Abstract
DNA promoter hypermethylation has been shown to be a functional mechanism of transcriptional repression. This epigenetic gene silencing is thought to involve the recruitment of chromatin-remodeling factors, such as histone deacetylases, to methylated DNA via a family of proteins called methyl-CpG binding proteins (MBD1 to -4). MBD1, a member of this family, exhibits transcription-repressive activity, but to this point no interacting protein partners have been identified. In this study, we demonstrate that MBD1 partners with the p150 subunit of chromatin assembly factor 1 (CAF-1), forming a multiprotein complex that also contains HP1α. The MBD1-CAF-1 p150 interaction requires the methyl-CpG binding domain of MBD1, and the association occurs in the C terminus of CAF-1 p150. The two proteins colocalize to regions of dense heterochromatin in mouse cells, and overexpression of the C terminus of CAF-1 p150 prevents the targeting of MBD1 in these cells without disrupting global heterochromatin structure. This interaction suggests a role for MBD1 and CAF-1 p150 in methylation-mediated transcriptional repression and the inheritance of epigenetically determined chromatin states.
The methylation of cytosines in promoter regions containing CpG islands leads to the transcriptional inactivation of the downstream coding sequence in vertebrate cells (3, 34). This epigenetic regulation of gene expression has been shown to play a role in the silencing of imprinted alleles, genes on the inactive X-chromosome, and a number of tumor suppressor genes (3, 4). It is thought that this method of transcriptional silencing can be mediated through the recruitment of chromatin-modifying factors, such as histone deacetylases (HDACs), to the methylated promoters (6, 27, 34, 44). The subsequent altering of the local chromatin structure can lead to gene silencing through the formation of a heritable heterochromatin state (5).
A family of proteins known as methyl-CpG binding proteins (MBD1 to -4), is thought to play an important role in methylation-mediated transcriptional silencing (6, 27, 29, 44). MeCP2 was the first member of this family to be characterized (20). It contains a methyl-CpG binding domain (MBD) and a transcriptional-repression domain (TRD), which facilitates an interaction with, and targets the Sin3A/HDAC complex to, methylated DNA (27). Like MeCP2, MBD1 to -3 have been shown to be potent transcriptional repressors (6, 13, 29, 30, 45, 46). MBD4 is a DNA glycosylase which repairs G:T mismatches (14). Each member of this family, with the exception of MBD3, forms complexes with methylated DNA in mammalian cells, and all but MBD1 and MBD4 have been placed in known chromatin-remodeling complexes (6, 13, 14, 29, 30, 45, 46). MBD2 is a component of the MeCP1 repressor complex and directly interacts with the Sin3A/HDAC members within this complex (6, 30). MBD3 has been shown to be a member of the Mi-2/NuRD complex (44, 46).
MBD1 (formally, PCM1) is the least characterized of the original methyl-CpG binding proteins. Deletion studies have demonstrated that MBD1 contains an N-terminal MBD that is required for binding to methylated DNA and a C-terminal TRD that mediates transcriptional repression through an association with HDAC activity (7, 11, 12, 29). The exact mechanism through which the TRD facilitates transcriptional repression, or how MBD1 itself initiates transcriptional silencing, is not completely understood, since MBD1 has not been placed in a known repressor complex. Unlike the other methyl-CpG binding proteins, which act as transcriptional repressors at methylated promoters, full-length MBD1 has been shown to repress unmethylated promoters in vitro (11, 12). This repression activity appears to be dependent on three cysteine-rich motifs, known as CXXC domains, similar to the CXXC domains seen in DNA methyltransferase 1 (DNMT1) and the mammalian trithorax protein, HRX1 (7, 11, 12). MBD1 is the only methyl-CpG binding protein to contain these domains. Alternative splicing within the C terminus and CXXC domains gives rise to five isoforms of MBD1 (12). These alternatively spliced variants show differences in both methylation-dependent and -independent repression (11, 12).
It is now apparent that particular chromatin states play critical roles in the transcriptional activity of the cell. One of the key players in the determination and inheritance of such chromatin states is chromatin assembly factor 1 (CAF-1) (25, 37, 38). CAF-1 is a complex of three subunits, p150, p60, and p48, and is responsible for the assembly of nucleosomes onto newly replicated DNA (16). The CAF-1 complex initiates nucleosome assembly following replication by recruiting acetylated histones H3 and H4 to the DNA (43). The p150 subunit is associated with the replication-coupled assembly activity and is known to interact with PCNA, linking its activity to DNA replication and DNA repair (22, 23, 36).
In Saccharomyces cerevisiae, CAF-1 is responsible for the inheritance of epigenetically determined chromatin states and subsequent gene silencing. Deletion mutants of the three CAF-1 subunits are defective in the stable inheritance of gene silencing at the mating-type loci and at telomeres (9, 10, 17, 24). The p150 subunit associates with transcriptionally silent heterochromatin through its interaction with HP1 proteins (25). In addition, recent evidence suggests that CAF-1 may also play a key role in methylation-mediated gene silencing. Expression of a truncated form of the p150 subunit of CAF-1, a fragment lacking a portion of the N terminus, was capable of relieving the repression of a methylated reporter gene without disrupting the promoter methylation (42).
In the present study, we show a direct interaction between MBD1 and the p150 subunit of CAF-1. We have determined that the C terminus of CAF-1 p150 and the MBD of MBD1 are essential for this interaction and that these two proteins form a complex in mammalian cells with HP1α. Furthermore, we show in nuclear localization studies the potential importance of the CAF-1 p150 subunit in the association of MBD1 with heterochromatin. This interaction now places MBD1 in a known chromatin assembly and structuring complex that is associated with transcriptional repression.
MATERIALS AND METHODS
DNA constructs.
All DNA sequences or fragments of these sequences used were created by PCR using the Platinum pfx proofreading DNA polymerase (Invitrogen). MBD1 (PCM1 variant [see Results]) was amplified from a previously described Integrated Molecular Analysis of Genomes and Their Expression (IMAGE) clone (no. 222390) (7). A human cDNA library was used to amplify all other desired gene products. Deletion of the MBD and TRD of MBD1 (PCM1 variant) and deletion of the N and C termini of CAF-1 p150 was carried out by PCR-directed mutagenesis. All sequences were constructed to have a 5′ SalI restriction site and a 3′ NotI site by PCR. The PCR products were inserted into the SalI and NotI restriction sites of the appropriate vectors, and positive clones were analyzed for sequence errors.
(i) Plasmids for yeast two-hybrid assays.
