Abstract
Death inducer-obliterator 1 (DIO-1) is a gene that is upregulated early in apoptosis. Here we report that in healthy cells, the DIO-1 gene product was located in the cytoplasm, where it formed oligomers. After interleukin-3 starvation or c-Myc-induced apoptosis in serum-free conditions, DIO-1 translocated to the nucleus, where it upregulated caspase levels and activity. A nuclear localization signal deletion mutant (DIO-1ΔNLS) was unable to translocate to the nuclear compartment in the absence of interleukin-3 and failed to upregulate procaspase levels or trigger cell death. In addition, cells stably expressing DIO-1ΔNLS were protected from apoptosis induced by interleukin-3 withdrawal. These results indicate that DIO-1 has a relevant role in regulating the early stages of cell death.
Apoptosis, or programmed cell death, has a major role in normal development, tissue homeostasis, defense against viral invasion, immune modulation, and, when dysregulated, modulation of autoimmune and clonal or neoplastic diseases (15, 17, 30). Apoptosis is characterized by cell shrinkage, chromatin condensation, internucleosomal DNA cleavage, membrane blebbing, and the formation of apoptotic bodies that are phagocytosed by other cells (8, 38). These morphological changes are orchestrated by the activity of a family of aspartate-specific proteases called caspases (4, 7, 34).
Caspases are produced in cells as catalytically inactive zymogens, or procaspases, composed of three subunits, a prodomain and two catalytic subdomains, known as the large and small subunits (1). Procaspases must be proteolytically processed to become active proteases. An effector caspase, for example caspase 3, is activated by an initiator caspase, such as caspase 9, through proteolytic cleavage at specific internal Asp residues to give rise to the two subunits of the mature caspase. Once activated, the effector caspases cleave a broad spectrum of cellular targets, leading ultimately to cell death (38). Activation of the initiator caspases is in turn regulated by upstream protein complexes. In the case of the so-called “extrinsic” pathway, activation of death receptors such as Fas/CD95 and tumor necrosis factor receptor 1 after binding of their respective ligands induces recruitment of caspase 8 (FLICE) via the adapter molecule FADD (Fas-associated protein with death domain) (27). Caspase 8 can activate effector caspases either directly (3, 27) or indirectly by cleaving Bid and inducing the release of mitochondrial cytochrome c (18, 25).
In the case of the “intrinsic” death receptor-independent pathway, apoptotic cell death is induced directly by death stimuli and is also regulated by adapter complexes. This is the case of one of the major routes to caspase activation pathways, triggered by cytochrome c release from the mitochondrial intermembrane space into the cytosol (23). Cytosolic cytochrome c promotes assembly of a protein complex called the apoptosome, which includes caspase 9 bound to the CED-4 homolog Apaf-1 (19, 42), inducing autoactivation of procaspase 9 (33, 37). Following activation, caspase 9 cleaves and activates procaspase 3 (19, 42), giving rise to a proteolytic cascade involving multiple caspases.
DIO-1 was identified by a differential display approach (21) in WOL-1 pre-B cells induced to undergo apoptosis by interleukin-7 (IL-7) starvation (9). Its predicted amino acid sequence showed transcriptional activation domains, a canonical bipartite nuclear localization signal (NLS), a PhD finger, and a carboxy-terminal lysine-rich region. DIO-1 mRNA was upregulated soon after apoptotic induction by several stimuli, including removal of IL-7, addition of dexamethasone or gamma interferon in WOL-1 cells, immunoglobulin M (IgM) receptor cross-linking in WEHI-231 cells, or c-myc activation under serum-free conditions in the absence of p53 expression in MEF(10.1)Val5MycER cells. Overexpression of DIO-1 in cells or misexpression in chick limbs induced massive apoptosis in the absence of any apoptotic stimuli; this could be inhibited by Bcl-2 overexpression or incubation with the general caspase inhibitor benzyloxycarbonyl-Val-Ala-Asp-fluoromethyl ketone (z-VAD-fmk). These results suggested that DIO-1-induced apoptosis requires caspase activation. Furthermore, overexpression of a DIO-1 deletion mutant lacking both NLSs failed to induce cell death, linking its lack of lethality to an inability to translocate.
Here we studied the mechanism by which DIO-1 induces apoptosis and the importance of its subcellular localization. We generated several tagged constructs and analyzed the subcellular distribution pattern of both wild-type and mutant DIO-1 in various apoptotic situations, revealing nuclear translocation as the main regulatory event in the DIO-1-activated apoptotic pathway. We provide evidence that DIO-1 translocation boosts the apoptotic machinery by upregulating protein levels of procaspase 3 and 9, which enhances their apoptosis-inducing activity.
MATERIALS AND METHODS
Expression plasmids.
DIO-1ΔNLSpcDNA3 was generated from DIO-1 cloned in pcDNA3 by reverse PCR with the circular plasmid as the template and the primers 5′-GAAGATTCTGCCGAAACTGGG-3′ and 5′-AAGTTCCTTCAACGTAAGG-3′, which span the NLSs and the connecting sequence; the PCR product was permitted to further self-ligate. Both N-terminally Flag-tagged DIO-1 and DIO-1ΔNLS were PCR amplified from their corresponding pcDNA3 constructs with a proof-reading polymerase and the primers 5′-GCGGATCCGATGATAAAGGGCACCTG-3′ and 5′-CGACCTCGAGTTACCAAGGCCTAAAACTG-3′ for in-frame reading. The PCR products were digested with BamHI and XhoI and subcloned into the pCMV-Tag 2B vector (Stratagene). DIO-1/Myc was generated similarly after PCR amplification with the primers 5′-TGGAATTCCACCATGGATGATAAAGGGCAC-3′ and 5′-GCTCTAGACCAAGGCCTAAAACTG-3′ from DIO-1pcDNA3, digested with EcoRI and XbaI, and subcloned into the pEF4/Myc-His A vector (Invitrogen). All constructs were verified by nucleotide sequencing.
Cell culture.
FL5.12 and MEF(10.1)Val5MycER cells were cultured as described previously (9). 293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and antibiotics.
Antibodies and reagents.
Production of the polyclonal antibody against murine DIO-1 amino acids 58 to 72 has been described (9). Polyclonal rabbit anti-mouse caspase 3 was the kind gift of T. Mak and R. Hakem (Ontario Cancer Institute, Toronto, Canada); anti-caspase 9 was the kind gift of D. R. Green and B. Wolf (39). Anti-Flag M2 monoclonal antibody (Sigma), anti-Myc 9E10 monoclonal antibody (Santa Cruz Biotechnologies), protein A-agarose (Sigma), protein G-agarose (Sigma), antiphosphoserine sampler kit (Biomol), antiphosphoserine monoclonal antibody (Calbiochem), and antiphosphothreonine monoclonal antibody (Calbiochem) were purchased as indicated. z-VAD-fmk was purchased from Bachem.
Establishment of FL5.12 cells stably overexpressing DIO-1ΔNLS.
