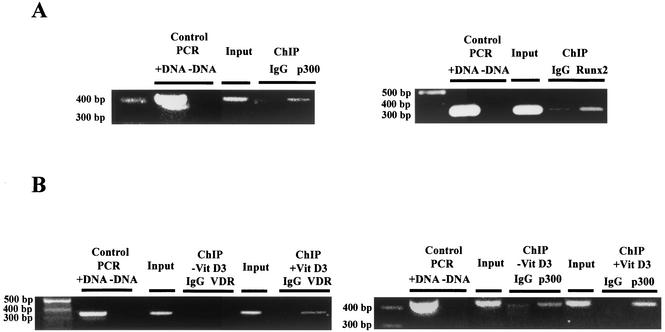
To assess whether the p300 coactivator is recruited, in association with other key transcriptional regulators, to the OC promoter, we performed ChIP assays using specific antibodies against p300, Runx2/Cbfa1, and VDR. These antibodies were used in whole-cell lysates to precipitate sonicated chromatin that was obtained after formaldehyde cross-linking of DNA to transcription factors in intact osteoblastic cells expressing OC. DNA fragments that coprecipitated with either p300, Runx2/Cbfa1, or VDR were purified upon reversal of protein-DNA cross-links and used as the templates for PCRs with gene-specific primers (see Materials and Methods). We adjusted the number of PCR cycles to remain within the linear range of PCR amplification. The resulting DNA products were fractionated by agarose electrophoresis and visualized by ethidium bromide staining. In each experiment, the specificity of the PCR procedure was assessed by appropriate positive- and negative-control reactions in which either specific primers or templates were omitted. Furthermore, the specificity of the recognition by the antibody was controlled by including samples immunoprecipitated with a nonrelated purified IgG.
As shown in Fig. 7, p300 is associated with the OC promoter in ROS 17/2.8 cells. This interaction occurs in the absence of vitamin D3 (Fig. 7A, left gel), condition in which these cells express basal levels of the OC gene. Similarly, we found that the Runx2/Cbfa1 transcription factor is associated with the OC promoter (Fig. 7A, right gel), further indicating that Runx2/Cbfa1 and p300 are regulating basal transcription of the OC gene. For a control, we show that no significant signal was detected in precipitates obtained with a nonrelated IgG antibody. We also determined that VDR is interacting with the distal OC promoter region only in cells that were treated with vitamin D3 for 4 h (Fig. 7B, left gel). This result is in agreement with previous observations, indicating that the VDR interacts with the OC VDRE in a ligand-dependent manner (22, 27). Interestingly, we reproducibly detect an increase in the PCR signal of chromatin samples from vitamin D3-treated ROS 17/2.8 cells that were precipitated with the anti-p300 antibody (Fig. 7B, right gel, compare ChIP −Vit D3 p300 and +Vit D3 p300 lanes). This result suggests an increased association of p300 with the OC promoter after vitamin D3 stimulation.
FIG. 7.
p300, Runx2/Cbfa1, and VDR interact with the OC promoter in intact osteoblastic cells expressing OC. The abilities of p300, Runx2/Cbfa1, and VDR to interact with the endogenous OC promoter in intact ROS 17/2.8 cells expressing OC were determined by ChIP analysis (see Materials and Methods). (A) ChIP assays performed with formaldehyde cross-linked chromatin isolated from ROS 17/2.8 cells cultured in the absence of vitamin D3 and antibodies against p300 (left gel) and Runx2/Cbfa1 (right gel). Both gels are ethidium bromide-stained agarose gels of the PCR products obtained with OC primers with sequence from positions −459 to −28 (left gel) and positions −459 to −118 (right gel) (32) in chromatin immunoprecipitates with the indicated antibodies (p300 or Runx2). Control PCRs were done with DNA (+DNA) and without DNA (−DNA). (B) ChIP assays performed with chromatin samples from ROS 17/2.8 cells cultured in the presence (+VitD3) or absence (−VitD3) of vitamin D3 and antibodies against the VDR (left gel) or p300 (right gel). The primers for VDR (positions −773 to −433) and p300 (positions −459 to −28) were used (32). The positions of molecular size markers (in base pairs) are indicated to the left of the gels. Samples were precipitated by the nonspecific antibody IgG as a control.
