Abstract
Studies of yeast DNA topoisomerase II with various alanine-substitution mutations provide strong biochemical support of a recent hypothesis that the type IA and IIA DNA topoisomerases act similarly in their cleavage and rejoining of DNA. DNA breakage and rejoining by either a type IA or a type IIA enzyme are shown to involve cooperation between a DNA-binding domain containing the active-site tyrosine and a Rossmann fold containing several highly conserved acidic residues. For a homodimeric type IIA enzyme, cooperation occurs in trans: the active-site tyrosine in the DNA-binding domain of one protomer cooperates with several residues in the Rossmann fold as well as other regions of the other protomer.
Keywords: enzyme mechanism, DNA–protein transesterification, alanine substitution mutagenesis, quinolone binding pocket
The DNA topoisomerases are enzymes that introduce transient breaks in DNA strands to permit their interpenetration (for a review, see ref. 1). These enzymes are divided into two types: the type I enzymes transiently break one DNA strand at a time for the passage of a second strand or perhaps a duplex DNA, and the type II enzymes transiently break both strands of one DNA double helix for the passage of a second DNA double helix. Extensive studies indicate that the type I enzymes can be further divided into two subfamilies, IA and IB, which are distinct in amino acid sequences and reaction mechanisms. The amino acid sequences of a large number of phylogenetically diverse type II DNA topoisomerases suggest that they can also be grouped into two subfamilies, IIA and IIB: the former includes eukaryotic DNA topoisomerase II, bacterial gyrase, and phage T4 DNA topoisomerase, and the latter, archaeal DNA topoisomerase VI (2).
In recent years, the three-dimensional structures of several large fragments of the type IA, IB, and IIA subfamilies of DNA topoisomerases have been determined. As expected from the amino acid sequences and mechanistic properties of these enzymes, the overall tertiary structures of enzymes of different subfamilies appear to be very different (reviewed in ref. 3). Recent comparisons of the type IA and IIA enzymes reveal, however, that there are common structural elements in both groups (4, 5). In the crystal structure of a 67-kDa fragment of a type IA enzyme, Escherichia coli DNA topoisomerase I (6), there are two domains that are similar to the DNA-binding domain of the E. coli catabolite activator protein (CAP). These CAP-like domains, one in domain III and the other in domain IV of the 67-kDa fragment, are related by a pseudo dyad axis (7). A CAP-like fold is also seen in the crystal structures of a 92-kDa fragment of yeast DNA topoisomerase II (8) and a 59-kDa fragment of the GyrA subunit of E. coli gyrase (9). Both of these type IIA enzyme fragments are dimeric in the crystals; therefore a pair of CAP-like domains related by a true dyad are present in each dimer. It was observed that the CAP-like folds in the type IA and IIA enzymes, as well as similar folds in several other DNA-binding proteins including histone H5, could be superposed reasonably well (4). Furthermore, the pair of CAP-like folds in the E. coli DNA topoisomerase I and in the GyrA crystal structures exhibit a similar spatial relationship (4).
An even more striking structural resemblance was noted between a Rossmann fold in E. coli DNA topoisomerase I, which makes up domain I of the protein (6), and a similar fold that makes up the bulk of the B′-fragment of yeast DNA topoisomerase II (4) corresponding to the C-terminal half of bacterial GyrB protein (10). When these folds were superposed, several highly conserved acidic residues assumed corresponding positions in space (4, 5). It was previously suggested that the Rossmann fold of E. coli DNA topoisomerase I might be involved in DNA breakage and rejoining. In the crystal structure, the domain III CAP-like fold and the domain I Rossmann fold are contacting each other (6). Recent mutagenesis studies of the enzyme indicate that Glu-9 of the Rossmann fold, which faces the active-site tyrosine Tyr-319 across the domain I and domain III interface, is a crucial side chain in DNA breakage (11, 12). Replacement of Glu-9 by alanine largely abolishes the DNA cleavage activity of the enzyme, and it has therefore been suggested that Glu-9 and Tyr-319 coordinate in the breakage and rejoining of a DNA strand (11, 12). Thus the structural similarity between the Rossmann folds of the type IA or IIA DNA topoisomerases suggested that the cleavage and subsequent rejoining of a DNA strand by either enzyme might involve both the CAP-like fold containing the active-site tyrosine and the Rossmann fold containing the acidic residues (4). Multiple alignment of amino acid sequences and structural modeling of a large number of different proteins, including the type IA, IIA, and IIB DNA topoisomerases, DNA primases, nucleases, and DNA repair proteins, have also implicated the Rossmann folds as a conserved catalytic domain (5).