FL5.12 cells (3 × 106) were electroporated with 10 μg of DIO-1ΔNLS construct or pcDNA3. Transfected cells were resuspended in 48 ml of complete medium and distributed in a 48-well plate. Selection was performed after 24 h by removing the medium and adding fresh complete medium containing 1 mg of G418 per ml. Selection was carried out for 30 days, and DIO-1ΔNLS expression was determined by reverse transcription-PCR and Western blotting.
Subcellular fractionation.
FL5.12 cells (2 × 107) were harvested and washed in ice-cold phosphate-buffered saline (PBS), and pellets were resuspended in 5 volumes of ice-cold buffer A (10 mM HEPES [pH 8.0], 0.5 M sucrose, 1 mM EDTA, 0.5 mM spermidine, 0.15 mM spermine, 15 mM KCl, 0.5 mM dithiothreitol, 10 μg of aprotinin per ml, 10 μg of pepstatin A per ml, 10 μg of leupeptin per ml, 1 mM phenylmethylsulfonyl fluoride [PMSF], 100 μM Na3VO4, and 10 mM NaF). After incubation on ice for 15 min, cells were lysed by three freeze-thaw cycles and centrifuged (1,000 × g, 10 min, 4°C), and the nuclear pellet was resuspended in buffer B (10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) [PIPES, pH 7.4], 80 mM KCl, 20 mM NaCl, 5 mM sodium EGTA, 250 mM sucrose, 1 mM dithiothreitol, 10 μg of aprotinin per ml, 10 μg of pepstatin A per ml, 10 μg of leupeptin per ml, 1 mM PMSF, 100 μM Na3VO4, and 10 mM NaF) at 8.5 × 107 nuclei/ml. Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions. Purity of the cytosolic and nuclear fractions was tested by Western blotting with compartment-specific antibodies to procyclic acidic repetitive protein and calpain-1 (not shown).
Transfections and immunoprecipitation.
Transient transfections in FL5.12 and MEF(10.1)Val5MycERcells were performed by electroporation as described previously (9). 293T cells were transiently transfected in six-well plates with Lipofectamine Plus (Life Technologies) following the manufacturer's protocol. Cytosolic extracts for immunoprecipitation were obtained after cell lysis in NP-40 buffer (40 mM Tris-HCl [pH 8], 500 mM NaCl, 0.1% NP-40, 6 mM EDTA, 6 mM EGTA, 10 μg of aprotinin per ml, 10 μg of pepstatin A per ml, 10 μg of leupeptin per ml, 1 mM PMSF, 100 μM Na3VO4, and 10 mM NaF) or a digitonin-containing buffer (10 mM triethanolamine [pH 8], 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% digitonin, 10 μg of aprotinin per ml, 10 μg of pepstatin A per ml, 10 μg of leupeptin per ml, 1 mM PMSF, 100 μM Na3VO4, and 10 mM NaF). Immunoprecipitation was performed by preclearing lysates with 30 μl of protein A/G-agarose (1 h, 4°C, with gentle rotation), followed by incubation with the appropriate antibody (10 μg/ml; 1 h, 4°C). After addition of protein A- or protein G-agarose (30 μl), beads were pelleted by brief centrifugation and washed three times in 50 mM Tris-HCl (pH 7.6) buffer, and the pellet was boiled in loading buffer and analyzed in Western blot.
Phosphatase treatment.
293T cells transiently transfected with Flag-tagged DIO-1 were harvested, washed in ice-cold PBS, and lysed in a digitonin-containing buffer (40 mM Tris-HCl [pH 8], 50 mM NaCl, 2 mM MnCl2, 1% digitonin, 10 μg of aprotinin per ml, 10 μg of pepstatin A per ml, 10 μg of leupeptin per ml, 1 mM PMSF). After removal of cellular debris by centrifugation, lysates were treated with 500 U of λ-phosphatase (Calbiochem) (30 min, 30°C). The reaction was terminated by addition of loading buffer and then analyzed in Western blot.
Western blot analysis.
Cells for analysis of procaspase expression were washed with PBS, and the pellet was suspended in lysis buffer (137 mM NaCl, 20 mM Tris-HCl [pH 8], 1 mM MgCl2, 1 mM CaCl2, 10% glycerol, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 10 μg of aprotinin per ml, 10 μg of pepstatin A per ml, 10 μg of leupeptin per ml, and 1 mM PMSF) for 30 min on ice. The protein content of the lysates was quantified with the Bio-Rad DC protein assay (Bio-Rad); after SDS-PAGE, proteins were transferred to nitrocellulose membranes (Bio-Rad). Equal protein loading was verified by Ponceau Red (Sigma) staining. Membranes were blocked overnight with 5% nonfat dry milk in TBS buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl); subsequent incubations and membrane washes were performed in TBS-T buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 0.2% Tween 20) containing 1% nonfat dry milk. After 2 h of antibody incubation and 1 h of washing, blots were developed with peroxidase-conjugated anti-rabbit or anti-mouse immunoglobulin antibodies (Dako), and proteins were detected by an enhanced chemiluminescence system (ECL; Amersham).
Fluorescence microscopy.
For immunofluorescence, cells were cultured on coverslips, washed in PBS, fixed in 4% paraformaldehyde (15 min, room temperature), washed three times in PBT (PBS with 0.1% Tween 20), incubated in 2% bovine serum albumin, and incubated for 1 h with anti-DIO-1 (1:100) or anti-Flag M2 (1:500) in PBT. After incubation, cells were washed three times in the same buffer and incubated for 1 h with indocarbocyanine-conjugated secondary antibodies (Jackson ImmunoResearch). After washing, samples were incubated with Sybr Green (Molecular Probes) in PBS for DNA staining. Serial Z-sections were obtained with an Ar-Kr laser and a TCS-NT Leica confocal imaging system.
Enzyme assay for caspase activity.
Cells were collected, washed with ice-cold PBS, and resuspended in extraction buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 0.5 mM EDTA, 10 mM NaH2PO4, 10 mM Na2HPO4, 1% Nonidet P-40, 0.4 mM Na3VO4, 1 mM PMSF, 10 μg of aprotinin per ml, 10 μg of pepstatin A per ml, 10 μg of leupeptin per ml). After incubation (30 min, on ice), the cell lysate was centrifuged (20,000 × g, 30 min), and the supernatant was used as the cytosolic extract. Five micrograms of cytosolic proteins, estimated by the bicinchoninic acid method (36), were diluted fivefold in assay buffer (25 mM HEPES [pH 7.5], 0.1% CHAPS, 10% sucrose, 10 mM dithiothreitol, and 0.1 mg of ovalbumin per ml) and incubated (1 h, 37°C) with 10 μM of the fluorescent substrate Ac-DEHD-AMC (acetyl-Asp-Glu-His-Asp-7-amino-4-methylcoumarin), Ac-DEVD-AMC (acetyl-Asp-Glu-Val-Asp-7-amino-4-methylcoumarin), Ac-VEID-AMC (acetyl-Val-Glu-Ile-Asp-7-amino-4-methylcoumarin), or Ac-LEHD-AMC (acetyl-Leu-Glu-His-Asp-7-amino-4-methylcoumarin) to measure caspase 2, caspase 3-like, caspase 6, and caspase 9 activity, respectively. The reaction was terminated by addition of high-pressure liquid chromatography (HPLC) buffer (water-acetonitrile [75:25], 0.1% trifluoroacetic acid). Cleaved substrate fluorescence was determined by C18 reverse-phase HPLC with fluorescence detection (338 nm excitation, 455 nm emission). Control experiments confirmed linearity with time and protein concentration of substrate release.