All “bait” plasmids were constructed by cloning the appropriate coding sequences into the pDBLeu vector (Invitrogen) downstream and in frame with the Gal4 DNA binding domain (DBD). The bait plasmids constructed were as follows: pDBLeu-MBD1 (PCM1), pDBLeu-CAF-1 p150, pDBLeu-MBD1ΔTRD, pDBLeu-MBD1ΔMBD, pDBLeu-mCAF-1 p150 clone, and pDBLeu-HP1α. All “prey” plasmids, including the library used for the initial screen, were created by cloning the sequences of interest into the pPC86 vector (Invitrogen) downstream and in frame with the Gal4 activation domain (AD). The following prey plasmids were constructed: pPC86-mCAF-1 p150 clone, pPC86-MBD1, and pPC86-HP1α.
(ii) Plasmids for mammalian expression.
The appropriate coding sequences were cloned into previously constructed pSVK3 vectors (Pharmacia Biotech) containing hemagglutinin (HA) and Flag (Sigma) epitope tags (35). The sequences were inserted downstream of and in frame with the appropriate epitope tags to create the following vectors: pSVK3-HA-MBD1 (PCM1), pSVK3-Flag-MBD1 (PCM1), pSVK3-Flag-CAF-1 p150, pSVK3-HA-MBD1ΔMBD, pSVK3-HA-MBD1ΔTRD, pSVK3-HA-HP1α, pSVK3-HA-CAF-1 p150 C terminus, and pSVK3-Flag-CAF-1 p150 N terminus. The CAF-1 p150 full-length and fragment coding sequences were then removed from the pSVK3 HA and Flag vectors by EcoRI and NotI digestion, maintaining the fused epitope tags, and inserted downstream of a cytomegalovirus promoter into the corresponding sites in the pcDNA 3.1 vector (Invitrogen) for enhanced expression to create the following vectors: pcDNA 3.1-Flag-CAF-1 p150, pcDNA 3.1-HA-CAF-1 p150 C terminus, and pcDNA 3.1-Flag-CAF-1 p150 N terminus.
Yeast two-hybrid assays.
All experiments were carried out in yeast MaV203 cells (Invitrogen). A mouse embryonic cDNA library (Invitrogen) described previously (35) was used as the prey in a screen with human MBD1, PCM1 sequence variant, as the bait. The screen was carried out with the ProQuest yeast two-hybrid system (Invitrogen) as suggested by the manufacturer. Putative interacting clones were isolated based on their abilities in conjunction with MBD1 to activate expression of the his3 selectable marker gene, thus producing growth on SD-His-Leu-Trp medium (Bio 101) supplemented with 3-aminotrizole (15 mM). The interacting clones were sequenced, and the corresponding mouse gene was identified by BLAST sequence analysis (National Center for Biotechnology Information). To analyze direct interactions between two proteins or protein fragments, the appropriate bait and prey constructs were cotransformed into yeast cells (as suggested by the manufacturer), and positive interacting clones were identified by His selection.
Tissue culture and transfections.
COS7 and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Gemini Bio-Products) and a mixture of penicillin and streptomycin (10 μl/ml; Invitrogen) at 37°C in 5% CO2. RKO cells were cultured in minimum essential medium (Invitrogen) supplemented with 10% FBS and antibiotics at 37°C in 5% CO2. Transient transfections of COS7 and RKO cells were carried out in 10-cm-diameter tissue culture dishes. COS7 cells were seeded at 2 × 106 per dish, and RKO cells were seeded at 10 × 106 per dish. The plated COS7 and RKO cells were allowed to grow overnight in DMEM (10% FBS) and minimum essential medium (10% FBS), respectively. The next day, the cells were washed with serum- and antibiotic-free medium and transfected with 12 μg of total DNA of the appropriate expression vectors using Lipofectamine Plus reagents (Invitrogen) as suggested by the manufacturer. The cells were washed with 1× phosphate-buffered saline (PBS; Bio Fluids) and harvested 48 h posttransfection for coimmunoprecipitation assays. NIH 3T3 cells were cultured in DMEM supplemented with 10% fetal calf serum (HyClone) and antibiotics at 37°C in 5% CO2. For transient transfections, 3 × 105 cells were seeded in six-well tissue culture dishes and allowed to grow overnight in serum containing medium. The next day, the cells were transfected with 3 μg of total DNA of the appropriate expression constructs in serum- and antibiotic-free DMEM using Lipofectamine Plus reagents. After 48 h, the cells were washed with 1× PBS and prepared for immunofluorescent staining.
Coimmunoprecipitations.
HeLa cells were harvested at 80% confluence, and the transfected cells were harvested as described above. The cells were lysed using radioimmunoprecipitation assay (RIPA) buffer (1× PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1 mM AEBSF [4-{2-aminoethyl}-benzenesulfonyl; Roche], 1 mM dithiothreitol [Invitrogen], 1× protease inhibitor cocktail [Roche]). All steps were carried out at 4°C. Five hundred micrograms of whole-cell protein extract was then precleared with 20 μl of protein A/G plus agarose beads (Santa Cruz Biotechnology) and 2.5 μg of preimmune mouse immunoglobulin G (IgG) antibody (Santa Cruz Biotechnology) for 1 h. The cleared extracts were then treated with the appropriate antibody overnight: 2 μg of anti-HA monoclonal mouse antibody (Santa Cruz Biotechnology), 2.5 μg of anti-Flag monoclonal mouse antibody (Sigma), 2.5 μg of preimmune mouse IgG antibody (Santa Cruz Biotechnology), 2.5 μg of anti-MBD1 monoclonal mouse antibody (Imgenex), or 2.5 μg of anti-CAF-1 p150 monoclonal mouse antibody (Zymed Laboratories Inc.). The immunocomplexes were precipitated by incubation with 40 μl of protein A/G plus agarose beads for 4 h. The beads were washed three times with RIPA buffer, and the immunoprecipitated samples were separated on sodium dodecyl sulfate-polyacrylamide gels and analyzed by Western blotting.
Immunofluorescence imaging.
NIH 3T3 cells were seeded and transfected, as described above, on 22- by 22-mm glass coverslips. After being harvested, the coverslips were washed once with 1× PBS (1 mM MgCl2) and then fixed in 3% paraformaldehyde for 10 min at room temperature (RT). The cells were washed three times with the PBS solution and then permeabilized by incubation for 15 min in 0.5% Triton X-100, made in 1× PBS containing MgCl2. After permeabilization, the cells were again washed three times with 1× PBS containing MgCl2 and then blocked for 30 min at RT with 3% donkey serum diluted in 1× PBS containing MgCl2. The blocking solution was removed, and the cells were incubated with a 1:500 dilution of primary antibodies, anti-Flag monoclonal antibody (MAb) (Sigma) and anti-HA rabbit polyclonal antibody (Santa Cruz Biotechnology), in 3% donkey serum overnight at 4°C. The following day, the coverslips were washed three times with 1× PBS containing MgCl2 and treated with secondary antibody for 1 h at RT. Fluorescein isothiocyanate (FITC)-conjugated anti-mouse and Cy5-conjugated anti-rabbit secondary antibodies (Jackson Immunoresearch) were diluted 1:100 and 1:250, respectively, in 1× PBS containing MgCl2. The coverslips were then washed five times with 1× PBS containing MgCl2 and mounted on glass slides using Prolong antifade reagent (Molecular Probes). Immunofluorescence images (×100) were taken of the cells using an Ultraview Confocal imaging system.
