Abstract
The reversible acetylation of the N-terminal tails of histones is crucial for transcription, DNA repair, and replication. The enzymatic reaction is catalyzed by large multiprotein complexes, of which the best characterized are the Gcn5-containing N-acetyltransferase (GNAT) complexes. GNAT complexes from yeast to humans share several conserved subunits, such as Ada2, Ada3, Spt3, and Tra1/TRRAP. We have characterized these factors in Drosophila and found that the flies have two distinct Ada2 variants (dAda2a and dAda2b). Using a combination of biochemical and cell biological approaches we demonstrate that only one of the two Drosophila Ada2 homologues, dAda2b, is a component of Spt-Ada-Gcn5-acetyltransferase (SAGA) complexes. The other Ada2 variant, dAda2a, can associate with dGcn5 but is not incorporated into dSAGA-type complexes. This is the first example of a complex-specific association of the Ada-type transcriptional adapter proteins with GNATs. In addition, dAda2a is part of Gcn5-independent complexes, which are concentrated at transcriptionally active regions on polytene chromosomes. This implicates novel functions for dAda2a in transcription. Humans and mice also possess two Ada2 variants with high homology to dAda2a and dAda2b, respectively. This suggests that the mammalian and fly homologues of the transcriptional adapter Ada2 form two functionally distinct subgroups with unique characteristics.
The packaging of DNA into chromatin by histones and nonhistone proteins creates substantial opportunities for the regulation of DNA replication, recombination, and repair and for gene transcription. Eukaryotic cells possess numerous large multiprotein complexes that exploit chromatin structure when controlling the functions of the genome (for reviews, see references 20, 34, and 52). Several multiprotein complexes function by covalently modifying the N-terminal tails of histones, which can create recognition sites for binding of activating or repressing protein complexes. The most studied covalent histone tail modification is the acetylation of specific lysines, and a large number of histone acetyltransferase enzymes (HATs) have been identified (for a review, see references 11, 26, and 34). The first identified HAT enzyme linked to transcription was the transcriptional coactivator Gcn5 (12). Gcn5 relatives have been found in virtually all eukaryotes and form a superfamily of Gcn5-related N-acetyltransferases (GNATs) (for a review, see references 11, 44, and 46). Recombinant Gcn5 mainly acetylates free histone H3, but it exhibits little HAT activity when using nucleosomal histones as substrates (30). This suggested that Gcn5 requires other factors to obtain the capability to acetylate nucleosomes and eventually led to the biochemical characterization of the Spt-Ada-Gcn5-acetyltransferase (SAGA) and ADA complexes from yeast (22). Within these complexes, Gcn5 acetylated an expanded set of lysines on both free and nucleosomal histone H3 (19, 23).
The 1.8-MDa yeast SAGA complex contains three classes of proteins that have been implicated in transcription control. The first group comprises the transcriptional adapter (Ada) proteins Ada1, Ada2, Ada3, and Ada5/Spt20 (22). The last has also been isolated in genetic screens for suppressors of transcriptional defects caused by transposable-element insertions at promoters (Spt genes) (51). Besides Ada5/Spt20, SAGA contains the Spt proteins Spt3, Spt7, and Spt8. These Spt proteins are thought to affect TATA-binding protein (TBP) function, and Spt3 was shown to directly interact with TBP (51). The third group of SAGA components includes a subset of TBP-associated factors (Tafs) (24). SAGA also contains Tra1, an ATM/PI-3 kinase-related homologue of the human TRRAP protein (25), which plays an important role in activator recruitment of SAGA (10). There are at least two other Gcn5-containing complexes in yeast. The 800-kDa ADA and the 200- to 500-kDa HAT A2 complexes share Gcn5, Ada2, and Ada3 with SAGA but lack the Spt, Taf, and Tra1 proteins (19, 22, 39, 41). In contrast to the ADA and HAT A2 complexes, SAGA interacts with both TBP and acidic activators and has coactivator functions beyond its HAT activity (17, 44, 49).