In the 92-kDa structure of yeast DNA topoisomerase II, however, the CAP-like domains and the Rossmann folds are far apart (8). Thus the notion that the type IA and IIA topoisomerases might act similarly in their breakage and rejoining of DNA would require large displacements of these domains from their locations in the crystal structure of the yeast enzyme. Structure-based model building suggested that juxtaposition of these from a pair of protomers in a type IIA enzyme could be accomplished in a stereochemically reasonable way (4).
In our efforts to elucidate the mechanism of reactions catalyzed by the type II DNA topoisomerases (for a review, see ref. 13), we have recently initiated site-directed mutagenesis studies of yeast DNA topoisomerase II (14). It is known that residues necessary for DNA cleavage and rejoining by this enzyme are located within a region spanning amino acid residues 410 to 1,166 of the enzyme (1). We have previously examined the plausible roles of residues within the C-terminal two thirds of this region (starting from residue 660), termed the A′ fragment, by alanine-substitution mutagenesis (14). We have now completed a similar study of the N-terminal remainder of this region, the B′ fragment spanning residues 410 to 660. Our results, which are presented below, provide strong biochemical evidence in support of the hypothesis that the type IA and IIA DNA topoisomerases act similarly in their breakage and rejoining of DNA.
MATERIALS AND METHODS
The experimental designs generally followed those previously reported (14) and significant modifications are described in the figure legends. Mutagenic oligodeoxyribonucleotides used in the construction of the alanine-substitution mutants are listed in Table 1.
Table 1.
Sequences of oligonucleotides used in alanine-substitution mutagenesis
| Mutant | Oligonucleotides, 5′ to 3′ | Restriction site alteration |
|---|---|---|
| K428A | CT AAT TAC CCT GCC CTG GAA GAT GCC | +BstNI |
| E449A | CT TTA GTT CTG ACA GCC GGG GAT TCC GCC TTG | +MspI |
| S452A | GTT CTG ACA GAA GGG GAT GCC GCC TTG TCA TTA GCT GTT GC | +Fnu4HI |
| N480A | CTT CGT GGT AAA ATG CTA GCC GTT AGA GAA GCT AGT GC | +NheI |
| D530A | CC GAT CAA GAT CAT GCC GGC TCG CAT ATT AAA GG | +MspI |
| D583A | TAC AAT ATG CCC GCC TAT GAA AAA TGG | −MspI |
| K603A | CAG AAG TAT TAT GCC GGA TTA GGG AC | +MspI |
| N619A | GTC CGA GAA TAT TTT TCG GCC TTG GAC AGA CAT TTG | +StyI |
| R650A | G GCA GAT GAC GCG AAA GAA TGG CTG | +BstUI |
| K651A | G GCA GAT GAC CGC GCA GAA TGG CTG AGA C | +BstUI |
The restriction site added (+ sign) or eliminated (− sign) by each mutagenic oligomer is underlined in the sequence and listed in the rightmost column.
RESULTS
Site-Directed Mutagenesis and Complementation Tests.
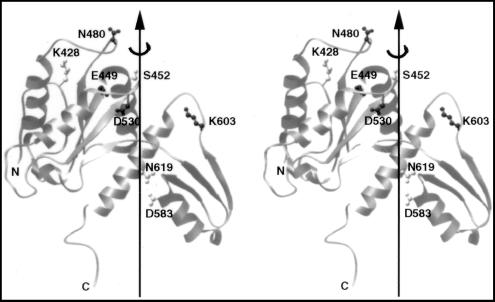
As reported previously (10, 14), we assumed that a residue participating in DNA breakage and rejoining was likely to possess a polar side chain and that these residues in all orthologues must possess side chains with a common chemical group. From the amino acid sequence alignment of Caron and Wang (10), 12 residues within the B′ fragment of the yeast enzyme, Lys-428, Glu-449, Asp-451, Ser-452, Arg-475, Lys-477, Asn-480, Asp-530, Lys-603, Asn-619, Arg-650, and Lys-651, were considered to fit the above selection criteria. Of these, alanine-substitution at Arg-475 and Lys-477, both of which are present in a highly conserved motif PLRGKILN (positions underlined), had previously been done in connection with a search of mutations that might lead to resistance to the antitumor agent amsacrine (15); alanine substitution at neither one of the adjacent pair of basic residues had a major effect on the catalytic activities of the enzyme. Asp-451, which lies within a highly conserved motif EGDSA (position underlined), was not selected because of the presence of a leucine at this position in Candida albican DNA topoisomerase II (16). Alanine substitution was carried out at each of the remaining nine positions. In addition, uncertainty in the sequence alignment around Asp-583 also led to its selection. Fig. 1 illustrates a stereo view of the locations of eight of the ten selected residues; Arg-650 and Lys-651 are located near the C terminus of the B′ fragment within the B′–A′ linker region, which is invisible in the crystal structure (8).
Figure 1.