The HAT activity of p300 is not required for up-regulation of OC promoter activity.
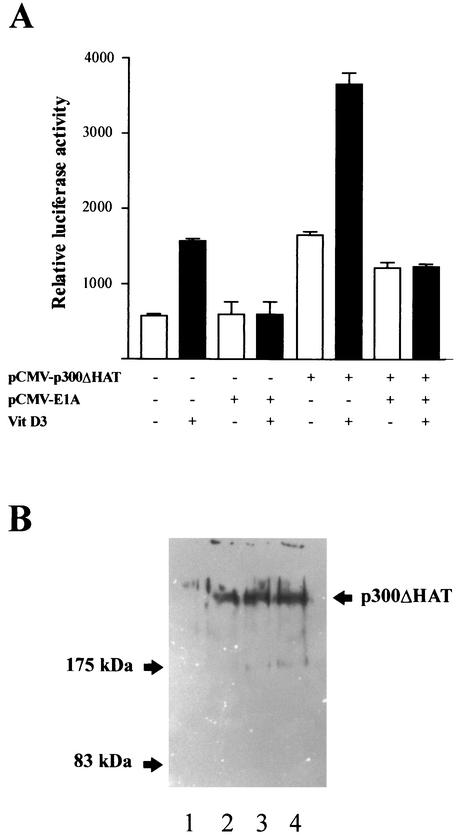
p300 has intrinsic HAT activity that may be essential for its function as a transcriptional coactivator of nuclear steroid receptors (6, 15, 33). We therefore analyzed whether the HAT activity of p300 is necessary to enhance OC promoter activity. As shown in Fig. 8A, forced expression of a mutant p300 protein which does not display HAT activity (16) up-regulates both basal (threefold) and vitamin D3-enhanced (two- to threefold) OC promoter activity to an extent similar to that of the wild-type p300 protein (Fig. 2A and 3A). Moreover, this effect is also inhibited by coexpression of E1A (Fig. 8A), indicating that as for the wild-type p300, the stimulation of OC promoter activity by the HAT-less p300 requires interaction with OC transcriptional regulators. For a control, we showed by Western blot analysis (Fig. 8B) that the overexpression of the HAT-less p300 generates high intracellular concentrations of this protein equivalent to those achieved for the wild-type p300 (Fig. 2B).
FIG. 8.
The intrinsic HAT activity of p300 is not required to up-regulate OC promoter activity. (A) An expression vector (3.0 μg) encoding a mutated p300 protein that does not display HAT activity (pCMV-p300ΔHAT) was transiently cotransfected with the pOC-LUC plasmid (1.0 μg) into ROS 17/2.8 cells cultured in the absence (white bars) or presence (black bars) of 10−8 M vitamin D3. The presence (+) or absence (−) of different plasmids and vitamin (Vit) D3 is indicated under the bars. After 24 h, the cells were harvested, and reporter activity was evaluated as described in Materials and Methods. In some transfections, the pCMV-E1A plasmid (1.0 μg) was also included. Pooled data from at least three independent experiments are presented. Each bar represents the mean ± standard error of the mean (n = 9; P < 0.05). (B) ROS 17/2.8 cells plated on 100-mm-diameter dishes were transiently transfected in parallel with either 1.0 μg of empty vector or increasing concentrations of the HAT-less p300 expression construct. Proteins (30 μg) were resolved by SDS-PAGE (8% polyacrylamide gels) and transferred to nitrocellulose membranes, and the presence of p300 was revealed by Western blotting. Lane 1, 1.0 μg of empty vector; lanes 2 to 4, 0.5 μg (lane 2), 2.0 μg (lane 3), and 4.0 μg (lane 4) of HAT-less p300 vector. The positions of molecular mass markers are shown to the left of the blot.