Homologues of yeast Gcn5-associated factors, such as Tafs, Ada2, Ada3, Spt3, and Tra1, have been identified in higher eukaryotes, suggesting that GNAT complexes from different organisms might have similar compositions and functions (for a review, see references 11, 35, and 46). However, the GNAT complexes from higher metazoans may be more diverse than those in yeast. For instance, mammalian cells have two distinct Gcn5 homologues, namely, Gcn5L and PCAF, which have been found in three different SAGA-like complexes (PCAF, STAGA, and TFTC). The PCAF complex contains PCAF as the catalytic subunit, Tafs, TRRAP, and homologues of Ada2, Ada3, and Spt3 (36). Gcn5L is the catalytic subunit of the human STAGA and TFTC complexes, which appear to have different compositions but partially overlapping functions (7, 8, 33). Recent studies have revealed that PCAF is dispensable for mouse development, whereas Gcn5L mutants die as embryos (53, 54). In contrast to PCAF, Gcn5L is ubiquitously expressed in the developing mouse embryo and may compensate for the loss of PCAF. The phenotype of Gcn5L-PCAF double-mutant animals, however, was more severe than that of Gcn5L single mutants. This suggested that both mouse Gcn5 relatives have distinct and partially overlapping functions during embryogenesis. Intriguingly, the defects in Gcn5L-defective mice were rather specific and affected dorsal mesoderm cells (53). It therefore appears likely that GNAT complexes play critical roles in the regulation of certain developmental processes in higher eukaryotes. However, the deletion of Gcn5L affected all classes of GNAT complexes, and it is not yet clear whether certain subtypes of GNAT complexes are more critical for the developing embryo. So far, only SAGA-type complexes have been described in mammals. It remains unclear whether higher eukaryotes also have ADA or HAT A2-type GNAT complexes.
To address the roles of GNAT complexes in higher eukaryotes, our laboratory has pursued GNAT complexes from Drosophila melanogaster. In contrast to mammals, Drosophila appears to have only one copy of Gcn5 (42), and we sought to identify the Drosophila homologues of dGcn5-associated factors. We characterized the Drosophila homologues of Ada2, Ada3, Spt3, and Tra1 and examined their relationships with dGcn5 and each other. We found that Drosophila has two distinct Ada2 genes. In a combination of biochemical and cell biological studies, we demonstrated that the two Drosophila Ada2 homologues are in functionally and structurally distinct multiprotein complexes. One of the Ada2 homologues, dAda2b, is associated with SAGA-type GNAT complexes. The second variant, dAda2a, is part of Gcn5-containing as well as Gcn5-independent complexes. Finally, we found that mice and humans also possess homologues of dAda2a and dAda2b, respectively. This suggests that mammals and insects have two distinct subgroups of Ada2, which characterize functionally distinct multiprotein complexes involved in transcriptional regulation.
MATERIALS AND METHODS
Characterization of GNAT subunits and generation of antisera.
DNA sequences with significant homology to known subunits of GNAT complexes were identified with the help of the BLASTN, TBLASTN, and PSI- and PHI-BLAST sequence alignment programs (3). Multiple sequence alignments were compiled with the help of CLUSTAL W version 1.8 (47). Phylogenetic studies were performed using PHYLIP (21). The accession numbers of sequences used for comparative studies are available upon request. The full-length open reading frames of dAda2a, dAda2b, dSpt3, and dAda3 were amplified with the help of the Prostar Ultra-HF reverse transcription (RT)-PCR kit (Stratagene) in three reactions using total RNAs from 4- to 8-h-old embryos, 12- to 16-h-old embryos, and third-instar larvae (the primer sequences are available upon request). Since the full-length open reading frame of dTra1 could not be amplified in total, we generated four partially overlapping clones from the same first-strand synthesis reaction. For dAda2b, the 3′ end of the open reading frame was obtained with help of the 3′-RACE kit (Roche). For the generation of antibodies against Drosophila HAT components, the full-length open reading frames (for dGcn5 and dAda3) or parts (for dAda2a, amino acids [aa] 175 to 518; for dAda2b, aa 1 to 241; for dSpt3, aa 63 to 426; for dTra1, aa 183 to 465) were cloned into the pQE12 or pQE31 vector (Qiagen). The resulting His6-tagged fusion polypeptides were expressed in bacteria and purified over nickel nitrilotriacetic acid-agarose (Qiagen) under denaturing conditions following the manufacturer's instructions. The proteins were solubilized by dialysis prior to immunization.