Apoptosis assay.
Apoptosis was evaluated by staining cellular DNA content with the DNA intercalator propidium iodide in a semiautomatic procedure (DNA-Prep Reagents; Coulter), followed by analysis on an Epics XL flow cytometer (Coulter). Briefly, cells (105 to 106) were recovered by centrifugation, resuspended in 100 μl of PBS, permeabilized, and stained by adding 100 μl of detergent reagent followed by 1 ml of propidium iodide solution. After mixing, samples were incubated (37°C, 1 h) and analyzed by flow cytometry.
RESULTS
DIO-1 nuclear translocation following apoptotic stimulation requires the NLS.
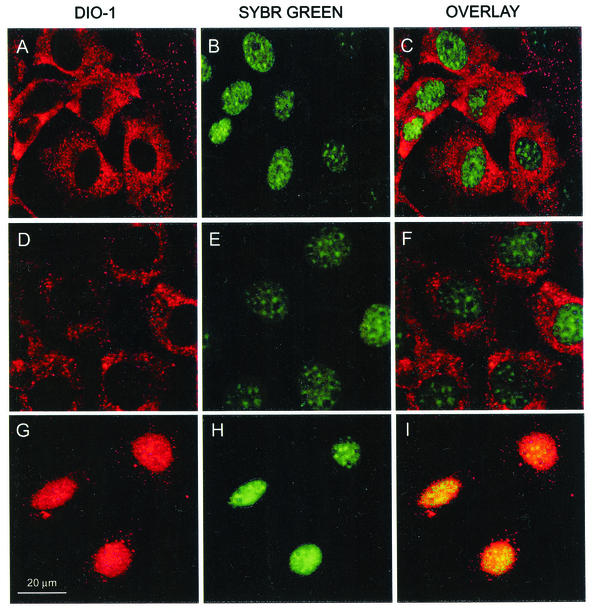
We previously reported differences in DIO-1 mRNA levels after induction of p53-dependent and -independent apoptosis in MEF(10.1)Val5MycER cells (9). DIO-1 transcripts were upregulated in apoptotic processes in a p53-independent fashion. The presence of two putative NLSs in the DIO-1 gene product suggested that it may be localized in the nucleus (9). To study its putative role as a transcription factor, we examined the subcellular localization pattern of the DIO-1 gene product and its NLS deletion mutant (DIO-1ΔNLS). Immunofluorescence microscopy of healthy MEF(10.1)Val5MycER cells with an affinity-purified rabbit antiserum specific for a DIO-1 peptide (9) indicated a clear cytoplasmic pattern for the DIO-1 protein, which was nearly absent in the nucleus (Fig. 1A to C). p53-mediated triggering of apoptosis showed no change in the DIO-1 localization pattern (Fig. 1D to F), although the cells underwent apoptosis (not shown). Cells cultured at 39°C to inactivate p53, then induced to apoptosis by 17β-estradiol addition and fetal bovine serum starvation, showed DIO-1 translocation from cytoplasm to the nucleus (Fig. 1G to I). This was observed before cell death was detectable by any method and in parallel to mRNA upregulation kinetics.
FIG. 1.
Immunolocalization of endogenous DIO-1 under apoptotic conditions. (A to C) Viable (nonapoptotic) MEF(10.1)Val5MycER cells were stained with anti-DIO-1 antibody (red); nuclei were stained with Sybr Green (green). (D to F) The same cells were induced to apoptosis by lowering the incubation temperature (32°C, 12 h) to activate p53 in the presence of 17β-estradiol (1 μM). Note the clear cytoplasmic pattern of DIO-1, although some cells have already begun the apoptotic program. (G to I) The same cell line, incubated at 39°C to inactivate p53, was 17β-estradiol treated (1 μM) and serum starved for 8 h, the time at which DIO-1 mRNA is upregulated. Note nuclear translocation of DIO-1 to the intact, nonapoptotic nuclei.
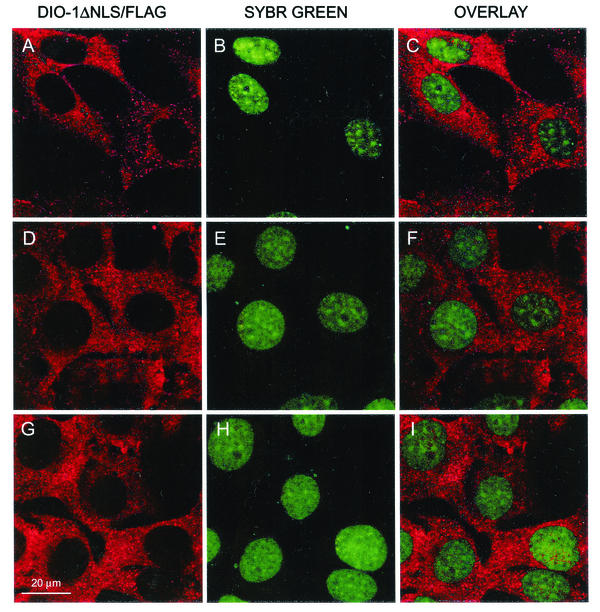
We examined the subcellular localization of the DIO-1ΔNLS mutant, but as the anti-DIO-1 antibody recognizes both wild-type and mutant proteins, the mutant was Flag tagged (DIO-1ΔNLS/Flag) and transiently transfected into MEF(10.1)Val5MycER cells. Apoptosis was induced 24 h posttransfection, and staining was performed at the same time points as for wild-type DIO-1. A clear cytoplasmic pattern was observed in healthy cells (Fig. 2A to C) and in those undergoing p53-induced apoptosis (Fig. 2D to F). DIO-1ΔNLS was unable to translocate to the nucleus under conditions that promoted translocation of the wild-type form (Fig. 2G to I), as predicted by its lack of NLS.
FIG. 2.
Immunolocalization of DIO-1ΔNLS in MEF(10.1)Val5MycER cells under apoptotic conditions. A DIO-1ΔNLS Flag-tagged construct was transiently transfected into the cells, which were analyzed 36 h posttransfection. A selected field of positive cells is shown. (A to C) Viable cells were stained with anti-Flag antibody (red) and Sybr Green (green). (D to F) Conditions as for Fig. 1D to F. (G to I) Conditions as for Fig. 1G to I. The protein did not translocate to the nucleus in the presence of the stimulus that induced wild-type DIO-1 translocation.
DIO-1 forms oligomers.