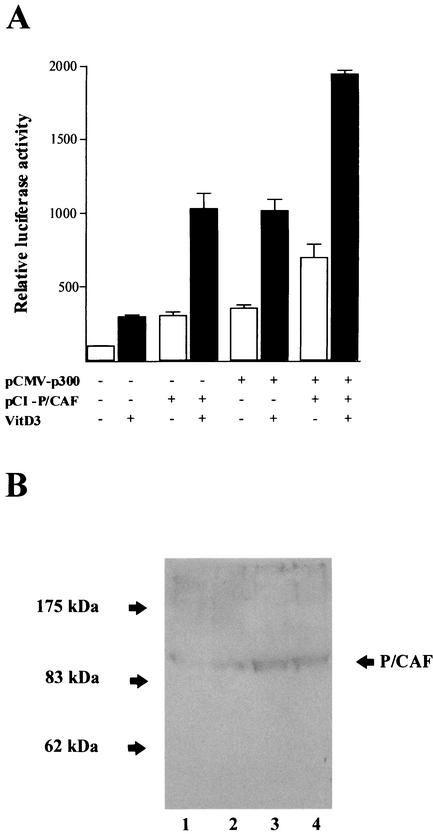
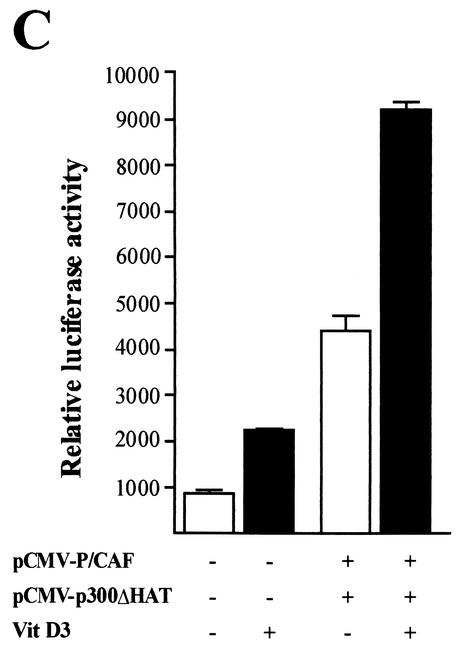
p300 interacts with P/CAF, which also contains a HAT domain, to form a protein complex with different HAT activities (6, 33). We evaluated the contribution of P/CAF to the stimulatory effect of p300 on the OC promoter in osteoblastic cells. As shown in Fig. 9A, forced expression of P/CAF in ROS 17/2.8 cells significantly enhances both basal (threefold) and vitamin D3-stimulated (three- to fourfold) OC promoter activity. For a control, we show by Western blotting that under our experimental conditions, significantly higher intracellular levels of P/CAF are obtained after transfection (Fig. 9B). Additionally, a marked increase in OC promoter activity is detected when p300 and P/CAF are coexpressed (six- to sevenfold), indicating that these two proteins functionally cooperate to up-regulate OC expression (Fig. 9A). Interestingly, similar additive effects of P/CAF and p300 on the OC promoter are found when the HAT-less p300 is overexpressed (Fig. 9C), further indicating that the HAT activity of p300 is not required for stimulation of OC promoter activity or for cooperation with P/CAF in osteoblastic cells.
FIG. 9.
p300 cooperates with P/CAF to stimulate rat OC gene promoter activity. Expression constructs (3.0 μg each) for the coactivators p300 (wild-type p300 [A] or HAT-deficient mutant p300 [C]) and P/CAF (pCI/P/CAF) were transiently cotransfected alone or in combination in ROS 17/2.8 cells. The effect on the pOC-LUC reporter plasmid (1.0 μg) was evaluated 24 h later. Cells were cultured in the presence (black bars) or absence (white bars) of 10−8 M vitamin D3. The presence (+) and absence (−) of different plasmids and vitamin D3 (VitD3) is shown under the bars. Pooled data from at least three independent experiments are presented. Each bar represents the mean ± standard error of the mean (n = 9; P < 0.05). (B) ROS 17/2.8 cells plated on 100-mm-diameter dishes were transiently transfected in parallel with 1.0 μg of empty vector or with increasing concentrations of the P/CAF expression construct. Proteins (30 μg) were resolved by SDS-PAGE (8% polyacrylamide gels) and transferred to nitrocellulose membranes, and the presence of P/CAF was revealed by Western blotting. Lane 1, 1.0 μg of empty vector; lanes 2 to 4, 0.5 μg (lane 2), 2.0 μg (lane 3), and 4.0 μg (lane 4) of P/CAF vector. The positions of the molecular mass markers are shown to the left of the blot.