Cell culture and generation of nuclear extracts.
Drosophila S2 cells (40) were grown in Schneider cell medium supplemented with 10% fetal calf serum (Gibco BRL). Nuclear extracts from 0- to 12-h-old Oregon R embryos were prepared as previously described (6). For the generation of S2 nuclear extracts, the cells were grown in 500-ml suspensions and were harvested at a density of 3 × 106 cells/ml. All steps were performed on ice or with ice-cold solutions. The cells were washed twice in 10 mM HEPES (pH 7.4)-1 mM dithiothreitol (DTT)-150 mM NaCl and once in cell lysis buffer (20 mM HEPES, pH 7.4, 0.1% Triton X-100, 1 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.1% [wt/vol] leupeptin, 0.2% [wt/vol] pepstatin A). The cells were lysed in 40 ml of lysis buffer by 30 to 40 strokes of a Dounce homogenizer (loose pestle; Wheaton). Cell lysis was monitored with the help of trypan blue staining. The nuclei were washed twice in cell lysis buffer by centrifugation at 2,000 × g for 10 min. The nuclei were extracted for 1 h in 2 ml of nuclear extraction buffer (20 mM HEPES, pH 7.4, 10% glycerol, 350 mM NaCl, 0.1% Triton X-100, 1 mM DTT, 0.5 mM PMSF, 0.1% [wt/vol] leupeptin, 0.2% [wt/vol] pepstatin A) at 4°C, and the extracts were cleared from the nuclear debris by centrifugation at 20,800 × g for 10 min at 4°C. The protein concentration was determined with the help of Bradford assays (Bio-Rad), and the nuclear extracts were adjusted with extraction buffer to a protein concentration of 5 mg/ml.
Chromatographic methods.
For anion-exchange chromatography, 5 mg of nuclear extract was adjusted to a salt concentration of 100 mM with ion-exchange buffer (20 mM HEPES, pH 7.8, 0.1% Triton X-100, 1 mM DTT, 1 mM PMSF, 0.1% [wt/vol] leupeptin, 0.2% [wt/vol] pepstatin A, 10% glycerol) and was applied to a MonoQ HR 5/5 column (Pharmacia). Bound proteins were eluted by a 25-ml linear gradient from 100 to 450 mM NaCl in ion-exchange buffer; 10 μl of each 500-μl fraction was used for Western blotting. Sephacryl HR S400 1.6/70 and Mini-Superose 6 PC 3.2/30 columns were used for size exclusion chromatography on ÄKTA and SMART systems (Pharmacia), respectively. The columns were calibrated with a molecular mass calibration kit (blue dextran 2000, 2 MDa; thyroglobulin, 669 kDa; ferritin, 440 kDa; catalase, 232 kDa; Pharmacia). For size exclusion chromatography on the Superose 6 column, the dAda2b/A peak fractions from the anion-exchange chromatography were pooled, dialyzed against size exclusion buffer (20 mM HEPES, pH 7.6, 350 mM NaCl, 0.1% Triton X-100, 1 mM DTT, 1 mM PMSF, 0.1% [wt/vol] leupeptin, 0.2% [wt/vol] pepstatin A, 5% glycerol, 4 μg of ethidium bromide/ml) and concentrated to a final volume of 50 μl with the help of Ultrafree-4 concentrators (Millipore). Twenty-five microliters of each 50-μl fraction was used for immunodetection assays, 5 mg of total nuclear extract was fractionated in size exclusion buffer on the S400 column, and 15 μl of each 750-μl fraction was used for immunodetection assays.
Protein interaction studies and Western blotting.