A stereo view of the crystal structure of a fragment of yeast DNA topoisomerase II spanning amino acid residues 420 to 633. Eight of the ten residues selected for mutagenesis are shown in ball-and-stick models; the other two, Arg-650 and Lys-651, are within a region invisible in the published crystal structure of the yeast enzyme (8). Dark or light shading of a selected amino acid residue indicates its essentiality, as explained in the text. Drawing is based of the coordinates reported in ref. 8, by using the program ribbon (24).
Alanine-substitution mutagenesis and functional tests of the mutant proteins in yeast strains carrying a temperature-sensitive top2–4 or a cold-sensitive top2–13 mutation in the chromosomal gene encoding DNA topoisomerase II were carried out as described previously (14). It was found that a mutant plasmid expressing K428A, S452A, D583A, or N619A, in which the designated amino acid had been substituted by an alanine, complemented well both top2–4 and top2–13 defects. These residues are represented in Fig. 1 by side chains in lightly shaded ball-and-stick models. Expression of the other mutant proteins did not rescue the top2 chromosomal defects, and the locations of the irreplaceable residues, other than Arg-650 and Lys-651, are represented by the more darkly shaded side chains in Fig. 1.
Because alanine substitution at Lys-428, Ser-452, Asp-583, or Asn-619 does not appear to inactivate the enzyme, these substitution mutants were not studied further. For the others, each alanine substitution was introduced into pGAL1Top2(1–1196)-HMK-(His)6, which was constructed for the overexpression of a catalytically active C-terminal truncation of yeast DNA topoisomerase II (17). In this plasmid, codons of residues 1197–1428 of the wild-type yeast enzyme are replaced by those for a heart-muscle kinase (HMK) phosphorylation site and a hexahistidine tag. The addition of a hexahistidine tag to the mutant polypeptides facilitates their purification (18), and the presence of the HMK tag allows radiolabeling of the polypeptide (19). Each of the six mutant proteins was purified to near homogeneity, and its relaxation of supercoiled DNA and formation of protein–DNA covalent adduct were examined.
Relaxation of Supercoiled DNA by Unsubstituted and Alanine-Substituted Mutant Yeast DNA Topoisomerase II.
Relaxation of a negatively supercoiled 3-kb plasmid by serial dilutions of the various purified enzymes was carried out as described previously (14). Two different buffers were used in these experiments. Both contained 50 mM Tris⋅HCl (pH 7.8)/10 mM MgCl2/1 mM DTT/100 μg/ml BSA/4.5% glycerol (vol/vol) and 1 mM ATP if present. In addition, the low-salt buffer contained 50 mM KCl and the high-salt buffer, 150 mM KCl. In the high-salt buffer, the unsubstituted enzyme was found to completely relax the input DNA at an enzyme to DNA molar ratio (E/D) of 1/8, and partial relaxation of the DNA at an E/D ratio as low as 1/128 was evident; none of the mutant proteins E449A, N480A, D530A, and R650A showed any detectable activity, however, at an E/D as high as 128. A large reduction in DNA relaxation activity was also seen in the low-salt buffer: complete relaxation of the DNA by the unsubstituted enzyme was seen at an E/D of about 2, and the same four mutant proteins showed no detectable relaxation activity at E/D ratios as high as 128.
The two mutant proteins K651A and K603A showed reduced DNA relaxation activity relative to the unsubstituted enzyme. With the K651A protein, nearly complete relaxation of the DNA was seen at E/D ratios of 8 and 16, respectively, in the high- and low-salt medium. Treatment with the K603A protein at the same E/D ratios led to partial relaxation of the DNA. The residual DNA relaxation activity of the K651A and K603A proteins indicates that Lys-651 and Lys-603 do not play a major role in the catalysis of DNA breakage and rejoining.
Covalent-Adduct Formation Between DNA and Unsubstituted and Alanine-Substituted Mutant DNA Topoisomerase II.