Nucleotide sequence accession number.
The MBD1 GenBank accession number is NM_015847.
RESULTS
MBD1 directly interacts with the p150 subunit of CAF-1.
In an effort to better understand the transcriptional repressor MBD1, we carried out a yeast two-hybrid screen to identify proteins that interact with MBD1. The PCM1 isoform of MBD1 was used in all following experimentation. This splice variant differs from the full-length isoform by lacking the first CXXC domain. PCM1 has been specifically shown to repress transcription in vitro when tethered to the Gal4 DBD (29). So far, no group has compared the repressive activity of the PCM1 variant with those of the other isoforms (11, 12, 29).
One of the interactors identified in our screen encoded the mouse p150 subunit of CAF-1 lacking the N-terminal 85 amino acids (Fig. 1A). The full-length human CAF-1 p150 gene was cloned and, like the truncated form isolated in the screen, was able to interact with MBD1 in the yeast system (Fig. 1A).
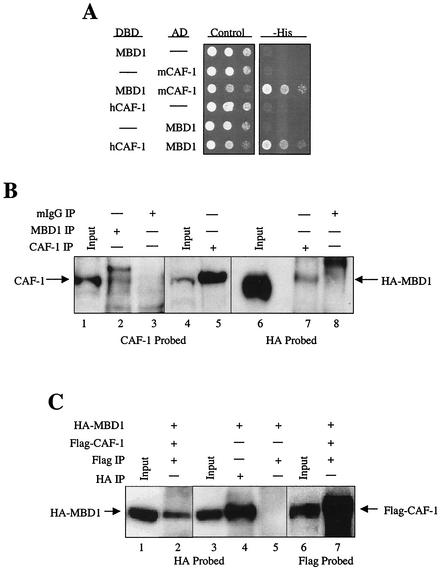
FIG. 1.
MBD1 interacts with the p150 subunit of CAF-1 in yeast and mammalian cells. (A) Both mouse and human homologues of CAF-1 p150 interact with MBD1 in yeast. A yeast two-hybrid screen was performed using human MBD1 fused to the Gal4 DBD as the bait to screen a mouse embryonic library fused to the Gal4 AD. An interaction was identified between MBD1 and a truncated form of the mouse CAF-1 p150 protein lacking the N-terminal 85 amino acids. Yeast cells were cotransformed with constructs expressing DBD-MBD1 and AD-mCAF-1 p150 clone and separately with constructs expressing human DBD-CAF-1 p150 and AD-MBD1. Controls for self-activating fusion proteins were carried out in all yeast two-hybrid assays by transforming particular expression constructs with a DBD or AD empty vector (dashes). Serial 1:10 dilutions of the transformed cells were plated on control medium that selects for cells expressing the two plasmids and on medium lacking histidine. Clones exhibiting a direct interaction between the two expressed fusion proteins were identified by cotransformed cells capable of growing on a medium lacking histidine. (B) Coimmunoprecipitations of endogenous CAF-1 p150 with endogenous MBD1 and overexpressed MBD1 with endogenous CAF-1 p150. Extracts made from HeLa cells were immunoprecipitated (IP) with (+) an anti-MBD1 antibody and a preimmune mouse IgG antibody (lanes 2 and 3, respectively). Input in lane 1 represents 10% of the total amount of HeLa cell extract used in the immunoprecipitations. Extracts from RKO cells expressing HA-MBD1 were immunoprecipitated with an anti-CAF-1 p150 antibody (lanes 5 and 7) and preimmune mouse IgG antibody (lane 8). Inputs in lanes 4 and 6 represent 10% of the total amount of RKO protein extract used in the immunoprecipitations with respect to protein concentration. Western blots were probed with a MAb against CAF-1 p150 (lanes 1 to 5) and a MAb against the HA epitope (lanes 6 to 8). (C) MBD1 complexes with CAF-1 p150 when both proteins are overexpressed. COS7 cells were cotransfected with constructs expressing MBD1 fused to an HA epitope tag and human CAF-1 p150 fused to a Flag epitope tag (lanes 1 and 2 and 6 and 7) and separately with HA-MBD1 alone (lanes 3 to 5). Extracts from cells expressing HA-MBD1 and Flag-CAF-1 p150 were immunoprecipitated with an anti-Flag MAb (lanes 2 and 7), and extracts from cells expressing HA-MBD1 alone were immunoprecipitated separately with anti-HA (lane 4) and anti-Flag (lane 5) MAbs. The input represents 10% of the total amount of protein extract used in the immunoprecipitations with respect to protein concentration. Western blots were probed with a MAb against the HA epitope (lanes 1 to 5) and a MAb against the Flag epitope (lanes 6 and 7).
To determine if MBD1 and CAF-1 p150 form a functional complex in mammalian cells, coimmunoprecipitation assays were conducted. Endogenous CAF-1 p150 was coimmunoprecipitated in HeLa cell extracts treated with an anti-MBD1 antibody. The immunoprecipitation produced two bands on a Western blot probed with an antibody against CAF-1 p150, with the lower band corresponding to endogenous CAF-1 p150 (Fig. 1B). The identity of the upper band, which has been seen in multiple experiments, is unknown. Due to the ineffectiveness of the MBD1 antibody mentioned above when used in Western blotting applications, HA-tagged MBD1 was overexpressed in RKO cells to conduct the reciprocal coimmunoprecipitations. HA-MBD1 was coimmunoprecipitated only when extracts from these cells expressing HA-MBD1 were immunoprecipitated with an anti-CAF-1 p150 antibody (Fig. 1B).
To provide further support for the notion that MBD1 and CAF-1 p150 form a cellular complex, HA-tagged MBD1 was either coexpressed with Flag-tagged CAF-1 p150 or expressed alone in COS7 cells (Fig. 1C). When extracts from both transfections were immunoprecipitated with an anti-Flag antibody, HA-tagged MBD1 was coimmunoprecipitated only when Flag-CAF-1 p150 was coexpressed (Fig. 1C). These results suggest that MBD1 and CAF-1 p150 associate in an in vivo setting.
The MBD of MBD1 is necessary for the interaction with CAF-1 p150.