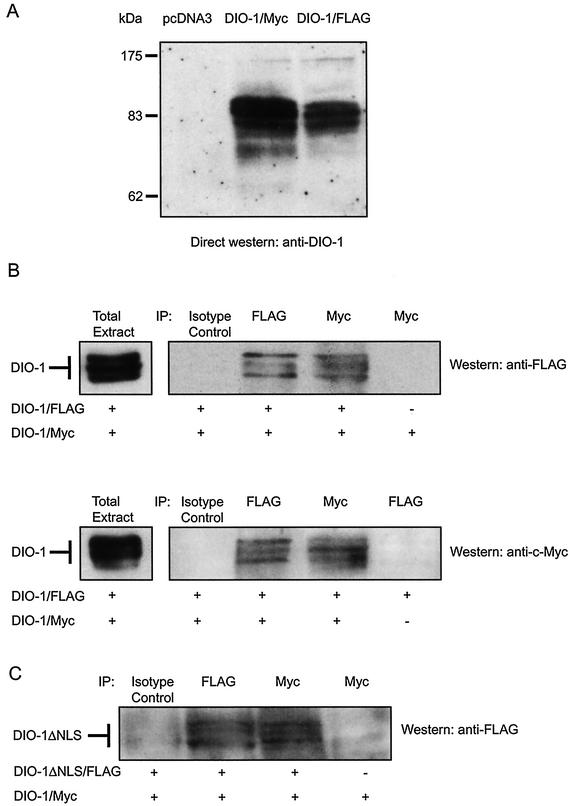
To test for the association state of DIO-1, we fused murine DIO-1 to N-terminal Flag (DIO-1/Flag) and C-terminal Myc (DIO-1/Myc) tags in different constructs. The constructs were coexpressed by transient transfection into human embryonic kidney 293T cells, and proteins were isolated under nondenaturing conditions with buffers that permitted cytosolic extraction. Western blot analysis with the anti-DIO-1 antibody (Fig. 3A) and antitag antibodies (Fig. 3B) confirmed expression in total lysates of cells transfected with the appropriate plasmids, but not in cells transfected with control plasmids. In all cases, a triplet was observed with very close bands. The triplets do not correspond to proteolytic degradation products, as they were detected equally well with antibodies to tags located at both ends of the protein, in the presence of large amounts of protease inhibitors. These multiple bands are more likely to represent posttranslational modifications of DIO-1.
FIG. 3.
DIO-1 forms oligomers. (A) Expression of DIO-1 gives rise to a three-band pattern in Western blot. Whole-cell extracts from 293T cells transfected with vector plasmid (pcDNA3) or expression plasmids encoding Myc- or Flag-tagged DIO-1 were analyzed in Western blot with the anti-DIO-1 antibody. Molecular size markers are indicated at the left. (B) 293T cells were cotransfected with constructs encoding Myc- and Flag-tagged DIO-1 except for the last lane, in which only one construct was transfected as a negative immunoprecipitation control. After 48 h, extracts were immunoprecipitated with anti-Flag or anti-Myc monoclonal antibody. An irrelevant isotype-matched antibody was used as a control. Immunoprecipitates were analyzed by SDS-PAGE and blotted with anti-Flag (upper panel) or anti-Myc antibody (lower panel). Total cell extracts were also analyzed by SDS-PAGE and blotted with the same antibodies (left). (C) 293T cells were cotransfected with constructs encoding Myc-tagged DIO-1 and Flag-tagged DIO-1ΔNLS except for the last lane, in which only DIO-1/Myc was transfected. The experimental procedure was identical to that for panel B, upper panel. All results are representative of three independent experiments.
Oligomerization was determined in an immunoprecipitation assay performed with anti-Myc and resolved in SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Subsequent Western blotting with an anti-Flag antibody showed that DIO-1/Flag coimmunoprecipitated with DIO-1/Myc, whereas negative controls gave no bands (Fig. 3B). The amount of coprecipitated material was distributed equally among the three bands observed. To verify this interaction, we performed reciprocal experiments with anti-Flag to immunoprecipitate DIO-1/Flag, followed by blotting with anti-Myc. Concurring with the reverse experiment, DIO-1/Myc coimmunoprecipitated specifically with DIO-1/Flag (Fig. 3B). Similar experiments with DIO-1/Myc and DIO-1ΔNLS/Flag showed that both constructs coimmunoprecipitated (Fig. 3C), as confirmed by the reciprocal experiment (not shown). These results strongly suggest that DIO-1 homo-oligomerizes and that deletion of both NLS regions and their interspace does not affect this interaction. In addition, the C-terminal end of the protein is not needed for oligomerization, as glutathione S-transferase fusions lacking the terminal 86 amino acids interacted with endogenous DIO-1 (not shown). The exact oligomerization site nonetheless remains to be determined. Based on computer predictions, the lysine-rich region, which resembles part of the c-myc dimerization site, may contribute to DIO-1 oligomerization; experiments are under way to analyze this possibility.
DIO-1 is present in multiple forms with distinct subcellular localizations.
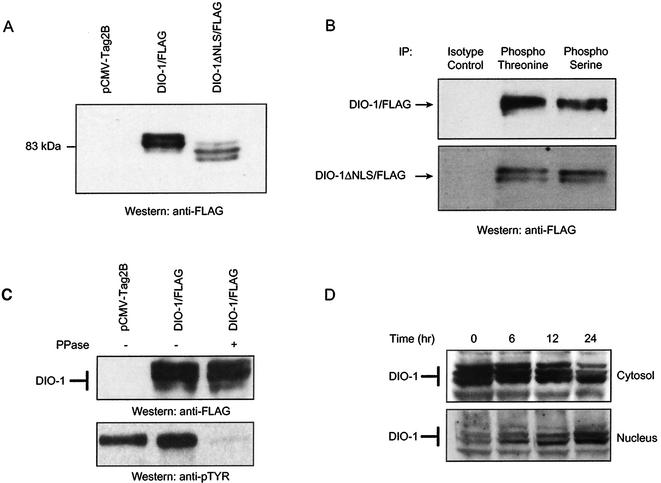
Triplet banding in Western blot, rather than a single band, may be produced by different protein phosphorylation states. To study this, Flag-tagged DIO-1 and DIO-1ΔNLS were transiently transfected in 293T cells and immunoprecipitated with antiphosphoserine, antiphosphothreonine, and antityrosine antibodies. Expression of both constructs showed the predicted three-band pattern (Fig. 4A), although only the two upper bands immunoprecipitated with antiphosphoserine and antiphosphothreonine (Fig. 4B). The lowest band was absent, suggesting that it corresponds to a protein that is not phosphorylated on serine/threonine, whereas the two upper bands appeared to correspond to phosphorylated forms. Antiphosphotyrosine antibodies did not immunoprecipitate DIO-1 (not shown). Phosphatase treatment of cell extracts did not result in the loss of the multiple bands (Fig. 4C), indicating that the mobility shift was not caused by phosphorylation alone. These results nonetheless show that DIO-1 is present in multiple forms that differ in electrophoretic mobility. Of these, the larger forms are phosphorylated on serine/threonine residues.
FIG. 4.