The following crude antisera were used for immunoblotting assays: anti-dGcn5 (rabbit, 1:4,000), anti-dAda3 (rabbit, 1:4,000), anti-dSpt3 (rabbit, 1:3,000), anti-dAda2b (rat, 1:1,500; rabbit, 1:2,000), rabbit anti-dAda2a (1:2,500), rabbit anti-dTra1 (1:2,000), anti-tubulin (mouse monoclonal, 1:2,000; Sigma), anti-phosphorylated RNA polymerase II (1:1,000; mouse monoclonal, clones H5 and H14; Covance); anti-dmTaf5 and anti-dmTaf9 (each 1:1,000; rabbit; generously provided by Y. Nakatani); and anti-dmTaf10 (1:400; rabbit; generously provided by L. Tora). For coimmunoprecipitation assays, 2 μl of the affinity-purified antibodies was conjugated to 10 μl of equilibrated protein A-Sepharose resin (Pharmacia). The conjugate was incubated with 300 μg of nuclear extract for 1 h at room temperature in 500 μl of immunoprecipitation buffer (20 mM HEPES, pH 7.6, 350 mM NaCl, 0.1% Triton X-100, 1 mM DTT, 1 mM PMSF, 0.1% [wt/vol] leupeptin, 0.2% [wt/vol] pepstatin A, 10% glycerol, 50 μg of ethidium bromide/ml). Low-stringency interaction studies (rabbit anti-Dmp53 antibodies, 1:1,000; generously provided by Exelixis, Inc.) were performed in immunoprecipitation buffer containing 175 mM NaCl. The immunoprecipitates were washed at least three times for 10 min at room temperature in immunoprecipitation buffer prior to immunoblotting or HAT assays. Glutathione (GSH) S-transferase (GST) fusion proteins were generated following standard protocols (Pharmacia). For interaction studies, 20 fmol of GST fusion protein was incubated with 300 μg of S2 nuclear extract in 500 μl of immunoprecipitation buffer for 1 h at room temperature; 2.5 μl of GSH-Sepharose beads (Pharmacia) was added to the solution, and the binding reaction was performed for a further 30 min at room temperature. The beads were washed at least three times for 10 min each time with 500 μl of immunoprecipitation buffer prior to Western blotting analysis. For developmental expression studies, Oregon R embryos were collected at room temperature and dechorionated to obtain lysates from the following developmental stages: blastoderm (0 to 3 h), gastrulation-germ band extension (0 to 7 h), germ band retraction (7 to 13 h), and late embryogenesis-differentiation (13 to 24 h). Embryos, larvae, pupae, and adult male and female flies were homogenized in sodium dodecyl sulfate (SDS) sample buffer. Aliquots of the lysates were separated by SDS-polyacrylamide gel electrophoresis (PAGE) followed by immunoblotting assays.
HAT assays.
HAT assays were performed essentially as previously published (18). In brief, the immunoprecipitates were split in half, and each half was incubated for 30 min at room temperature with 1 μg of histones in 50 μl of reaction buffer containing 0.05 μCi of [3H]acetyl coenzyme A (7.2 Ci/mmol; Amersham). For HAT assays with core histones (ch) as substrates, a ch-HAT buffer (50 mM Tris-HCl, pH 7.5, 50 mM KCl, 5% glycerol, 1 mM DTT, 0.5 mM PMSF, 0.1% [wt/vol] leupeptin, 0.2% [wt/vol] pepstatin A, 0.1 mM trichostatin A) was used. HAT assays with nucleosomes (nuc) were performed using a nuc-HAT buffer (20 mM HEPES, pH 7.6, 50 mM KCl, 5% glycerol, 1 mM DTT, 0.5 mM PMSF, 0.1% [wt/vol] leupeptin, 0.2% [wt/vol] pepstatin A, 0.1 mM trichostatin A); 25 μl of each sample was subjected to liquid scintillation counting, and the remaining samples were separated in SDS-PAGE on 18% polyacrylamide gels. The gels were treated with En3Hance (NEN Research Products) prior to exposure on X-ray films. Total nuclear extract (0.5 to 1 μg) and 200 ng of C-terminally His6-tagged recombinant dGcn5 (rdGcn5) were used as positive controls.
Immunohistochemical methods.