Next, we wanted to determine what region of MBD1 was responsible for its interaction with CAF-1 p150. MBD1 has the following identified domains: an N-terminal MBD, central CXXC domains, and a C-terminal TRD (26). We independently deleted the MBD (MBD1ΔMBD) and the TRD (MBD1ΔTRD) of MBD1 (PCM1 variant) to determine if either domain was required for the interaction with CAF-1 p150 (Fig. 2A). The two deletion constructs of MBD1 were cotransformed with CAF-1 p150 in yeast two-hybrid assays. MBD1ΔTRD, but not MBD1ΔMBD, was able to interact with mCAF-1 p150, suggesting that the MBD of MBD1 is required for interaction with CAF-1 p150 (Fig. 2B).
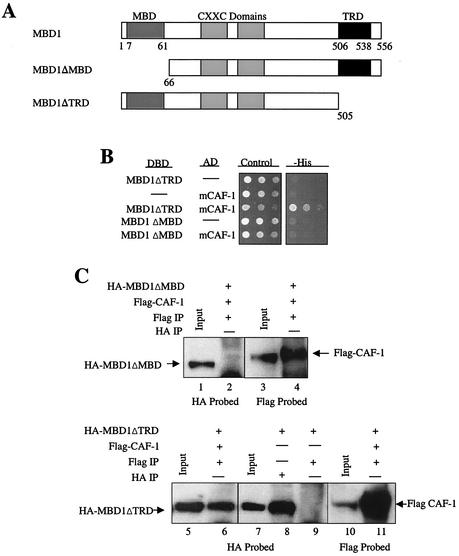
FIG. 2.
The MBD of MBD1, not the TRD, is necessary for the direct interaction between MBD1 and CAF-1 p150. (A) Map of MBD1 and deletions of the MBD and TRD. MBD1 (transcriptional variant PCM1) contains an N-terminal MBD, two central cysteine-rich CXXC domains, and a C-terminal TRD. MBD1 truncations of the MBD (MBD1ΔMBD) and TRD (MBD1ΔTRD) were created by deleting the N-terminal 65 amino acids and the C-terminal 51 amino acids, respectively. (B) The MBD, not the TRD, of MBD1 is required for the direct interaction of MBD1 and CAF-1 p150 in yeast cells. The mouse CAF-1 p150 truncation clone isolated in the yeast two-hybrid screen was cotransformed separately with constructs expressing the MBD1ΔMBD and MBD1ΔTRD truncations. The transformed cells were spotted in serial 1:10 dilutions on control and histidine selection media. Cells positive for direct interactors were identified as in Fig. 1. Dashes, empty vectors. (C) MBD1ΔMBD fails to associate with CAF-1 p150, but MBD1ΔTRD does coimmunoprecipitate with CAF-1 p150. HA-MBD1ΔMBD and Flag-CAF-1 p150 expressing COS7 whole-cell extracts were immunoprecipitated (IP) with (+) an anti-Flag antibody (lanes 2 and 4). Protein extracts from COS7 cells expressing HA-MBD1ΔTRD and Flag-CAF-1 p150 were immunoprecipitated with an anti-Flag antibody (lanes 6 and 11). Extracts from cells expressing HA-MBD1ΔTRD alone were immunoprecipitated separately with anti-HA and anti-Flag antibodies (lanes 8 and 9, respectively). Western blots were probed with the anti-HA (lanes 1 and 2 and 7 to 9) and anti-Flag (lanes 3 and 4 and 10 and 11) antibodies. The input amount is 10% of the total amount of extract used for immunoprecipitations.
The MBD1 deletions were then used in coimmunoprecipitation assays to examine if either truncated form of MBD1 failed to coimmunoprecipitate with CAF-1 p150. HA-MBD1ΔMBD failed to immunoprecipitate with Flag-CAF-1 p150 when extracts from COS7 cells coexpressing the two proteins were immunoprecipitated with the anti-Flag antibody (Fig. 2C). In contrast, HA-MBD1ΔTRD coimmunoprecipitated with Flag-CAF-1 p150, indicating that the two proteins were able to form a complex (Fig. 2C). When HA-MBD1ΔTRD was expressed alone in COS7 cells, the protein failed to immunoprecipitate with the anti-Flag antibody (Fig. 2C). These results suggest that the MBD of MBD1, not the TRD, is necessary for mediating an intracellular association with CAF-1 p150.
The C terminus of CAF-1 p150 interacts with MBD1.
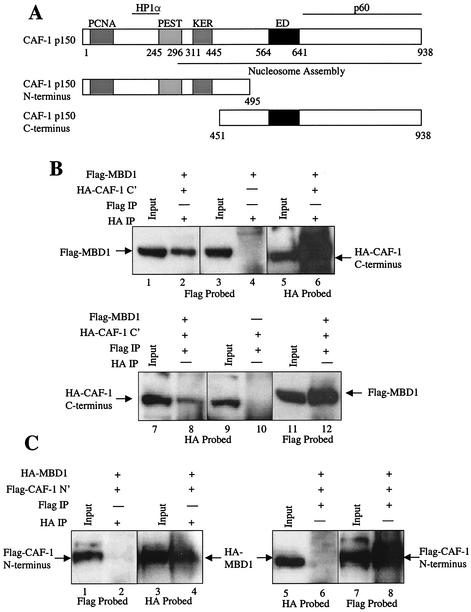
We next evaluated what region of CAF-1 p150 is responsible for interacting with MBD1. Our previous results indicated that this interaction does not involve the extreme N terminus of CAF-1 p150, since the mouse CAF-1 p150-interacting clone identified in the yeast two-hybrid screen lacked the N-terminal 85 amino acids. The N terminus of CAF-1 p150 contains the PCNA and HP1α binding sites, as well as a PEST domain, which is associated with proteins that undergo rapid proteolysis, and a charged KER domain. Much of the C terminus of CAF-1 p150 is required for nucleosome assembly, including the p60 binding domain along with the ED and KER domains (Fig. 3A) (16, 33). We split CAF-1 p150 into N- and C-terminal regions (Fig. 3A). These regions, along with MBD1, were expressed in COS7 cells to conduct coimmunoprecipitation assays to determine which half of CAF-1 p150 associates with MBD1 (Fig. 3B and C). Flag-MBD1 could be coimmunoprecipitated with an HA antibody only when the HA-tagged C terminus of CAF-1 p150 was coexpressed in COS7 cells (Fig. 3B). Extracts from cells coexpressing Flag-MBD1 and HA-CAF-1 p150 C terminus and from cells expressing HA-CAF-1 p150 C terminus alone were immunoprecipitated with an anti-Flag MAb. A small amount of HA-CAF-1 p150 C terminus coimmunoprecipitated when Flag-MBD1 was also present in the extract but not when the protein extract lacked Flag-MBD1 (Fig. 3B). Similar coimmunoprecipitations were done with extracts from COS7 cells coexpressing HA-MBD1 and Flag-CAF-1 p150 N terminus (Fig. 3C). Flag CAF-1 p150 N terminus failed to coimmunoprecipitate with the HA antibody in these experiments, and HA-MBD1 failed to coimmunoprecipitate with the Flag antibody (Fig. 3C). These results indicate that MBD1 interacts with the C-terminal half of the CAF-1 p150 protein.