DIO-1 is present in several forms with distinct subcellular distribution. (A) Western blot of lysates from 293T cells transfected with empty vector (pCMV-Tag2B), Flag-tagged DIO-1, or Flag-tagged DIO-1ΔNLS. The membrane was blotted with anti-Flag antibody. A representative result of five independent experiments is shown. Note the lower apparent molecular size of the deletion mutant and the three-band pattern. (B) 293T cells were transiently transfected with the indicated Flag-tagged constructs. Lysates were immunoprecipitated with antiphosphothreonine, antiphosphoserine, or an irrelevant control antibody and then blotted with anti-Flag monoclonal antibody. Only the upper and middle bands were detected. (C) 293T cells were transiently transfected with Flag-tagged DIO-1. Untreated or λ-phosphatase-treated lysates were separated by SDS-PAGE and then blotted with anti-Flag monoclonal antibody. Phosphatase treatment did not cause a mobility shift (upper panel). Dephosphorylation of total cellular proteins was verified by blotting with antiphosphotyrosine antibody (lower panel). (D) FL5.12 cells deprived of IL-3 for the times indicated were separated into cytosolic and nuclear fractions (Materials and Methods). Extracts were separated by SDS-PAGE and analyzed by Western blotting with the anti-DIO-1 antibody.
To determine the role of the different DIO-1 forms in apoptosis, we examined their subcellular distribution and appearance under apoptotic conditions. Apoptosis was induced in FL5.12 cells by IL-3 withdrawal. Samples were taken at several time points, and the cytosolic and nuclear fractions were separated by centrifugation, resolved in SDS-PAGE, and analyzed in Western blots with the anti-DIO-1 antibody (Fig. 4D). Healthy cells (lanes labeled 0 h) showed higher DIO-1 protein levels in the cytosol than in the nucleus, as observed in the confocal images of MEF(10.1)Val5MycER cells; the three bands were present in the cytosolic fraction, whereas the upper band was nearly absent in the nucleus. With time, more cells entered apoptosis, and a progressive decrease was observed in the upper and middle cytosolic bands. At the same time, the nuclear fraction showed a clear increase in the lower band and a decrease in the middle band. These results suggest that the cytosolic form of DIO-1 is phosphorylated on serine/threonine and is predominant in healthy cells, whereas the unphosphorylated nuclear form is found under apoptotic conditions.
DIO-1 overexpression upregulates procaspase levels, leading to increased caspase activity.
As discussed above, several lines of evidence suggest that nuclear translocation of DIO-1 is a step that activates the apoptotic machinery. Although the exact details of the programmed cell death pathways remain to be fully determined, the essential role of caspases at various stages of the apoptotic process has been established (3, 4, 38). The majority of apoptotic stimuli that signal through a pathway engage the common cell death machinery at the caspase level. We thus explored whether DIO-1 is also associated with caspase activation by examining procaspase levels and activity after transient expression of DIO-1 in FL5.12 cells.
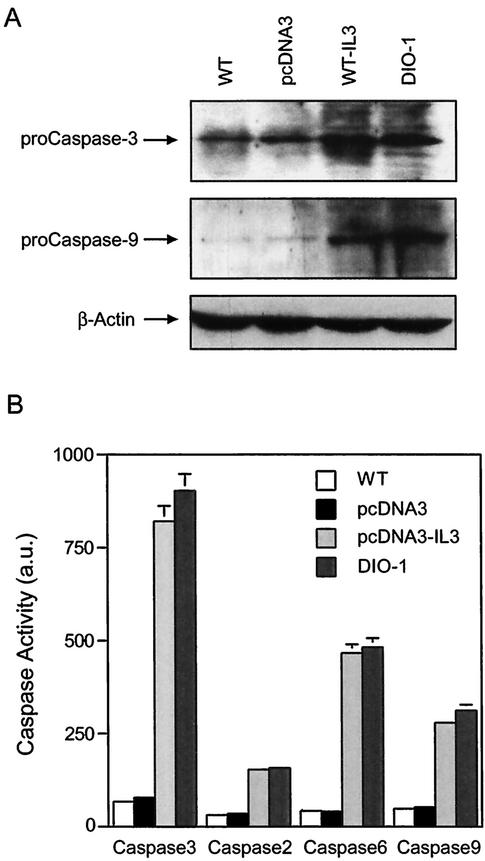
Cells were initially treated with z-VAD-fmk to block proteolytic processing of the procaspases, facilitating accurate protein measurement in Western blot. Procaspases 3 and 9 were upregulated in wild-type cells induced to undergo apoptosis by IL-3 starvation. Similarly, DIO-1-transfected cells showed an increase in both proteins in the presence of IL-3, whereas transfection of the empty vector had no effect (Fig. 5A). In a fluorescence assay, protein lysates from cells not treated with caspase inhibitors were assayed for caspase activity. The previously observed increase in procaspase levels correlated with mature caspase activity (Fig. 5B). Caspase 3 showed an increase in activity following IL-3 starvation or DIO-1 expression compared to wild-type or mock-transfected cells. Caspases 6 and 9 were also activated, whereas caspase 2 showed lower activation levels. Caspase 8 was not implicated in IL-3-induced apoptosis and showed no variation after DIO-1 overexpression (not shown).
FIG. 5.
DIO-1 upregulates and increases activation of caspases. (A) DIO-1 overexpression upregulates procaspase levels. FL5.12 cells were cultured in the presence of IL-3 (WT) or IL-3-starved for 24 h (WT-IL3). A plasmid vector (pcDNA3) or a DIO-1-expressing construct (DIO-1) were transiently transfected into FL5.12 cells and analyzed 24 h posttransfection. All cells were incubated with z-VAD-fmk (100 μM) to block proteolytic processing of the procaspase, facilitating accurate quantitation of protein levels. Procaspase 3 and 9 levels were analyzed in Western blots with anti-caspase 3 (upper panel) or anti-caspase 9 antibodies (middle panel). Loading was controlled with β-actin (lower panel). (B) Conditions as for panel A, but apoptosis was induced by IL-3 removal in pcDNA3-transiently transfected FL5.12 cells. Cells were cultured in the absence of z-VAD-fmk to measure caspase activity. Caspase 3-like, caspase 2, caspase 6, and caspase 9 activities were determined by fluorescence emission of the cleaved substrates (Materials and Methods). Data are given as the mean ± standard deviation for at least three independent experiments.
DIO-1ΔNLS is a dominant negative mutant that protects cells from apoptosis.