Polytene chromosomes were dissected from actively moving third-instar Oregon R larvae; they were either fixed for 3 min in fixative A (4% formaldehyde, 45% acetic acid) or prefixed for 30 s in fixative B (4% formaldehyde in phosphate-buffered saline [PBS], pH 7.4) followed by fixation for 3 min in fixative A. No substantial differences between the different fixation methods were observed. The chromosomes were spread on poly-l-lysine-coated microscope slides. The slides were blocked for 1 h at room temperature in PBS (pH 7.4)-0.3% Triton X-100- 2% bovine serum albumin (PBTB). The following affinity-purified antibodies were used: dAda3 (rabbit, 1:50), dAda2b (rat, 1:7.5; rabbit, 1:10), dAda2a (rabbit, rat, 1:7.5), and dGcn5 (rabbit, 1:25; rat, 1:15) and anti-phosphorylated RNA-polymerase II (mouse monoclonal, 1:40; combination of clones H5 and H14; Covance). The supernatants of the affinity purifications or preimmunization bleeds did not show any nonspecific reactivity in control stainings of polytene chromosomes. We were not able to detect dSpt3 and dTra1 on polytene chromosomes, presumably because these epitopes might be inaccessible upon chromatin association of dSAGA complexes. Primary antibodies were incubated in PBTB at 4°C overnight or for 1 h at room temperature. The DNA was labeled with 4-6-diamidino-2-phenylindole dihydrochloride (DAPI; stock solution, 10 μg/ml in methanol; final concentration, 1:2,000 in PBS-0.3% Triton X-100) for 5 min at room temperature. The slides were washed two times for 15 min each time in PBS-0.3% Triton X-100 followed by incubation of the secondary antibodies in PBTB (fluorescein isothiocyanate- or Texas Red-conjugated goat anti-rabbit immunoglobulin G [IgG; 1:100] and horse anti-mouse IgG [1:100] [Vector] antisera, Cy2-conjugated goat anti-rabbit IgG and donkey anti-rat IgG antibodies [1:100; Jackson], and Alexa 594-conjugated goat anti-rabbit IgG [1:250] and goat anti-rat IgG [1:150] [Molecular Probes] antibodies). The secondary antibodies were incubated for 1 h at room temperature, followed by two washes for 15 min each time in PBS (pH 7.4)-0.5% Tween 20- 0.5% NP-40- 300 mM NaCl followed by two washes in PBS (pH 7.4)-0.5% Tween 20- 0.5% NP-40- 400 mM NaCl. The preparations were embedded in Vectashield mounting medium (Vector) prior to microscopy on an Axioplan II microscope (Zeiss, Thornwood, N.Y.). Photographs were taken with an Axiocam digital color camera using Axiovision software (Zeiss). In situ detection of proteins or mRNA in whole-mount embryos was performed as previously described (31).
Nucleotide sequence accession numbers.
The GenBank accession numbers of the Drosophila factors are as follows: AY 142213 (dAda2b), AY 142214 (dAda3), AY 142215 (dSpt3), AY 142216 (dAda2a), and AY 142217 (dTra1).
RESULTS
Characterization of the dGcn5-associated factors dAda3, dAda2a, dAda2b, dSpt3, and dTra1.
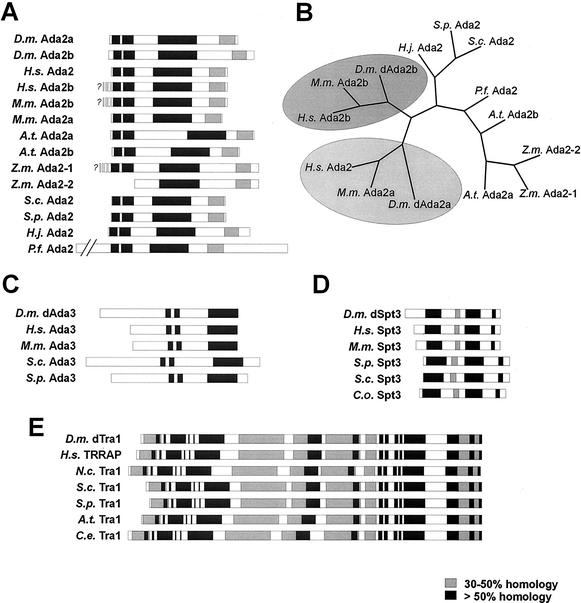
In an attempt to characterize putative SAGA components from flies, we searched Drosophila genomic and expressed sequence tag (EST) databases for coding regions with homology to human Ada3, Ada2, Spt3, and TRRAP. The corresponding open reading frames were obtained by RT-PCR. Intriguingly, two distinct genomic loci (accession numbers AA698620 [BACR clone 48I13] and AC006495 [EST LD24527]), which we refer to as dAda2a and dAda2b, respectively, showed high homology to human Ada2 (Fig. 1A). The dAda2a 1,467-base open reading frame encodes a 557-aa polypeptide with a predicted molecular mass of 59.9 kDa. The dAda2b open reading frame consists of 1,668 bases and codes for a 556-aa protein with a molecular mass of 62 kDa. In protein database searches, each of the translated protein sequences retrieved Ada2 proteins from other organisms as the strongest matches. Both fly Ada2 variants show high conservation within three regions that are conserved among other Ada2 proteins (Fig. 1A). In addition, the multiple sequence alignment revealed that all Ada2 homologues show weak homology within the carboxy terminus. When we