FIG. 3.
MBD1 associates with the C terminus of CAF-1 p150. (A) Map of CAF-1 p150 and C-terminal and N-terminal truncations. CAF-1 p150 contains N-terminal PCNA- and HP1α-interacting domains and a C-terminal CAF-1 p60 subunit-interacting domain. A large region of the protein is associated with nucleosomal assembly activity. An N-terminal fragment of CAF-1 p150 was created which lacks the C-terminal p60 binding domain, the ED motif, and the majority of the nucleosomal assembly domain. The C-terminal fragment lacks the PCNA and HP1α binding sites and the PEST and KER domains. (B) MBD1 and the C terminus of CAF-1 p150 coimmunoprecipitate in mammalian cells. Protein extracts from COS7 cells coexpressing both Flag-MBD1 and HA-CAF-1 p150 C terminus were immunoprecipitated (IP) with (+) an anti-HA MAb (lanes 2, 4, and 6) and an anti-Flag MAb (lanes 8, 10, and 12). Extracts from cells expressing Flag-MBD1 alone and HA-CAF-1 p150 C terminus alone were immunoprecipitated with the anti-HA MAb (lane 4) and anti-Flag MAb (lane 10), respectively. Western blots were probed with the anti-Flag MAb (lanes 1 to 4 and 11 and 12) and the anti-HA MAb (lanes 5 and 6 and 7 to 10). The input represents 10% of the total protein extract used for the immunoprecipitations. (C) MBD1 does not complex with the N terminus of CAF-1 p150 in mammalian cells. COS7 cells were cotransfected with HA-MBD1 and Flag-CAF-1 p150 N terminus. Extracts from these cells were immunoprecipitated with an anti-HA MAb (lanes 2 and 4) and an anti-Flag MAb (lanes 6 and 8). Western blots were probed with the anti-Flag (lanes 1 and 2 and 7 and 8) and anti-HA (lanes 3 to 6) MAbs. Ten percent of the total extract used in the immunoprecipitations was used as the input.
MBD1 and CAF-1 p150 colocalize to regions of heterochromatin in mouse cells.
We next used immunofluorescence confocal imaging to investigate whether MBD1 and CAF-1 p150 colocalize within cells. Mouse cells were used in these experiments because the pericentromeric chromosomal regions of mouse cells contain extensive expanses of dense heterochromatin in which the DNA is hypermethylated. These pericentromeric heterochromatin regions produce a very strong punctate immunofluorescent signal when stained with antibodies to detect the cellular localization of proteins associated with heterochromatin (12, 25, 28). We used HP1α as a marker of heterochromatin in this case. CAF-1 p150 has been shown to target areas of heterochromatin in mouse cells by colocalizing with HP1α (25), while MBD1 has also been shown to target regions of heterochromatin in both human and mouse cells (1, 12). We transiently expressed HA-MBD1 and Flag-CAF-1 p150 in mouse NIH 3T3 cells. Immunofluorescent confocal images show that both proteins colocalize to very distinct foci in all cotransfected cells (Fig. 4A). Flag-MBD1 and Flag-CAF-1 p150 were separately coexpressed with HA-HP1α to determine if the cellular localization seen in Fig. 4A was indeed pericentromeric heterochromatin. Both Flag-MBD1 (Fig. 4B) and Flag-CAF-1 p150 (Fig. 4C) target to foci containing HP1α, suggesting that MBD1 and CAF-1 p150 form a cellular complex at regions of heterochromatin in mouse cells.
FIG. 4.
MBD1 and CAF-1 p150 colocalize to regions of heterochromatin in mouse fibroblasts. NIH 3T3 cells grown on coverslips were cotransfected with the corresponding expression constructs and incubated with anti-Flag MAb and anti-HA polyclonal rabbit primary antibodies. The cells were then washed and incubated with anti-mouse FITC-conjugated and anti-rabbit Cy5-conjugated secondary antibodies. Confocal images of the treated cells were taken to detect FITC (green) and Cy5 (red) immunofluorescence. The two images were merged, and regions of colocalization are indicated by a yellow signal. (A) Coexpressed Flag-CAF-1 p150 and HA-MBD1 colocalize in confocal images of NIH 3T3 cells. (B) Flag-MBD1 colocalizes with HA-HP1α to regions of pericentromeric heterochromatin. (C) Flag-CAF-1 p150 and HA-HP1α colocalize to areas of heterochromatin in mouse cells.
HP1α associates with the MBD1/CAF-1 p150 complex through its interaction with CAF-1 p150.
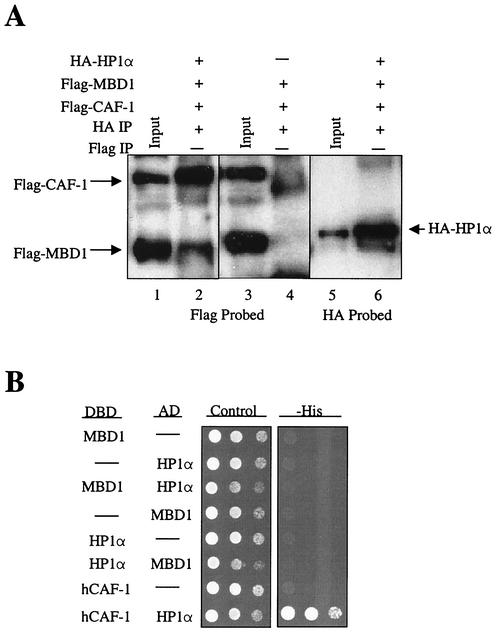
HP1α is known to be an important factor in the formation of heterochromatin and has been shown to directly interact with CAF-1 p150 (8, 25). In light of this evidence and our results shown in Fig. 4 demonstrating that MBD1 and CAF-1 p150 colocalize with HP1α, we hypothesized that MBD1 could be in a cellular complex not only with CAF-1 p150 but also with HP1α. To test this, we conducted coimmunoprecipitation experiments where Flag-MBD1 and Flag-CAF-1 p150 were coexpressed with and without HA-HP1α in COS7 cells (Fig. 5A). Protein extracts from these cells were immunoprecipitated with the HA antibody. Both Flag-MBD1 and Flag-CAF-1 p150 coimmunoprecipitated with HA-HP1α but failed to immunoprecipitate in the absence of HA-HP1α, showing that these proteins do complex in vivo (Fig. 5A). To determine if HP1α associates with MBD1 and CAF-1 p150 through its interaction with CAF-1 p150, rather than a direct interaction with MBD1, yeast two-hybrid assays were carried out with both MBD1 and CAF-1 p150 against HP1α (Fig. 5B). While HP1α did not interact with MBD1 in the yeast assay, HP1α did interact with CAF-1 p150, providing further evidence for an association between these two proteins (25) (Fig. 5B). These results suggest that CAF-1 p150 mediates the association between HP1α and MBD1.