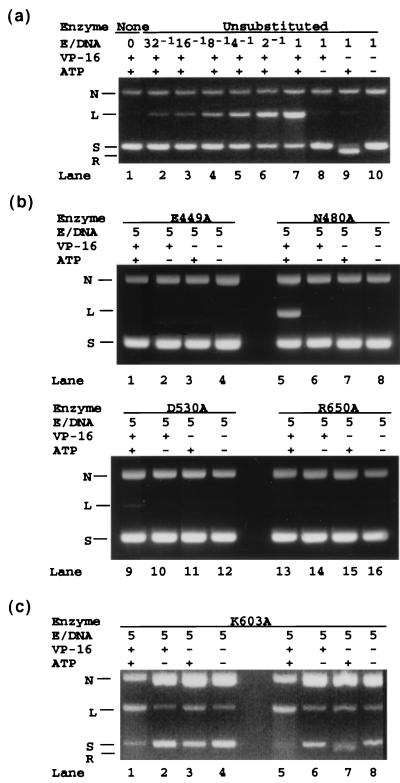
Lanes 1–10 of Fig. 2a show the results of a control experiment on covalent adduct formation between DNA and unsubstituted yeast DNA topoisomerase II lacking the C-terminal domain. The samples analyzed in lanes 1–7 were treated in the low-salt buffer in the presence of ATP and the drug etoposide, which stabilizes the covalent adduct (20). The lane 1 sample contained no enzyme, and the E/D ratios for the next six samples varied from 1/32 to 1 in 2-fold increments. In the lanes 8–10 samples, the E/D ratios were maintained at 1; ATP was omitted in the lane 8 sample and etoposide was omitted in the lanes 9 and 10 samples (see signs above the lanes; + indicates presence and −, absence). DNA cleavage was insignificant in sample 10 in which both etoposide and ATP were omitted, but was nearly quantitative in the presence of saturating concentrations of etoposide and ATP (lanes 2–7), as expected (21). As reported previously (20), the trapping of the protein–DNA covalent adduct by etoposide is strongly ATP dependent (compare the products of the samples in lanes 7 and 8). A similar set of samples for monitoring DNA cleavage by the unsubstituted enzyme in the high-salt buffer gave identical results (data not shown).
Figure 2.
Covalent adduct formation between DNA and unsubstituted and alanine-substituted yeast DNA topoisomerase II. Formation of a covalent bond between a DNA and a topoisomerase, which is accompanied by the breakage of a DNA phosphodiester bond, was monitored by the cleavage of a plasmid DNA about 3 kb in length (see ref. 14 for reaction conditions). (a) DNA cleavage by unsubstituted yeast DNA topoisomerase II. The enzyme used contained the first 1,196 amino acid residues of the intact 1,428 amino acid residue protein. (b) DNA cleavage by E449A, N480A, D530A, and R650A mutant enzymes, all of which showed no activity in the relaxation of a supercoiled DNA. (c) DNA cleavage by mutant enzyme K603A. For each reaction mixture, the molar ratio of enzyme to plasmid DNA (E/D), and the presence (+) or absence (−) of ATP and the drug VP-16 (etoposide) that stabilizes the covalent enzyme–DNA complex are indicated above the lane containing the sample. N, nicked DNA ring; L, linear 3-kb DNA; S, input negatively supercoiled DNA; R, S after relaxation by the topoisomerase. Purification of the various proteins was done as described previously (14).
DNA cleavage in the low-salt medium by mutant proteins that showed no DNA relaxation activity is shown in Fig. 2b. Two of the four proteins, E449A and R650A, also showed no detectable DNA cleavage activity for all combinations of etoposide and ATP (see the quartets of samples run in lanes 1–4 and lanes 13–16 of Fig. 2b). For the remaining two, in the presence of ATP and etoposide E480A at an E/D of 5 yielded linearized product comparable in amount to that of the unsubstituted enzyme at an E/D ratio between 1/8 and 1/4 (compare lane 5 of Fig. 2b with lanes 4 and 5 of Fig. 2a); under the same reaction conditions, D530A gave a trace of linear DNA (the linear DNA band in lane 9 of Fig. 2b was barely visible, and its amount was less than that seen in the sample treated with the unsubstituted enzyme at an E/D ratio of 1/32). Covalent-adduct formation between DNA and these mutant proteins in the high-salt buffer generally followed the patterns described above for the low-salt buffer. For the N480A protein, however, no activity was detectable in the high-salt buffer.
As expected, mutant proteins that showed some proficiency in DNA relaxation were found to exhibit readily detectable levels of DNA cleavage activity. In the low-salt buffer, the extent of DNA cleavage by the K651A protein was not significantly different from that by the unsubstituted enzyme; in the high-salt buffer, however, DNA cleavage by this mutant protein was reduced by several-fold (results not shown). Cleavage of DNA by the K603A protein was also evident in either the low-salt (Fig. 2c, lanes 1–4) or high-salt buffer (Fig. 2c, lanes 5–8). DNA cleavage by this mutant protein appeared unusual, however, in that it occurred even in the absence of both ATP and etoposide (Fig. 2c, lanes 4 and 8). In the absence of the drug, ATP, or both, a significant fraction of the input supercoiled DNA was also converted by the K603A mutant protein to the nicked form (Fig. 2c, lanes 2–4 and 6–8 ). This DNA nicking reaction might reflect a less favorable equilibrium for covalent adduct formation. Under these conditions, intermediates with nicked DNA strands and end products with both DNA chains severed would coexist (22).
Cooperation in Trans Between Active-Site Residues in the Cleavage of DNA.