Taken together, our results suggest that p300 has an important regulatory role in basal and vitamin D3-enhanced OC expression. This role requires interaction with key regulatory transcription factors that bind to the distal and proximal OC promoter sequences and does not involve its intrinsic HAT activity.
DISCUSSION
We designed experiments to determine whether the coactivator protein p300 has a regulatory role in transcription of the rat OC gene in osteoblastic cells. p300 can interact with several transcription factors to form large multimolecular protein complexes that regulate transcription of eukaryotic genes (6, 33). Among these factors are the nuclear steroid receptors which bind to p300 upon ligand activation. This interaction has been shown to be important for ligand-dependent transactivation by steroid receptors (5, 12, 35). Here we show that forced expression of p300 results in a dose-dependent increase of basal and vitamin D3-enhanced OC promoter activity. This stimulatory effect of p300 requires the binding sites for the key OC regulatory transcription factors Runx2/Cbfa1 and the VDR complex. Moreover, we determined by coimmunoprecipitation analysis that within nuclear extracts from osteoblastic cells expressing the OC gene, Runx2/Cbfa1 and p300 are components of the same protein complexes. Coexpression experiments in nonosseous cells and GST pull-down analysis also indicate that Runx2/Cbfa1 and p300 interact. Similarly, previous studies have shown that upon ligand activation, VDR interacts with p300 through the SRC-1 coactivator (31). Together, these results provide a basis for postulating that p300 stimulates OC basal expression, as it is recruited to the promoter through its interaction with Runx2/Cbfa1. After ligand activation, p300 is able to interact functionally with the VDR-RXR heterodimer bound to the VDRE and further enhances OC transcription. The recruitment of p300 to the OC promoter was confirmed by our ChIP analysis, which demonstrated that Runx2/Cbfa1 and p300 are associated with this promoter in intact osteoblastic cells that are expressing basal and vitamin D3-enhanced OC levels.
In support of this proposal, we have recently shown that mutation of Runx2/Cbfa1 sites A and B (which flank the VDRE in the distal OC promoter region) eliminates vitamin D3 responsiveness and that mutation of all three Runx2/Cbfa1 sites (sites A, B, and C) inhibits chromatin remodeling and significantly reduces OC transcriptional activity in bone cells (11). Because binding of the VDR-RXR heterodimer to the OC VDRE requires chromatin remodeling (22, 27), our results indicate that Runx2/Cbfa1 interaction is a requirement for subsequent vitamin D3-dependent enhancement of the rat OC promoter.
Kitabayashi et al. have shown that in myeloid cells the transcription factor Runx1/Cbfa2, a homologue of the bone-specific Runx2/Cbfa1, interacts with p300 and together these two factors regulate myeloid-specific genes (13). They established that a region located C terminal of the Runt DNA binding domain in Runx1/Cbfa2 is critical for its interaction with p300. In agreement with this report, our results suggest that in osteoblastic cells Runx2/Cbfa1 interacts with p300 through an amino acid region also located C terminal of the Runt domain. Considering the high degree of homology between these two members of the Runx/Cbfa family of transcription factors, it is possible that the structural features that determine the ability of Runx/Cbfa to interact with p300 are conserved.
p300 interacts with other proteins that also contain intrinsic HAT activity. Among these are P/CAF and the coactivators SRC-1 and ACTR, which together with p300 form large complexes containing different HAT activities (6, 33). It appears, however, that the HAT activity of p300 may not always be required for its function as a coactivator. Thus, the HAT activity of P/CAF but not of p300 is essential for the transcriptional activation mediated by the RAR complex (15). In contrast, Li and colleagues have recently reported that the ability of p300 to enhance hormone-dependent activation by the thyroid receptor-RXR complex in the context of chromatin requires p300 HAT activity (19). We find that the stimulatory effect of p300 on the OC promoter does not require its intrinsic HAT activity, as a HAT-deficient p300 mutant up-regulates the OC gene promoter and cooperates with P/CAF to further enhance this promoter to the same extent that the wild-type p300 protein does. Together, these results indicate that p300 HAT activity may be dependent on the steroid receptor.