FIG. 5.
MBD1 and CAF-1 p150 form a cellular complex with HP1α. (A) Immunoprecipitation of a cellular complex containing MBD1, CAF-1 p150, and HP1α. Protein extracts from COS7 cells expressing HA-HP1α, Flag-MBD1, and Flag-CAF-1 p150 and cells expressing only Flag-MBD1 and Flag-CAF-1 p150 were immunoprecipitated (IP) with (+) an anti-HA MAb (lanes 2, 4, and 6). The input was 10% of the total extract used in the immunoprecipitations. Western blots were probed with antibodies against the Flag (lanes 1 to 4) and HA (lanes 5 and 6) epitopes. (B) Human HP1α interacts with human CAF-1 (hCAF-1) p150 but not MBD1 in yeast-two hybrid assays. Yeast cells were cotransformed with the following expression constructs: DBD-MBD1 and AD-HP1α, DBD-HP1α and AD-MBD1, and DBD-hCAF-1 and AD-HP1α. Cells expressing direct interactors were identified as in Fig. 1. Dashes, empty vectors.
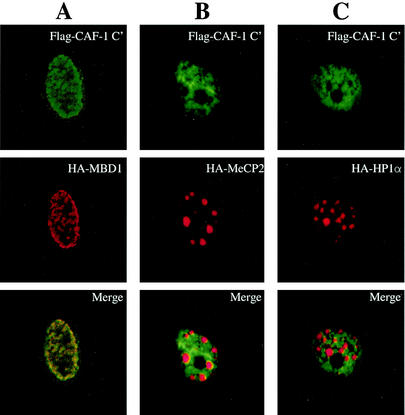
Expression of the C terminus of CAF-1 p150 in mouse cells disrupts the heterochromatin targeting of MBD1.
Recent evidence has shown that CAF-1 p150 may play a direct role in methylation-mediated transcriptional silencing (42). In this study, Tchénio et al. showed that the expression of a clone of CAF-1 p150 lacking the N-terminal 380 amino acids led to the reactivation of a methylated gene (42). This N-terminally deleted clone of CAF-1 p150 significantly overlaps with the C-terminal region that we have shown to be responsible for the interaction with MBD1 (Fig. 3). Knowing that MBD1 interacts with CAF-1 p150 at regions of heterochromatin, we wanted to determine whether our C-terminal CAF-1 p150 truncated protein could disrupt the targeting of MBD1 to sites of heterochromatin. We analyzed, by immunofluorescence imaging, the localizations of MBD1, MeCP2 (another methyl-CpG binding protein known to target to heterochromatin) (1, 28), and HP1α when coexpressed with our CAF-1 p150 C terminus construct. Unlike full-length CAF-1 p150, CAF-1 p150 C terminus exhibits a diffuse staining pattern and does not appear to be associated with heterochromatin (Fig. 6). Importantly, when Flag-CAF-1 p150 C terminus is expressed with HA-MBD1, the latter protein also fails to target to heterochromatin foci (Fig. 6A). In contrast, expression of Flag-CAF-1 p150 C terminus does not affect the normal targeting of HA-MeCP2 to heterochromatin foci (Fig. 6B) or the localization of HA-HP1α to heterochromatin (Fig. 6C). We can conclude from these results that the expression of CAF-1 p150 C terminus prevents the targeting of MBD1 to regions of heterochromatin. Presumably, this occurs because the truncated protein (Fig. 3A) cannot localize to heterochromatin but does bind to MBD1, thus siphoning off MBD1 from its normal heterochromatin location. Importantly, since the CAF-1 fragment has no affect on the localization of other heterochromatin components, like MeCP2 and HP1α, the results indicate a relatively selective role for CAF-1 p150 in the association of MBD1 with heterochromatin.
FIG. 6.
Expression of the C terminus of CAF-1 p150 in mouse fibroblasts disrupts the targeting by MBD1 of regions of heterochromatin without affecting the formation of heterochromatin complexes. NIH 3T3 cells grown on coverslips were cotransfected with the appropriate expression constructs and incubated with an anti-Flag MAb and an anti-HA polyclonal rabbit primary antibody. The cells were then washed and incubated with anti-mouse FITC-conjugated and anti-rabbit Cy5-conjugated secondary antibodies. Confocal images of the treated cells were taken to detect FITC (green) and Cy5 (red) immunofluorescence separately. The two images were merged to detect protein colocalization as in Fig. 4. (A) Flag-CAF-1 p150 C terminus and HA-MBD1 fail to target to heterochromatin foci in mouse cells. (B) Flag-CAF-1 p150 C terminus expression has no affect on the targeting of HA-MeCP2. (C) Expression of CAF-1 C terminus in mouse cells does not alter heterochromatin complexes at pericentromeric heterochromatin, as determined by normal HA-HP1α targeting.
DISCUSSION
The mechanisms through which DNA methylation leads to transcriptional silencing have been an area of intense study over the past decade. Methyl-CpG binding proteins play a critical role in this process, and the dynamics through which they carry out their repressive activities are proving increasingly complex. Many of the members of this family associate with complexes containing HDACs, but in some cases it is now apparent that mechanisms in addition to HDAC activity may facilitate their transcription-repressive effects (18, 44). In an attempt to identify protein partners of the methyl-CpG binding protein MBD1, we employed a yeast two-hybrid screen and identified a direct interaction between MBD1 and the p150 subunit of CAF-1. This interaction is potentially significant, since it places MBD1 in a known complex involved in the formation of heterochromatin. The CAF-1 complex plays an important role in the inheritance of chromatin states, first, by initiating chromatin formation on newly replicated DNA, and second, by being involved in the structuring of chromatin into dense transcriptionally silent heterochromatin, perhaps through its interaction with HP1 proteins (25, 37, 38). Our present results indicate a possible mechanism underlying the way in which MBD1 may participate in the formation of a repressive chromatin structure on methylated DNA by complexing with CAF-1 p150 and HP1α. Both MBD1 and CAF-1 p150 colocalize to regions of heterochromatin in mouse cells. The C terminus of CAF-1 p150 interacts with MBD1, and when overexpressed in mouse cells, the C terminus disrupts the targeting of MBD1 to heterochromatin through a direct interaction but does not affect the targeting of MeCP2 or HP1α.