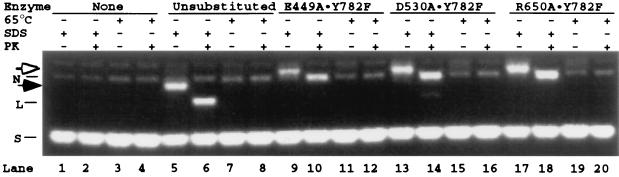
The results described in the above sections indicate that among residues in the B′ fragment, Glu-449, Asp-530, and Arg-650 of yeast DNA topoisomerase II are likely to assist Tyr-782 in DNA cleavage and rejoining. To test whether these residues cooperate with Tyr-782 of the same protomer (cooperation in cis) or the other protomer (cooperation in trans), the cleavage of DNA by heterodimeric proteins containing a Y782F mutation in one protomer and an E449A, D530A, or R650A mutation in the other protomer was examined. In lanes 5–20 of Fig. 3, samples for each protein preparation were analyzed in a set of four lanes: the pair of lanes on the left contained the DNA cleavage products without and with proteinase K treatment, as indicated in the margin above the lanes, and the next pair of lanes contained the corresponding protein–DNA mixtures that had been heated at 65°C for 30 min before the addition of SDS to trap the topoisomerase-DNA covalent intermediates. In lanes 1–4, control samples containing no DNA topoisomerase were analyzed.
Figure 3.
Covalent adduct formation between DNA and yeast DNA topoisomerase II heterodimers in which one protomer contains an alanine-substitution mutation and the other, a Tyr-782 to phenylalanine (Y782F) mutation. The leftmost quartet of lanes contained the no-enzyme controls and the next quartet, samples treated with unsubstituted yeast DNA topoisomerase II. The heterodimeric enzymes used in the other quartets of lanes are specified in the upper margin (across the row marked Enzyme). Heat treatment of the reaction mixtures is indicated by + in the row marked 65°C, and subsequent SDS and proteinase K treatments are similarly marked (rows denoted SDS and PK, respectively). Other symbols are the same as those shown in Fig. 2. Each reaction mixture contained 1 pmol of a 3-kb plasmid DNA and, with the exception of the no-enzyme controls, about 2 pmol of the unsubstituted enzyme or the heterodimeric enzyme in a volume of 200 μl of 50 mM Tris⋅HCl (pH 7.8)/50 mM KCl/10 mM MgCl2/0.5 mM etoposide/1 mM ATP/7 mM β-mercaptoethanol/100 μg/ml BSA/4.5% glycerol (vol/vol)/3% dimethyl sulfoxide (vol/vol). After 20 min at 30°C, each reaction mixture was divided into two equal portions. Ten μl of 2% SDS was introduced to terminate one portion of the reaction. The other portion was heated at 65°C for 30 min before termination. Each of the pair of samples was further divided into two equal portions, one of which was treated with proteinase K. Stock solutions of 5 M NaCl (5.5 μl), DNA loading dye solution, and water were added to each sample to give a final volume of 125 μl, of which 25 μl was loaded for gel electrophoresis (3 h at 4 V/cm in 100 mM Tris⋅borate/2 mM EDTA/0.1% SDS; see ref. 25).
With unsubstituted yeast DNA topoisomerase II, DNA cleavage yielded primarily linear products, as expected (lanes 5 and 6); the mobility of the cleavage product in the lane 5 sample, which was not treated with proteinase K (band marked by a solid arrowhead in the left margin of the figure), was slower than that of the proteinase K-treated sample run in lane 6 (position marked by “L” in the left margin), as expected for the presence of polypeptides at the 5′ ends of the untreated samples. Heating of the enzyme–DNA incubation mixture before the addition of SDS reversed covalent complex formation (compare the patterns shown in lanes 7 and 8 with the corresponding unheated samples run in lanes 5 and 6).
The DNA cleavage products of all three heterodimers Y782F⋅E449A, Y782F⋅D530A, and Y782F⋅R650A were found to efficiently introduce single-stranded nicks (Fig. 3, lanes 9–20). The major cleavage products were nicked DNA rings in the proteinase K-treated samples (the band marked by N in Fig. 3, lanes 10, 14, and 18). When proteinase treatment was omitted, the mobility of the cleavage products was reduced from that of the nicked DNA ring (band marked by an open arrowhead in lanes 9, 13, and 17), which corresponded to that of a nicked DNA ring with a polypeptide attached to the 5′ end of the nicked strand. Heat treatment again reversed the cleavage reaction (Fig. 4, lanes 11, 12, 15, 16, 19, and 20). In the experiment with the Y782F⋅D530A heterodimer, a trace of double-stranded cleavage product was also visible (see Fig. 4, lane 14). This species is most likely the product of inefficient double-stranded DNA cleavage by the D530A⋅D530A homodimer, which was present as a contaminant in the Y782F⋅D530A heterodimer preparation.