We have recently shown a functional linkage of histone H3 and H4 acetylation in the OC promoter to tissue-specific and vitamin D enhancement of OC gene expression (32). By using ChIP assays, we found that acetylated histones H3 and H4 are associated with the OC promoter only when the gene is transcriptionally active and that the acetylation status is relatively uniform across the OC locus under basal conditions. Interestingly, acetylation of H4 at the OC gene is selectively increased after vitamin D3 enhancement of OC gene transcription, with the most prominent changes occurring in the vicinity of the VDRE (32). It appears reasonable to suggest that p300 functions as a coactivator within the context of the OC gene promoter (although its intrinsic HAT activity is not required) and that other p300-associated proteins containing HAT activity are responsible for the increased histone H3 and H4 acetylation found in this promoter when the OC gene is transcriptionally active. Hence, we propose that p300 may have an architectural role at the OC gene promoter by interacting with key transcription regulators (e.g., Runx2/Cbfa1 and VDR upon ligand stimulation) and, at the same time, by connecting factors bound to distal and proximal promoter sequences which control basal tissue-specific and vitamin D3-enhanced OC gene transcription.
Acknowledgments
We thank Neil Perkins and the members of his laboratory (University of Dundee, Dundee, Scotland) for the p300 expression plasmid and for their generous help in performing the p300 immunoprecipitation experiments. We also thank W. L. Krauss (Cornell University) for the HAT-less p300 expression plasmid and Y. Nakatani (Harvard University) for the P/CAF expression vector. We thank José Gutiérrez and Fabián Arenas for helpful discussions and critically reading the manuscript.
This work was supported in part by grants from FONDECYT 1000361 (to M.M.) and 2000140 (to J.S.), NIH-FIRCA TW00990 (to G.S.S. and M.M.), DIUC 201.031.090-1.4 (to M.M.), and NIH AR48818 (to G.S.S.) and DE12528 (to J.B.L.). J.S. and A.V. were supported by MECESUP UCH9903.
The contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Aslam, F., L. McCabe, B. Frenkel, A. van Wijnen, G. S. Stein, J. B. Lian, and J. Stein. 1997. AP-1 and vitamin D receptor (VDR) signaling pathways converge at the rat osteocalcin VDR element: requirement for the internal activating protein-1 site for vitamin D-mediated trans-activation. Endocrinology 140:63-70. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee, C., S. Hiebert, J. Stein, J. Lian, and G. Stein. 1996. An AML-1 consensus sequence binds an osteoblast-specific complex and transcriptionally activates the osteocalcin gene. Proc. Natl. Acad. Sci. USA 93:4968-4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee, C., L. McCabe, J. Choi, S. Hiebert, J. Stein, G. Stein, and J. Lian. 1997. Runt homology domain proteins in osteoblast differentiation: AML3/CBFA1 is a major component of a bone-specific complex. J. Cell. Biochem. 66:1-8. [DOI] [PubMed] [Google Scholar]
- 4.Breen, E., A. van Wijnen, J. Lian, G. Stein, and J. Stein. 1994. In vivo occupancy of the vitamin D responsive element in the osteocalcin gene supports vitamin D-dependent transcriptional upregulation in intact cells. Proc. Natl. Acad. Sci. USA 91:12902-12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakravarti, D., V. J. La Morte, M. C. Nelson, T. Nakajima, I. G. Schulman, H. Juguilon, M. Montminy, and R. M. Evans. 1996. Role of CBP/p300 in nuclear receptor signalling. Nature (London) 383:9-13. [DOI] [PubMed] [Google Scholar]