The results of our experiments with deletions of the MBD and TRD of MBD1 in interaction assays suggest that it is the MBD, not the TRD, that is necessary for the association of MBD1 with CAF-1 p150. In repression assays, the TRD of MBD1 has been shown to be responsible for transcriptional repression in an HDAC-dependent manner (29). Intuitively, one would think that any interaction of MBD1 with a known chromatin-modifying complex would occur through the TRD, as is the case with MeCP2 (27). However, recent studies suggest that mechanisms not involving HDAC activity may lead to transcriptional repression by members of the methyl-CpG binding protein family (18, 44). Transcriptional repression studies with Drosophila cells show that the domain of MeCP2 responsible for repression in this setting is the N-terminal portion of the protein, including the methyl-CpG binding domain, and not the C-terminal TRD (18). Notably, the TRD of MBD2, which mediates an interaction with the Sin3A/HDAC complex, overlaps with the N-terminal MBD of the protein (6). The functions of MBD1 may not be completely HDAC dependent and may involve an alternative, or additive, mechanism for the formation of transcriptionally silent heterochromatin on methylated DNA following DNA replication.
The preservation of heterochromatin domains in the genome through successive rounds of DNA replication is critical for epigenetic inheritance (4, 34). The CAF-1 complex is recruited to replication foci through the interaction of the p150 subunit with PCNA, where it initiates chromatin formation by assembling nucleosomes on the newly replicated DNA (36, 43). The CAF-1 complex then remains on the newly replicated DNA following nucleosome assembly and may possibly act as a “gatekeeper” determining whether the DNA remains in an open euchromatin state or is compacted into dense heterochromatin (33, 40). The next step in compacting the DNA into heterochromatin is the covalent modification of the histone tails, for example, by HDACs and histone methyltransferases (33, 34, 39). Intense research has been done recently on the role of histone methylation in the determination of chromatin states. Briefly, methylation of the lysine 9 residue on histone H3 is associated with the formation of transcriptionally silent heterochromatin domains (21, 31, 32). To finish the compaction process, heterochromatin-associating proteins, such as HP1α, condense the chromatin further into closed heterochromatin domains. CAF-1 is thought to play a role in this final step, since it is associated with regions of dense heterochromatin, perhaps through its interaction with HP1α (25).
More work is needed to explore the precise functional role of the interaction between MBD1 and CAF-1 p150. Evidence is mounting to support the notion that CAF-1 may serve as a initiating factor in the formation of heterochromatin, perhaps at regions of DNA methylation, and thus as an important factor in the inheritance of epigenetically determined chromatin states (33). Previous results, and the results presented in this paper, suggest that CAF-1 p150 may act to initiate heterochromatin assembly by interacting with chromatin-associated proteins, like HP1α and MBD1. HP1 proteins are known to play a critical role in the formation of heterochromatin (8). Recent evidence suggests that this occurs by the interaction of the chromo domains of HP1 proteins with histone H3 proteins methylated at the lysine 9 residue (2, 19). It is thought that this action of HP1 proteins provides the mechanism for epigenetically determined heterochromatin formation directed by histone methylation. In this regard, our finding that HP1α associates with the MBD1/CAF-1 p150 complex may suggest a synergistic effect of DNA methylation and histone methylation to direct heterochromatin formation by the association of MBD1 with these two proteins at regions of both DNA methylation and histone H3 lysine 9 methylation. It has yet to be shown if histone methylation plays a role in DNA methylation-mediated transcriptional silencing. Evidence from studies of Neurospora crassa and Arabidopsis thaliana suggest a molecular link between the two processes in which establishment of a histone methylation pattern is required for DNA methylation to occur (15, 41). It will be interesting to see what role the MBD1/CAF-1 p150/HP1α complex plays in mediating the functions of these two epigenetic modifications.
Further support for the notion that CAF-1 plays a role in methylation-mediated repression through its interaction with MBD1 comes from a report that the expression of an N-terminal truncation of CAF-1 p150 in mammalian cells led to the reexpression of a gene silenced by DNA methylation (42). Cells expressing this C-terminal fragment of CAF-1 p150 showed increased chromatin accessibility around the promoter of the reexpressed gene through hypersensitivity to nuclease digestion. It was suggested that this mutant clone of CAF-1 p150 has this effect by binding the endogenous p60 CAF-1 subunit and therefore acting as a dominant negative for the whole CAF-1 complex, which could lead to defects in epigenetic inheritance. Our reported results may provide an alternative, or additive, mechanism to explain how CAF-1 p150 C terminus expression led to reactivation of a methylated gene. It does not appear that expression of the C terminus leads to disruption of global chromatin structure, considering that there was no alteration of heterochromatin complexes containing MeCP2 and HP1α in our experiments, but it did alter the targeting of MBD1 (Fig. 6). Thus, if MBD1 is responsible for the silencing of the methylated promoter in the previous study, the overexpression of the CAF-1 p150 C terminus could lead to the reactivation of a methylated gene by preventing MBD1 from binding at the methylated promoter.
In conclusion, we have demonstrated that MBD1 interacts directly with CAF-1 p150, which interacts directly with HP1α, forming a complex of the three proteins. Our identification of the MBD1/CAF-1 p150/HP1α complex places MBD1 in a known chromatin-forming complex and should warrant continued studies of the functional significance of this complex in the formation and maintenance of transcriptionally repressive chromatin on methylated DNA.
Acknowledgments
We thank K. Schuebel, K. Jair, R. Casero, J. Herman, and J. Bender for advice. We also thank D. Murphy and M. Delanoy for immunofluorescent confocal microscopy advice.
This work was supported by National Institutes of Health-National Cancer Institute grant NCI-CA43318.