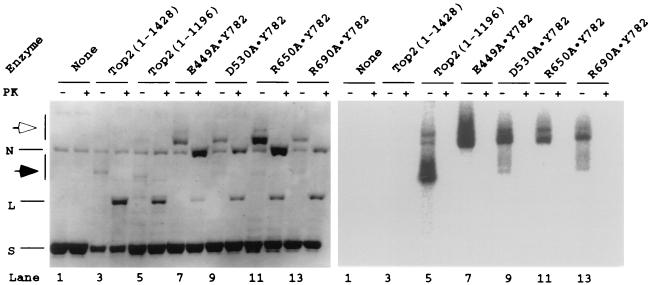
Figure 4.
DNA cleavage by unsubstituted yeast DNA topoisomerase II and heterodimeric enzymes containing one mutant protomer and one wild-type protomer. With the exception of the no-enzyme controls, each pair of reaction mixtures analyzed in the gel shown started with 1.5 pmol of a 3-kb plasmid DNA and about 3 pmol of enzyme in a volume of 150 μl of the same buffer described in the legend to Fig. 3. After a 20-min incubation at 30°C, 15 μl of 2% SDS was added to effect DNA cleavage. Each reaction was divided into three equal portions. Two were kept on ice and the third was processed for radiolabeling of the protein as follows. The sample was first desalted by passage through a Sephadex G-50 column (Pharmacia Nick) equilibrated with 1 mM Tris⋅HCl (pH 8.0)/0.1 mM EDTA. About 400 μl of the same buffer was then added, and the eluate containing the DNA–protein covalent complex was passed through a Sephadex G-25 column (Pharmacia NAP-10) for further removal of SDS. Each eluted sample (1.5 ml) was concentrated at room temperature in an evaporator (Savant Speed Vac) to about 40 μl. Stock solutions of protein kinase buffer, γ-32P ATP (≈10 μCi) and protein kinase were then added to a final volume of 50 μl. After 20-min incubation at 30°C, EDTA was added to 20 mM to stop the reaction. Unincorporated nucleotides were removed with a Sephadex G-50 column as described above. Ten percent of the 32P-labeled protein (40 μl) was added to each of the samples kept on ice, and one of the pair was treated with proteinase K. DNA-loading dye stock and 5 M NaCl were added to the pair of proteinase treated and untreated samples (plus and minus sign across the row marked PK) to a final NaCl concentration of 220 mM. About 25 μl of each sample was loaded in the sample well of a 1% agarose gel slab for electrophoresis (see legend to Fig. 3). The gel was dried onto DEAE filter paper (Whatman) and exposed to an image plate (Fuji) overnight. The dried gel was then soaked for 3 h in H2O containing ethidium bromide (0.6 μg/ml) and photographed over a UV light source. N, L, and S, nicked, linear, and supercoiled plasmid DNA, respectively.
The above results provide strong evidence that Glu-449, Asp-530, and Arg-650 cooperate with Tyr-782 in trans in the catalysis of both DNA cleavage and rejoining. To provide a more stringent test of the participation of Glu-449, Asp-530, and Arg-650 in trans in the transesterification reactions, a series of experiments using heterodimers between wild-type yeast DNA topoisomerase II and its alanine-substitution derivative were carried out. Substitution of a catalytically important residue by alanine in one but not the other protomer of a homodimeric enzyme should abolish the cleavage of one DNA strand, even though both active-site tyrosines are present. The use of heterodimers with one unsubstituted yeast DNA topoisomerase II polypeptide, however, introduced a technical complication: homodimers with two unsubstituted polypeptides would efficiently introduce double-stranded DNA breaks, and therefore any linear product could be the result of cleavage either by the heterodimer of interest or by contaminating homodimers of the unsubstituted protein. To circumvent this complication, we resorted to the presence of the HMK tag in the mutant but not the wild-type polypeptide. Treatment of the cleavage products with HMK, in the presence of γ-32P-ATP, would only radiolabel the HMK-tagged mutant polypeptides (17); the products of double-stranded cleavage of DNA by wild-type homodimers would not be radiolabeled. The results of this series of experiments are depicted in Fig. 4.