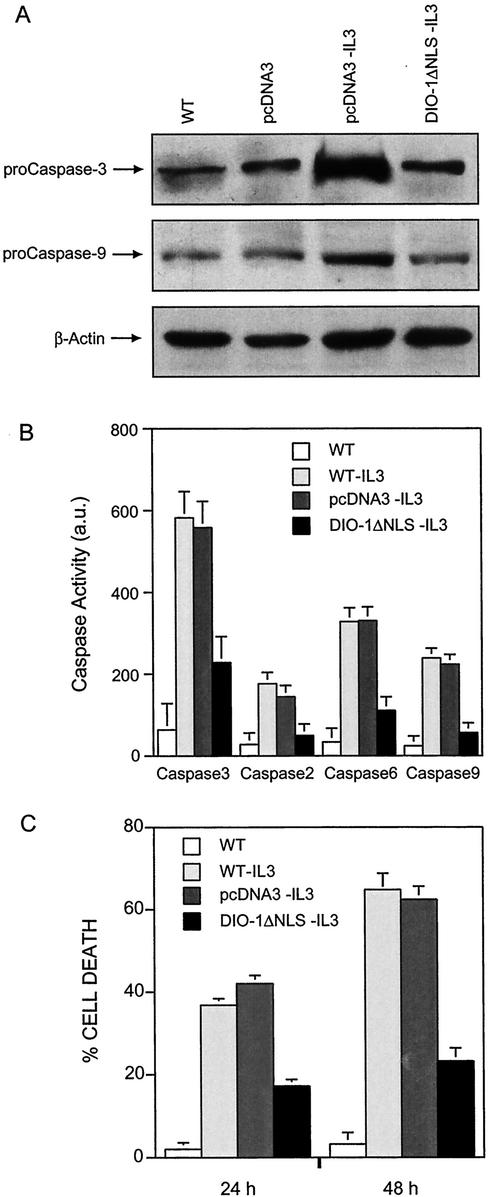
To confirm that DIO-1 upregulates procaspases after its translocation to the nucleus, we performed similar experiments with the NLS deletion mutant, predicting failure to upregulate caspases due to its inability to translocate to the nucleus. We generated FL5.12 cells stably expressing DIO-1ΔNLS as well as a control bearing the empty vector. IL-3-starved empty vector cells showed procaspase 3 and 9 upregulation, whereas IL-3-starved cells expressing DIO-1ΔNLS did not upregulate procaspase levels (Fig. 6A). In addition, caspase activity in IL-3-starved DIO-1ΔNLS-expressing cells was clearly below the levels observed in DIO-1-transfected cells (Fig. 6B; compare to Fig. 5B). These low activity levels remained constant even after 48 h of IL-3 deprivation, when more than 60% of wild-type cells were apoptotic (not shown). The mutation thus failed to alter basal caspase levels. Taken together, these data suggest that DIO-1 translocation to the nucleus is essential to induce upregulation of several procaspases, to increase their activity, and to trigger the apoptotic pathway.
FIG. 6.
DIO-1ΔNLS mutant is unable to upregulate and activate caspases. (A) FL5.12 cells were untreated (WT) or stably transfected with an empty vector (pcDNA3) or a DIO-1ΔNLS-expressing construct. Where indicated, IL-3 was removed from culture medium, and analysis was performed after 24 h. All cells were cultured in the presence of z-VAD-fmk (100 μM). Lysates were resolved by SDS-PAGE, followed by Western blotting with anti-caspase 3 (upper panel) or anti-caspase 9 (middle panel) antibodies. Loading was controlled with β-actin (lower panel). (B) Analysis of caspase activation. Samples were collected from FL5.12 wild-type (WT) cells cultured in the presence or absence of IL-3 and from the indicated stable FL5.12 cell lines in the absence of IL-3. Caspase activity was measured as in Fig. 5B. Results are expressed as the mean ± standard deviation for at least three independent experiments. a.u., arbitrary units. (C) DIO-1ΔNLS is a dominant negative mutant that protects cells from growth factor deprivation-induced apoptosis. FL5.12 wild-type cells (WT) and the FL5.12pcDNA3, and FL5.12DIO-1ΔNLS stable cell lines were induced to undergo apoptosis by IL-3 removal. Cells were collected at the times indicated, permeabilized, and propidium iodide stained (Materials and Methods). Apoptosis corresponds to the amount of fragmented DNA in the hypoploid sub-G0/G1 peak of the cell cycle. Values are expressed as percentages and represent the mean ± standard deviation for at least three independent experiments.
Given that the NLS mutation does not alter caspase levels, we tested whether it prevents apoptosis. FL5.12 wild-type cells were IL-3 starved, and apoptosis was measured by propidium iodide staining at 24 and 48 h postinduction (Fig. 6C). The cells underwent apoptotic cell death that increased with time, and similar results were obtained in FL5.12 pcDNA3 cells after IL-3 withdrawal. When FL5.12 cells stably expressing DIO-1ΔNLS (FL5.12 DIO-1ΔNLS) were IL-3 starved, we observed protection from apoptosis even after 48 h without the survival factor, with a reduction in the percentage of apoptotic cells of approximately 50% at 24 h and 66% at 48 h compared to cells expressing the empty vector. This behavior corresponds to a dominant negative mutation that is able to block the apoptotic signal transmitted by the wild-type form.
DISCUSSION
We report that DIO-1 translocation from the cytoplasm to the nucleus is an important early event in activating the apoptotic machinery. DIO-1 is present in the cell in multiple forms, which can be distinguished on the basis of their electrophoretic mobilities and their distinct subcellular localizations. We previously showed that DIO-1 mRNA and protein levels were upregulated by serum withdrawal from MEF(10.1)Val5MycER cells independently of p53-induced apoptosis (9). Here we show that, in the same cell system and with the same apoptotic stimulus, the DIO-1 gene product translocated to the nuclear compartment, whereas p53-induced cell death did not provoke alterations in its subcellular localization. The DIO-1 mutant lacking both NLSs (DIO-1ΔNLS) was unable to translocate to the nucleus or to trigger apoptosis, even in the presence of physiological death signals, and prevented IL-3 deprivation-induced cell death. The observation that DIO-1 but not DIO-1ΔNLS overexpression upregulated procaspase 3 and 9 levels and increased the activity of their mature forms establishes a link between the increase in DIO-1 levels and apoptosis induction (9). Although transcriptional activation by DIO-1 remains to be demonstrated, the need for nuclear translocation suggests that DIO-1 acts through the induction of caspase promoters rather than exerting a direct effect on cytoplasmic caspases. These findings are consistent with earlier studies reporting that transcriptional activation of caspases may be an important regulatory mechanism in programmed cell death (6, 12, 22, 29, 40).
One important model for caspase 9 activation involves its recruitment to Apaf-1 in the presence of ATP or dATP, following cytochrome c release from mitochondria (19, 42). Other authors propose that caspase 9 is activated and apoptosis can take place in the absence of cytochrome c (5, 10, 20, 32) after overexpression of the adaptor molecule Apaf-1 (16, 28), Nod-1 (14), TMS-1 (26), or CED-4 (35, 41). Furthermore, Apaf-1 mutants lacking the WD-40 repeats (WDR) constitutively self-associate and activate procaspase 9 independently of cytochrome c and dATP (13, 37). The Caenorhabditis elegans Apaf-1 homolog CED-4 lacks the WDR, implying that it may constitutively activate the procaspase 9 homolog CED-3 and suggesting that cytochrome c may not be required for CED-4-mediated CED-3 activation.