searched the human genome databases using either of the Drosophila Ada2 variants, we found a second open reading frame with 40% amino acid sequence identity to dAda2b (Fig. 1A, H.s. Ada2b). The novel human Ada2b-like locus is located on chromosome 4, whereas the initially characterized Ada2 locus was mapped to chromosome 17 (16). The human Ada2 and fly dAda2a are ∼33% identical in their amino acid sequences but are <25% identical to the dAda2b variants from flies and humans. Mice also have two Ada2 variants with high homology to either of the fly Ada2 proteins (Fig. 1A, M.m. Ada2a [33% identical to dAda2a] and M.m. Ada2b [39% identical to dAda2b]). Maize appears to have two Ada2 variants, too (Fig. 1A and B, Z.m. Ada2-1 and Z.m. Ada2-2). It has recently been demonstrated that Arabidopsis possesses two distinct Ada2 variants (46). In contrast to the human and murine Ada2 variants, the plant Ada2 paralogues do not exhibit significantly higher similarity to either of the fly Ada2 proteins. Taken together, plants, flies, mice, and humans possess two Ada2 variants, suggesting that higher eukaryotes possess two distinct Ada2 proteins.
FIG. 1.
Homology plots of the Drosophila GNAT subunits dAda2a and dAda2b (A), dAda3 (C), dSpt3 (D), and dTra1 (E). (B) Unrooted phylogenetic tree generated with the CLUSTAL W (47) and PHYLIP (21) programs of the Ada2 homologues from various species (bootstrap value, 100; 15 repetitions). The distinct Ada2 variants from human, mouse, and Drosophila lie within the same branch (shaded ovals). Abbreviations: D.m., D. melanogaster; H.s., Homo sapiens; M.m., Mus musculus; A.t., Arabidopsis thaliana; Z.m., Zea mais; S.c., Saccharomyces cerevisiae; S.p., Schizosaccharomyces pombe; P.f., Plasmodium falciparum; C.o., Clavispora opuntiae; C.e., C. elegans; H.j., Hypocrea jecorina.
Amino acid sequence homology comparisons usually do not reveal subtle similarities between proteins with rather broad homology, such as the Ada2 proteins. We therefore performed phylogenetic comparisons between various Ada2 proteins in order to determine whether they might fall into different subgroups based on subtle sequence similarities (21). As shown in Fig. 1B, the second human Ada2 variant (H.s. Ada2b), as well as the mouse Ada2b, falls within the same branch of a phylogenetic tree as the fly dAda2b variant. This suggests that these three Ada2 proteins might form a distinct subgroup. Intriguingly, the initially characterized human Ada2 and the mouse Ada2a protein group with dAda2a. The distinct Ada2 variants from plants do not show higher homology to either of the two Ada2 variants from metazoans.
We also found a fly homologue of Ada3 (dAda3, 557 aa, 59.9 kDa; EST LD27205, AA941883; BACR clone 48205, AC011614) (Fig. 1C) which shows homology in three conserved domains with Ada3 homologues from fungi and mammals. Furthermore, we identified the fly Spt3 homologue dSpt3 (427 aa, 48.1 kDa; BACR06O005, AC007692) (Fig. 1D) and the highly conserved fly TRRAP/Tra1 homologue dTra1 (3,804 aa, 436.2 kDa; EST LD40728, AW941980; genomic clone AC005712) (Fig. 1E). RT-PCRs from three different stages of development (4- to 8- and 12- to 16-h-old embryos and third-instar larvae) did not display any significant differences in signal intensities, suggesting that the mRNAs for dAda2b, dAda2a, dTra1, dSpt3, and dAda3 are constitutively and ubiquitously expressed (not shown). Furthermore, in situ hybridizations of whole-mount embryos with digoxigenin-labeled cDNA probes suggested that mRNAs coding for these factors are maternally loaded and ubiquitously expressed throughout embryonic development (not shown). Taken together, the existence of the transcriptional adapter proteins Ada2 and Ada3 suggests that Drosophila might possess GNAT complexes similar to the ones from yeast and humans. Furthermore, the existence of homologues of Spt3 and Tra1 in Drosophila indicates that flies might have SAGA-type GNAT complexes, since these two proteins are characteristic subunits of SAGA-type complexes from yeast and humans.