- 6.Collingwood, T. W., F. D. Urnov, and A. Wolffe. 1999. Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J. Mol. Endocrinol. 23:255-275. [DOI] [PubMed] [Google Scholar]
- 7.Ducy, P., and G. Karsenty. 1995. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol. Cell. Biol. 15:1858-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ducy, P., R. Zhang, V. Geoffroy, A. Ridall, and G. Karsenty. 1997. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747-754. [DOI] [PubMed] [Google Scholar]
- 9.Goldman, R., and R. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 10.Gutierrez, S., A. Javed, D. K. Tennat, M. van Rees, M. Montecino, G. S. Stein, J. L. Stein, and J. B. Lian. 2002. CCAAT/enhancer-binding proteins (C/EBP) β and δ activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J. Biol. Chem. 277:1316-1323. [DOI] [PubMed] [Google Scholar]
- 11.Javed, A., S. Gutierrez, M. Montecino, A. van Wijnen, J. Stein, G. Stein, and J. Lian. 1999. Multiple Cbfa/AML sites in the rat osteocalcin promoter are required for basal and vitamin D-responsive transcription and contribute to chromatin organization. Mol. Cell. Biol. 19:7491-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamei, Y., L. Xu, T. Heinzel, J. Torchia, R. Kurokawa, B. Gloss, S. C. Lin, R. A. Heyman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1996. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85:403-414. [DOI] [PubMed] [Google Scholar]
- 13.Kitabayashi, I., A. Yokohama, K. Shimuzu, and M. Ohki. 1998. Interaction and functional cooperation of the leukemia-associated factor AML-1 and p300 in myeloid cell differentiation. EMBO J. 17:2994-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komori, T., H. Yagi, S. Nomura, A. Yamaguchi, K. Sasaki, K. Deguchi, Y. Shimizu, R. T. Bronson, Y. H. Gao, M. Inada, M. Sato, R. Okamoto, Y. Kitamura, S. Yoshiki, and T. Kishimoto. 1997. Targeted disruption of Cbfa1 results in complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755-764. [DOI] [PubMed] [Google Scholar]
- 15.Korzus, E., J. Torchia, D. W. Rose, L. Xu, R. Kurokawa, E. McInerney, R.-M. Mullen, C. K. Glass, and M. G. Rosenfeld. 1998. Transcription factor-specific requirements for co-activators and their acetyltransferase functions. Science 279:703-707. [DOI] [PubMed] [Google Scholar]
- 16.Krauss, L. W., E. T. Manning, and J. Kadonaga. 1999. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol. 19:8123-8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurokawa, R., D. Kalafus, M.-H. Ogliastro, C. Kioussi, L. Xu, J. Torchia, M. G. Rosenfeld, and C. K. Glass. 1998. Differential use of CREB binding protein-coactivator complexes. Science 279:700-703. [DOI] [PubMed] [Google Scholar]
- 18.Lemon, B., and L. Freedman. 1999. Nuclear receptor cofactors as chromatin remodelers. Curr. Opin. Gen. Dev. 9:499-504. [DOI] [PubMed] [Google Scholar]
- 19.Li, J., B. W. O'Malley, and J. Wong. 2000. p300 requires its acetyltransferase activity and SRC-1 interaction domain to facilitate thyroid hormone receptor activation in chromatin. Mol. Cell. Biol. 20:2031-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lian, J., G. Stein, J. Stein, and A. van Wijnen. 1999. Regulated expression of the bone-specific osteocalcin gene by vitamins and hormones. Vitam. Horm. 55:443-509. [DOI] [PubMed] [Google Scholar]
- 21.Majeska, R., S. Rodan, and G. Rodan. 1980. Parathyroid hormone responsive clonal lines from osteosarcoma. Endocrinology 107:1494-1503. [DOI] [PubMed] [Google Scholar]