In our system, apoptosis induction via a DIO-1-triggered increase in procaspase 9 levels concurs with previous reports that procaspase 9 overexpression in vivo results in cell death, overriding the need for cytosolic cytochrome c (2, 14, 37). We observed that in MEF(10.1)Val5MycER and FL5.12 cells induced to apoptosis as well as in DIO-1-transfected cells, cytochrome c release was preceded by DIO-1 nuclear translocation and caspase upregulation (not shown). Caspase 9 activation is thus mediated by dimerization, and cytochrome c-induced recruitment by Apaf-1 creates high local caspase 9 concentrations that allow dimer-induced activation (31). This explains the caspase 9 activation observed prior to cytochrome c release from mitochondria.
Change in subcellular distribution is an important regulatory event for many proteins involved in apoptosis, cell cycle, or transcriptional regulation. Here we show that DIO-1 also changes its localization early in apoptosis induction and that this change is accompanied by an electrophoretic mobility shift. Nonetheless, the exact nature of the signal leading to these changes remains to be established. Coprecipitation of differentially tagged proteins showed that DIO-1 forms homo-oligomers in vivo. As homo-oligomers were detected for all forms, the oligomerization state is unlikely to differ following DIO-1 translocation to the nucleus. This has also been found for p53, which appears to be transported across the nuclear membrane in a tetrameric form (11). Proteolytic processing can also be ruled out as a regulatory mechanism, since all forms were detected with tags on either end of the protein. Although only the forms with lower electrophoretic mobility appeared to be phosphorylated, phosphorylation alone is not the basis of the mobility change. Additional modifications may thus be needed for the change in DIO-1 localization. In a similar case, MDM-2-mediated nuclear export of p53 depends on multiple signals that include the DNA-binding domain, conformational change, and C-terminal ubiquitination (11, 24); ubiquitinated p53 is subsequently degraded by proteasomes in the cytosol. DIO-1 localization may be regulated in a similar fashion. DIO-1 would thus be continuously modified and exported from the nucleus in healthy cells, appearing as cytosolic localization. In apoptotic cells, loss of the export signal would result in a net accumulation of the unmodified product in the nucleus.
DIO-1-mediated apoptosis requires caspase activation.
In these experiments, neither procaspase upregulation nor caspase activation was detected after DIO-1ΔNLS overexpression, indicating that this mutant acts in a dominant negative manner, since it inhibits IL-3 withdrawal-induced apoptosis. This allows us to propose a model that explains the mechanism of DIO-1 activation and induction of cell death. In healthy cells, the DIO-1 transcript and protein are expressed at low basal levels, and the protein is found in the cytosol. Following an appropriate apoptotic stimulus, such as IL-3 starvation or c-myc induction in serum-free conditions, DIO-1 translocates to the nucleus before the appearance of any classical indicators of apoptotic machinery activation, such as caspase activation, alteration of cell membrane polarity, nuclear disruption, or DNA laddering. This indicates that DIO-1 activation is a very early step in apoptosis induction.
In our model, the presence of DIO-1 in the nucleus leads to an increase in procaspase 3 and 9 levels, resulting in caspase 9 activation. Activated caspase 9 can then activate procaspase 3. The processing of the large amounts of procaspase 3 gives rise to rapid accumulation of mature caspase 3, which acts as another amplification step due to caspase 3 feedback onto procaspase 9 Asp-330 (37). Subsequent cytochrome c release from mitochondria would induce Apaf-1 oligomerization, amplifying the apoptotic signal. The DIO-1ΔNLS mutant could form stable oligomers with wild-type DIO-1 and block nuclear translocation of the entire complex, preventing DIO-1-mediated gene upregulation and inhibiting the cell death program.
Acknowledgments
We thank B. B. Wolf, D. R. Green, T. W. Mak, and R. Hakem for generous gifts of reagents, A. Ruiz-Vela for excellent and continuous advice, and A. Fütterer and M. Torres, A. Ruiz-Vela, K. van Wely, and M. Campanero for critical reading of the manuscript. We also thank I. López-Vidriero and M. C. Moreno-Ortiz for help with flow cytometry, all technical members of the department who aided with cell culture and general reagents, and C. Mark for editorial assistance.
D.G.D. is the recipient of a fellowship from the Spanish Ministerio de Educación y Ciencia. This work was supported by grants from the Ministerio de Ciencia y Tecnología and the European Union (QLG1-CT-2001-01536). The Department of Immunology and Oncology was founded and is supported by the Spanish Council for Scientific Research (CSIC) and the Pharmacia Corporation.
REFERENCES
- 1.Alnemri, E. S., D. J. Livingston, D. W. Nicholson, G. Salvesen, N. A. Thornberry, W. W. Wong, and J. Yuan. 1996. Human ICE/CED-3 protease nomenclature. Cell 87:171.. [DOI] [PubMed] [Google Scholar]
- 2.Bertin, J., W. J. Nir, C. M. Fischer, O. V. Tayber, P. R. Errada, J. R. Grant, J. J. Keilty, M. L. Gosselin, K. E. Robison, G. H. Wong, M. A. Glucksmann, and P. S. DiStefano. 1999. Human CARD4 protein is a novel CED-4/Apaf-1 cell death family member that activates NF-κB. J. Biol. Chem. 274:12955-12958. [DOI] [PubMed] [Google Scholar]
- 3.Boldin, M. P., T. M. Goncharov, Y. V. Goltsev, and D. Wallach. 1996. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell 85:803-815. [DOI] [PubMed] [Google Scholar]
- 4.Budihardjo, I., H. Oliver, M. Lutter, X. Luo, and X. Wang. 1999. Biochemical pathways of caspase activation during apoptosis. Annu. Rev. Cell Dev. Biol. 15:269-290. [DOI] [PubMed] [Google Scholar]
- 5.Chauhan, D., T. Hideshima, S. Rosen, J. C. Reed, S. Kharbanda, and K. C. Anderson. 2001. Apaf-1/cytochrome c-independent and Smac-dependent induction of apoptosis in multiple myeloma (MM) cells. J. Biol. Chem. 276:24453-24456. [DOI] [PubMed] [Google Scholar]
- 6.Dai, C., and S. B. Krantz. 1999. Interferon gamma induces upregulation and activation of caspases 1, 3, and 8 to produce apoptosis in human erythroid progenitor cells. Blood 93:3309-3316. [PubMed] [Google Scholar]
- 7.Earnshaw, W. C., L. M. Martins, and S. H. Kaufmann. 1999. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 68:383-424. [DOI] [PubMed] [Google Scholar]