The left panel (Fig. 4) shows an ethidium-stained agarose gel slab in which the HMK-treated products of the DNA cleavage reaction, without or with subsequent proteinase K treatment, were analyzed by electrophoresis. The right panel shows an autoradiogram of the same gel. From the left side, the first pair of lanes contained the no-enzyme controls and the next pair samples treated with wild-type yeast DNA topoisomerase II. Linearization of the plasmid DNA by the wild-type enzyme was evident. Because the wild-type protein contained no HMK site, no radiolabeling of the DNA-linked protein was observed in the sample before or after proteinase K treatment. For the next pair of samples, unsubstituted yeast DNA topoisomerase II containing amino acids 1–1,196, with HMK and hexahistidine tags added to the C terminus, was used. Double-stranded cleavage of DNA was apparent (lanes 5 and 6; a faint ladder of topoisomers was also seen in these lanes because of partial relaxation of the input supercoiled DNA). Furthermore, in the sample without proteinase K treatment, intense radiolabeling of the linearized DNA was observed (Fig. 4 Right, lane 5). In the sample treated with the heterodimer between the E449A mutant polypeptide and the wild-type polypeptide, the major DNA cleavage product was nicked DNA rings, but the presence of a small amount of linear DNA was also evident in the proteinase K-treated sample (Fig. 4 Left, lane 8). Significantly, the autoradiogram shows that without proteinase K treatment the nicked DNA band was intensely labeled, but no radioactivity was detectable at the expected positions of covalent complexes between the protein and linear DNA (Fig. 4 Right, lane 7 ). Thus the low amount of linear DNA in lane 8 of the left panel was most likely the product of double-stranded DNA cleavage by the contaminating homodimeric wild-type enzyme. A similar result was observed for the heterodimer between the R650A mutant polypeptide and the wild-type polypeptide (Fig. 4 Left, lanes 11–12 and Right, lane 11).
For the heterodimer between the D530A mutant polypeptide and the unsubstituted polypeptide, the nicked DNA band in the sample without proteinase K treatment (Fig. 4 Right, lane 9 ) was intensely radiolabeled. A low level of radioactivity was observed, however, in a broad region between the linear and the nicked DNA bands. The presence of this broad radiolabeled band suggests that the heterodimer had a low level of double-stranded DNA cleavage activity, in agreement with the observation that even the homodimeric D530A mutant protein exhibited a very low but detectable level of DNA cleavage activity (see Fig. 2). The broad and faint band of radiolabeled material might also contain linear DNA with more than two covalently attached DNA polypeptides. A plasmid with a homodimer of unsubstituted and a heterodimer of D530A and unsubstituted protein would yield, for example, a linear cleavage product with unsubstituted polypeptides attached to its ends and a D530A polypeptide, which would become radiolabeled, attached to an internal nick. In the experiment shown in Fig. 4, we had also examined a heterodimer between unsubstituted and mutant polypeptide R690A that was studied previously (14). Homodimeric R690A mutant protein showed no detectable DNA cleavage activity, but a heterodimer with one R690A mutant polypeptide and one Y782F polypeptide was found to efficiently nick DNA (14). For the heterodimer between wild-type and R690A polypeptide, the predominant DNA cleavage product was again nicked DNA rings; a broad smear of faintly radiolabeled species was also observed, however, similar to the DNA cleavage reaction with the heterodimer between a D530A and an unsubstituted polypeptide.
DISCUSSION
The present work identifies several residues in the B′ fragment of yeast DNA topoisomerase II, which corresponds to the C-terminal half of the bacterial GyrB protein, that are important in its catalysis of DNA cleavage and rejoining. In combination with our previous mutagenesis studies of the A′ fragment of the same enzyme corresponding to the N-terminal two thirds of bacterial GyrA protein, a picture of the pair of catalytic pockets for DNA cleavage and rejoining begins to emerge. Each of the pockets appears to be constituted of residues contributed by both halves: one protomer contributes Glu-449 and Asp-530 of the B′ fragment, Arg-690 of the A′ fragment, and Arg-650 in the linker region between A′ and B′; the other protomer contributes the active-site tyrosyl residue Tyr-782 in the A′ fragment. A number of other residues, including Lys-700 and Arg-781 in the A′ fragment, are probably involved in DNA binding near the scissile sites but appear to be less critical in covalent catalysis (14). Because of the strict conservation of these residues in all type IIA DNA topoisomerases, inferences from results drawn from studies of the yeast enzyme can be readily extended to all type IIA enzymes.
Glu-449 and Asp-530, two of the B′ residues that are likely to participate directly in the catalysis of DNA breakage and rejoining, are located in the Rossmann fold. The position of Glu-449 of the yeast enzyme in the Rossmann fold corresponds to that of Glu-9 in a similar fold in E. coli DNA topoisomerase I. Asp-530 in the yeast enzyme forms an acidic triad with Asp-526 and Asp-528; its counterpart in the E. coli enzyme, Glu-115, forms a similar triad with Asp-111 and Asp-113. Therefore the present results that Glu-449 and Asp-530 of the yeast enzyme cooperate with the active-site tyrosine in trans, together with previous findings establishing the importance of Glu-9 of E. coli DNA topoisomerase I (11, 12), add credence to the model that catalysis of DNA breakage and rejoining by both type IA and IIA DNA topoisomerases involve the coordinated action of the CAP-like fold containing the active-site tyrosine and the Rossmann fold containing the key acidic residues (4).