REFERENCES
- 1.Bachman, K. E., M. R. Rountree, and S. B. Baylin. 2001. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 276:32282-32287. [DOI] [PubMed] [Google Scholar]
- 2.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 3.Baylin, S. B., and J. G. Herman. 2000. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 4:168-174. [DOI] [PubMed] [Google Scholar]
- 4.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 5.Bird, A. P., and A. Wolffe. 1999. Methylation-induced repression—belts, braces, and chromatin. Cell 99:451-454. [DOI] [PubMed] [Google Scholar]
- 6.Boeke, J., O. Ammerpohl, S. Kegel, U. Moehren, and R. Renkawitz. 2000. The minimal repression domain of MBD2b overlaps with the methyl-CpG binding domain and binds directly to Sin3A. J. Biol. Chem. 275:34963-34967. [DOI] [PubMed] [Google Scholar]
- 7.Cross, S. H., R. R. Meehan, X. Nan, and A. Bird. 1997. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat. Genet. 16:256-259. [DOI] [PubMed] [Google Scholar]
- 8.Eissenberg, J. C., and S. C. Elgin. 2000. The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 10:204-210. [DOI] [PubMed] [Google Scholar]
- 9.Enomoto, S., and J. Berman. 1998. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 12:219-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enomoto, S., P. D. McCune-Zierath, M. Gerami-Nejad, M. A. Sanders, and J. Berman. 1997. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 11:358-370. [DOI] [PubMed] [Google Scholar]
- 11.Fujita, N., N. Shimotake, I. Ohki, T. Chiba, H. Saya, M. Shirakawa, and M. Nakao. 2000. Mechanism of transcriptional regulation by methyl-CpG binding protein MBD1. Mol. Cell. Biol. 20:5107-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita, N., S. Takebayashi, K. Okumura, S. Kudo, T. Chiba, H. Saya, and M. Nakao. 1999. Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol. Cell. Biol. 19:6415-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrich, B., and A. Bird. 1998. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 18:6538-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrich, B., U. Hardeland, H.-H. Ng, J. Jiricny, and A. Bird. 1999. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature 401:301-304. [DOI] [PubMed] [Google Scholar]
- 15.Jackson, J. P., A. M. Lindroth, X. Cao, and S. E. Jacobsen. 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416:556-560. [DOI] [PubMed] [Google Scholar]
- 16.Kaufman, P. D., R. Kobayashi, N. Kessler, and B. Stillman. 1995. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell 81:1105-1114. [DOI] [PubMed] [Google Scholar]
- 17.Kaufman, P. D., R. Kobayashi, and B. Stillman. 1997. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor I. Genes Dev. 11:345-357. [DOI] [PubMed] [Google Scholar]
- 18.Kudo, S. 1998. Methyl-CpG-binding protein MeCP2 represses Sp1-activated transcription of the human leukosialin gene when the promoter is methylated. Mol. Cell. Biol. 18:5492-5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 20.Lewis, J. D., R. R. Meehan, W. J. Henzel, I. Maurer-Fogy, P. Jeppesen, F. Klein, and A. Bird. 1992. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69:905-914. [DOI] [PubMed] [Google Scholar]
- 21.Litt, M. D., M. Simpson, M. Gaszner, C. D. Allis, and G. Felsenfeld. 2001. Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science 293:2453-2455. [DOI] [PubMed] [Google Scholar]
- 22.Martini, E., D. M. Roche, K. Marheineke, A. Verreault, and G. Almouzni. 1998. Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J. Cell Biol. 143:563-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moggs, J. G., P. Grandi, J.-P. Quivy, Z. O. Jónsson, U. Hübscher, P. B. Becker, and G. Almouzni. 2000. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol. Cell. Biol. 20:1206-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monson, E. K., D. de Bruin, and V. A. Zakian. 1997. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc. Natl. Acad. Sci. USA 94:13081-13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murzina, N., A. Verreault, E. Laue, and B. Stillman. 1999. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Cell 4:529-540. [DOI] [PubMed] [Google Scholar]
- 26.Nakao, M., S. Matsui, S. Yamamoto, K. Okumura, M. Shirakawa, and N. Fujita. 2001. Regulation of transcription and chromatin by methyl-CpG binding protein MBD1. Brain Dev. 1(Suppl.):S174-S176. [DOI] [PubMed] [Google Scholar]
- 27.Nan, X., H.-H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386-389. [DOI] [PubMed] [Google Scholar]
- 28.Nan, X., P. Tate, E. Li, and A. Bird. 1996. DNA methylation specifies chromosomal localization of MeCP2. Mol. Cell. Biol. 16:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng, H.-H., P. Jeppesen, and A. Bird. 2000. Active repression of methylated genes by the chromosomal protein MBD1. Mol. Cell. Biol. 20:1394-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng, H.-H., Y. Zhang, B. Hendrich, C. A. Johnson, B. M. Turner, H. Erdjument-Bromage, P. Tempst, D. Reinberg, and A. Bird. 1999. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 23:58-61. [DOI] [PubMed] [Google Scholar]
- 31.Noma, K., C. D. Allis, and S. I. Grewal. 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293:1150-1155. [DOI] [PubMed] [Google Scholar]
- 32.Peters, A. H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Saure, C. Schöfer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, S. Opravil, M. Doyle, M. Sibilia, and T. Jenuwein. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323-337. [DOI] [PubMed] [Google Scholar]
- 33.Ridgway, P., and G Almouzni. 2000. CAF-1 and the inheritance of chromatin states: at the crossroads of DNA replication and repair. J. Cell Sci. 113:2647-2658. [DOI] [PubMed] [Google Scholar]
- 34.Rountree, M. R., K. E. Bachman, J. G. Herman, and S. B. Baylin. 2001. DNA methylation, chromatin inheritance, and cancer. Oncogene 20:3156-3165. [DOI] [PubMed] [Google Scholar]
- 35.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269-277. [DOI] [PubMed] [Google Scholar]
- 36.Shibahara, K., and B. Stillman. 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96:575-585. [DOI] [PubMed] [Google Scholar]
- 37.Smith, S., and B. Stillman. 1989. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication. Cell 58:15-25. [DOI] [PubMed] [Google Scholar]
- 38.Stillman, B. 1986. Chromatin assembly during SV40 DNA replication in vitro. Cell 45:555-565. [DOI] [PubMed] [Google Scholar]
- 39.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 40.Taddei, A., D. Roche, J.-B. Sibarita, B. M. Turner, and G. Almouzni. 1999. Duplication and maintenance of heterochromatin domains. J. Cell Biol. 147:1153-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamaru, H., and E. U. Selker. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414:277-283. [DOI] [PubMed] [Google Scholar]
- 42.Tchénio, T., J.-F. Casella, and T. Heidmann. 2001. A truncated form of the human CAF-1 p150 subunit impairs the maintenance of transcriptional gene silencing in mammalian cells. Mol. Cell. Biol. 21:1953-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verreault, A., P. D. Kaufman, R. Kobayashi, and B. Stillman. 1996. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87:95-104. [DOI] [PubMed] [Google Scholar]
- 44.Wade, P. A. 2001. Methyl-CpG-binding proteins and transcriptional repression. Bioessays 23:1131-1137. [DOI] [PubMed] [Google Scholar]
- 45.Wade, P. A., A. Gegonne. P. L. Jones, E. Ballestar, F. Aubry, and A. P. Wolffe. 1999. Mi-2 complex couples DNA methylation to chromatin remodeling and histone deacetylation. Nat. Genet. 23:62-66. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, Y., H.-H. Ng, H. Erdjument-Bromage, P. Tempst, A. Bird, and D. Reinberg. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]