- 22.Montecino, M., B. Frenkel, A. van Wijnen, J. Lian, G. Stein, and J. Stein. 1999. Chromatin hyperacetylation abrogates vitamin D-mediated transcriptional upregulation of the tissue-specific osteocalcin gene in vivo. Biochemistry 38:1338-1345. [DOI] [PubMed] [Google Scholar]
- 23.Montecino, M., J. Lian, G. Stein, and J. Stein. 1996. Changes in chromatin structure support constitutive and developmentally regulated transcription of the bone-specific osteocalcin gene in osteoblastic cells. Biochemistry 35:5093-5102. [DOI] [PubMed] [Google Scholar]
- 24.Montecino, M., S. Pockwinse, J. Lian, G. Stein, and J. Stein. 1994. DNase I hypersensitive sites in promoter elements associated with basal and vitamin D dependent transcription of the bone-specific osteocalcin gene. Biochemistry 33:348-353. [DOI] [PubMed] [Google Scholar]
- 25.Otto, F., A. P. Thronelkt, T. Crompton, A. Denzel, K. C. Gilmour, I. R. Rosewell, G. W. H. Stamp, R. S. P. Beddington, S. Nundlos, B. R. Olsen, P. B. Selby, and M. J. Owen. 1997. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome is essential for osteoblast differentiation and bone-development. Cell 89:765-771. [DOI] [PubMed] [Google Scholar]
- 26.Owen, T., M. Aronow, V. Shalhoub, L. Barone, L. M. Wilming, M. S. Tassinari, M. B. Kennedy, S. Pockwinse, J. B. Lian, and G. S. Stein. 1990. Progressive development of the rat phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. J. Cell. Physiol. 143:420-430. [DOI] [PubMed] [Google Scholar]
- 27.Paredes, R., J. Gutierrez, S. Gutierrez, L. Allisson, M. Puchi, M. Imschenetzky, A. van Wijnen, J. Lian, G. Stein, J. Stein, and M. Montecino. 2002. Interaction of the 1α,25-dihydroxyvitamin D3 receptor at the distal promoter region of the bone-specific osteocalcin gene requires nucleosomal remodeling. Biochem. J. 363:667-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rachez, C., B. Lemon, Z. Suldan, V. Bromleigh, M. Gamble, A. Naar, H. Erdjument-Bromage, P. Tempst, and L. Freedman. 1999. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature (London) 398:824-828. [DOI] [PubMed] [Google Scholar]
- 29.Rachez, C., Z. Suldan, J. Ward, C.-P. Chang, D. Burakov, H. Erdjument-Bromage, P. Tempst, and L. Freedman. 1998. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 12:1787-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rachez, C., M. Gamble, C.-P. B. Chang, G. B. Atkins, M. Lazar, and L. P. Freedman. 2000. DRIP complex and SRC-1/p160 coactivators share similar nuclear receptor binding determinants but constitute functionally distinct complexes. Mol. Cell. Biol. 20:2718-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rachez, C., and L. P. Freedman. 2000. Mechanisms of gene regulation by vitamin D3 receptor: a network of coactivator interactions. Gene 246:9-21. [DOI] [PubMed] [Google Scholar]
- 32.Shen, J., M. Montecino, J. B. Lian, G. S. Stein, A. van Wijnen, and J. Stein. 2002. Histone acetylation in vivo at the osteocalcin locus is functionally linked to vitamin D-dependent bone tissue-specific transcription. J. Biol. Chem. 277:20284-20292. [DOI] [PubMed] [Google Scholar]
- 33.Spencer, V. A., and J. Davie. 1999. Role of covalent modifications of histones in regulating gene expression. Gene 240:1-12. [DOI] [PubMed] [Google Scholar]
- 34.Yang, X. J., V. V. Ogryzco, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature (London) 382:319-324. [DOI] [PubMed] [Google Scholar]
- 35.Yao, T. P., S. P. Oh, M. Fuchs, N. D. Zhou, L. E. Ch'ng, D. Newsome, R. T. Bronson, E. Li, D. M. Livingston, and R. Eckner. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93:361-372. [DOI] [PubMed] [Google Scholar]