- 8.Ellis, R. E., J. Y. Yuan, and H. R. Horvitz. 1991. Mechanisms and functions of cell death. Annu. Rev. Cell Biol. 7:663-698. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Domingo, D., E. Leonardo, A. Grandien, P. Martinez, J. P. Albar, J. C. Izpisua-Belmonte, and C. Martinez-A. 1999. DIO-1 is a gene involved in onset of apoptosis in vitro, whose misexpression disrupts limb development. Proc. Natl. Acad. Sci. USA 96:7992-7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross, A., J. Jockel, M. C. Wei, and S. J. Korsmeyer. 1998. Enforced dimerization of BAX results in its translocation, mitochondrial dysfunction and apoptosis. EMBO J. 17:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu, J., L. Nie, D. Wiederschain, and Z. M. Yuan. 2001. Identification of p53 sequence elements that are required for MDM2-mediated nuclear export. Mol. Cell. Biol. 21:8533-8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta, S., V. Radha, Y. Furukawa, and G. Swarup. 2001. Direct transcriptional activation of human caspase-1 by tumor suppressor p53. J. Biol. Chem. 276:10585-10588. [DOI] [PubMed] [Google Scholar]
- 13.Hu, Y., M. A. Benedict, L. Ding, and G. Nuñez. 1999. Role of cytochrome c and dATP/ATP hydrolysis in Apaf-1-mediated caspase-9 activation and apoptosis. EMBO J. 18:3586-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inohara, N., T. Koseki, L. del Peso, Y. Hu, C. Yee, S. Chen, R. Carrio, J. Merino, D. Liu, J. Ni, and G. Nuñez. 1999. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-κB. J. Biol. Chem. 274:14560-14567. [DOI] [PubMed] [Google Scholar]
- 15.Jacobson, M. D., M. Weil, and M. C. Raff. 1997. Programmed cell death in animal development. Cell 88:347-354. [DOI] [PubMed] [Google Scholar]
- 16.Kamarajan, P., N. K. Sun, C. L. Sun, and C. C. Chao. 2001. Apaf-1 overexpression partially overcomes apoptotic resistance in a cisplatin-selected HeLa cell line. FEBS Lett. 505:206-212. [DOI] [PubMed] [Google Scholar]
- 17.Kerr, J. F., A. H. Wyllie, and A. R. Currie. 1972. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 26:239-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, H., H. Zhu, C. J. Xu, and J. Yuan. 1998. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94:491-501. [DOI] [PubMed] [Google Scholar]
- 19.Li, P., D. Nijhawan, I. Budihardjo, S. M. Srinivasula, M. Ahmad, E. S. Alnemri, and X. Wang. 1997. cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91:479-489. [DOI] [PubMed] [Google Scholar]
- 20.Li, P. F., R. Dietz, and R. von Harsdorf. 1999. p53 regulates mitochondrial membrane potential through reactive oxygen species and induces cytochrome c-independent apoptosis blocked by Bcl-2. EMBO J. 18:6027-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang, P., and A. B. Pardee. 1992. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257:967-971. [DOI] [PubMed] [Google Scholar]
- 22.Liu, W., G. Wang, and A. G. Yakovlev. 2002. Identification and functional analysis of the rat caspase-3 gene promoter. J. Biol. Chem. 277:8273-8278. [DOI] [PubMed] [Google Scholar]
- 23.Liu, X., C. N. Kim, J. Yang, R. Jemmerson, and X. Wang. 1996. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86:147-157. [DOI] [PubMed] [Google Scholar]
- 24.Lohrum, M. A., D. B. Woods, R. L. Ludwig, E. Balint, and K. H. Vousden. 2001. C-terminal ubiquitination of p53 contributes to nuclear export. Mol. Cell. Biol. 21:8521-8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo, X., I. Budihardjo, H. Zou, C. Slaughter, and X. Wang. 1998. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94:481-490. [DOI] [PubMed] [Google Scholar]
- 26.McConnell, B. B., and P. M. Vertino. 2000. Activation of a caspase-9-mediated apoptotic pathway by subcellular redistribution of the novel caspase recruitment domain protein TMS1. Cancer Res. 60:6243-6247. [PubMed] [Google Scholar]
- 27.Muzio, M., A. M. Chinnaiyan, F. C. Kischkel, K. O'Rourke, A. Shevchenko, J. Ni, C. Scaffidi, J. D. Bretz, M. Zhang, R. Gentz, M. Mann, P. H. Krammer, M. E. Peter, and V. M. Dixit. 1996. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell 85:817-827. [DOI] [PubMed] [Google Scholar]
- 28.Perkins, C., C. N. Kim, G. Fang, and K. N. Bhalla. 1998. Overexpression of Apaf-1 promotes apoptosis of untreated and paclitaxel- or etoposide-treated HL-60 cells. Cancer Res. 58:4561-4566. [PubMed] [Google Scholar]
- 29.Pohl, D., P. Bittigau, M. J. Ishimaru, D. Stadthaus, C. Hubner, J. W. Olney, L. Turski, and C. Ikonomidou. 1999. N-Methyl-d-aspartate antagonists and apoptotic cell death triggered by head trauma in developing rat brain. Proc. Natl. Acad. Sci. USA 96:2508-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raff, M. 1998. Cell suicide for beginners. Nature 396:119-122. [DOI] [PubMed] [Google Scholar]
- 31.Renatus, M., H. R. Stennicke, F. L. Scott, R. C. Liddington, and G. S. Salvesen. 2001. Dimer formation drives the activation of the cell death protease caspase 9. Proc. Natl. Acad. Sci. USA 98:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz-Vela, A., G. Gonzalez de Buitrago, and C. Martinez-A. 1999. Implication of calpain in caspase activation during B cell clonal deletion. EMBO J. 18:4988-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salvesen, G. S., and V. M. Dixit. 1999. Caspase activation: the induced-proximity model. Proc. Natl. Acad. Sci. USA 96:10964-10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salvesen, G. S., and V. M. Dixit. 1997. Caspases: intracellular signaling by proteolysis. Cell 91:443-446. [DOI] [PubMed] [Google Scholar]
- 35.Shaham, S., and H. R. Horvitz. 1996. Developing Caenorhabditis elegans neurons may contain both cell-death protective and killer activities. Genes Dev. 10:578-591. [DOI] [PubMed] [Google Scholar]
- 36.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein with bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 37.Srinivasula, S. M., M. Ahmad, T. Fernandes-Alnemri, and E. S. Alnemri. 1998. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol. Cell 1:949-957. [DOI] [PubMed] [Google Scholar]
- 38.Thornberry, N. A., and Y. Lazebnik. 1998. Caspases: enemies within. Science 281:1312-1316. [DOI] [PubMed] [Google Scholar]
- 39.Wolf, B. B., J. C. Goldstein, H. R. Stennicke, H. Beere, G. P. Amarante-Mendes, G. S. Salvesen, and D. R. Green. 1999. Calpain functions in a caspase-independent manner to promote apoptosis-like events during platelet activation. Blood 94:1683-1692. [PubMed] [Google Scholar]
- 40.Yakovlev, A. G., K. Ota, G. Wang, V. Movsesyan, W. L. Bao, K. Yoshihara, and A. I. Faden. 2001. Differential expression of apoptotic protease-activating factor-1 and caspase-3 genes and susceptibility to apoptosis during brain development and after traumatic brain injury. J. Neurosci. 21:7439-7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan, J., and H. R. Horvitz. 1992. The Caenorhabditis elegans cell death gene CED-4 encodes a novel protein and is expressed during the period of extensive programmed cell death. Development 116:309-320. [DOI] [PubMed] [Google Scholar]
- 42.Zou, H., W. J. Henzel, X. Liu, A. Lutschg, and X. Wang. 1997. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90:405-413. [DOI] [PubMed] [Google Scholar]