In addition to Glu-449 and Asp-530 of the B′ fragment of yeast DNA topoisomerase II, Arg-650 was also found to have a crucial role in DNA breakage and rejoining. Arg-650 is located in a highly basic region, and the proximity of this region to DNA was first implicated in protein-footprinting studies of the yeast enzyme (17). Binding of the yeast enzyme to DNA was found to protect one or more of the lysines between Lys-644 and Lys-651 against citraconylation (17). It is likely that Arg-650 is present in the catalytic pocket and is interacting directly with the scissile phosphate.
The proximity between the Rossmann fold in the B′ fragment and the active-site tyrosine in the A′ fragment in some conformational states of the type IIA DNA topoisomerases also lends support to an earlier suggestion based on the locations of quinolone-resistance mutations in E. coli DNA gyrase (23). The identification of three clusters of such mutations in E. coli DNA gyrase, in the Ser-83 region of GyrA and within the EGDSA and PLRGKILN motifs of GyrB, led to the suggestion that these regions might be close to each other to form a quinolone binding pocket (23).
Acknowledgments
This work was supported by National Institutes of Health Grants GM24544 and CA47958.
ABBREVIATIONS
- CAP
catabolite activator protein
- GyrA and GyrB
the A and B subunit of bacterial DNA gyrase, respectively
- E/D
molar ratio of enzyme to plasmid DNA
- HMK
heart-muscle kinase
References
- 1.Wang J C. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 2.Bergerat A, de Massy B, Varoutas P C, Nicolas A, Forterre P. Nature (London) 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 3.Berger J M. Biochim Biophys Acta. 1998;1400:3–18. doi: 10.1016/s0167-4781(98)00124-9. [DOI] [PubMed] [Google Scholar]
- 4.Berger J M, Fass D, Wang J C, Harrison S C. Proc Natl Acad Sci USA. 1998;95:7876–7881. doi: 10.1073/pnas.95.14.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aravind L, Leipe D D, Koonin E V. Nucleic Acids Res. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lima C D, Wang J C, Mondragón A. Nature (London) 1994;367:138–146. doi: 10.1038/367138a0. [DOI] [PubMed] [Google Scholar]
- 7.Merzin A G. Curr Opin Struct Biol. 1994;4:441–449. [Google Scholar]
- 8.Berger J M, Gamblin S J, Harrison S C, Wang J C. Nature (London) 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 9.Cabral J H M, Jackson A P, Smith C V, Shikotra N, Maxwell A, Liddington R C. Nature (London) 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 10.Caron P, Wang J C. In: DNA Topoisomerases and Their Applications in Pharmacology. Liu L F, editor. San Diego: Academic; 1994. pp. 271–297. [DOI] [PubMed] [Google Scholar]
- 11.Chen S-J, Wang J C. J Biol Chem. 1998;273:6050–6056. doi: 10.1074/jbc.273.11.6050. [DOI] [PubMed] [Google Scholar]
- 12.Zhu C X, Roche C J, Papanicolaou N, DiPietrantonio A, Tse-Dinh Y C. J Biol Chem. 1998;273:8783–8789. doi: 10.1074/jbc.273.15.8783. [DOI] [PubMed] [Google Scholar]
- 13.Wang J C. Q Rev Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 14.Liu Q, Wang J C. J Biol Chem. 1998;273:20252–20260. doi: 10.1074/jbc.273.32.20252. [DOI] [PubMed] [Google Scholar]
- 15.Wasserman R A, Wang J C. J Biol Chem. 1994;269:20943–20951. [PubMed] [Google Scholar]
- 16.Keller B A, Patel S, Fisher L M. Biochem J. 1997;324:329–339. doi: 10.1042/bj3240329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, Wang J C. J Biol Chem. 1997;272:31190–31195. doi: 10.1074/jbc.272.49.31190. [DOI] [PubMed] [Google Scholar]
- 18.Hochuli E, Bannwarth W, Döbeli H, Gentz R, Stüber D. Bio/Technology. 1988;6:1321–1325. [Google Scholar]
- 19.Kennelly P J, Krebs E G. J Biol Chem. 1991;266:15555–15558. [PubMed] [Google Scholar]
- 20.Chen G L, Yang L, Rowe T C, Halligan B D, Tewey K M, Liu L F. J Biol Chem. 1984;259:13560–13566. [PubMed] [Google Scholar]
- 21.Worland S T, Wang J C. J Biol Chem. 1989;264:4412–4416. [PubMed] [Google Scholar]
- 22.Lee M P, Sander M, Hsieh T S. J Biol Chem. 1989;264:13510–13518. [PubMed] [Google Scholar]
- 23.Yoshida H, Bogaki M, Nakamura M, Yamanaka L M, Nakamura S. Antimicrob Agents Chemother. 1991;35:1647–1650. doi: 10.1128/aac.35.8.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlson M. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 25.Been M D, Champoux J J. Nucleic Acids Res. 1980;8:6129–6142. doi: 10.1093/nar/8.24.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]