dAda3, dAda2a, dAda2b, dSpt3, and dTra1 are associated with dGcn5.
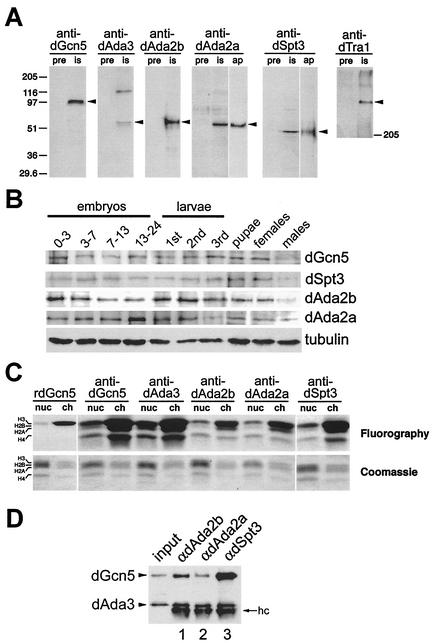
To study the relationship between dGcn5 and its putative interaction partners, we raised antisera against dGcn5, dAda3, dSpt3, dTra1, and the two distinct dAda2 variants (Fig. 2). The antisera were successfully tested for their specificities on nuclear extracts from an embryonic cell line (S2 cells [40]) and recognized proteins of the predicted molecular masses (Fig. 2A). The antisera against dAda2a and anti-dSpt3, respectively, appeared to recognize a cross-reactive band of similar size (Fig. 2A, lanes is). This band was not detectable in blots probed with the affinity-purified antibodies (Fig. 2A, lanes ap).
FIG. 2.
Characterization of the Drosophila GNAT subunits dAda2a, dAda2b, dAda3, dSpt3, and dTra1. (A) The antisera (is, immune serum) recognize proteins with the predicted molecular masses of dGcn5, dAda2b, dAda2a, dAda3, dSpt3, and dTra1 (arrowheads). Each lane contains 25 μg of nuclear extract. pre, preimmunization bleeds. Crude antisera against dAda2a and Spt3 detect a cross-reactive band of similar molecular mass, which is not recognized by affinity-purified antibodies (ap). (B) Developmental expression of dGcn5, dSpt3, dAda2a, and dAda2b. Lysates from Oregon R animals were electrophoresed on an SDS-10% polyacrylamide gel and analyzed by Western blotting. Embryonic lysates: 0-3 (blastoderm), 0-7 (gastrulation), 7-13 (germ band retraction), and 13-24 (differentiation). Larval stages: first, second, and third instars. Antibodies against tubulin served as loading controls. (C) HAT activity of the immunoprecipitates from 300 μg of S2 nuclear extract on nucleosomes (nuc) and core histones (ch), respectively. dAda3, dAda2a, dAda2b, and dSpt3 are associated with a dGcn5-specific HAT activity. Bacterially expressed rdGcn5 was used as a control. Top, fluorogram; bottom, photography of the Coomassie brilliant blue-stained SDS-polyacrylamide gel. (D) Western blot probed for dAda3 and dGcn5 (arrowheads). Input, 30 μg of nuclear extract; lane 1, immunoprecipitates from anti-dAda2b antibodies; lane 2, immunoprecipitates from anti-dAda2a; lane 3, immunoprecipitates from anti-dSpt3. hc, IgG heavy chain.
The embryonic mRNA coding for dGcn5, dSpt3, dAda3, dAda2a, dAda2b, and dTra1 was maternally deposited and ubiquitously expressed. In order to confirm and extend these results in more detailed analyses, we immunoblotted total cell lysates from different developmental stages for dGcn5, dSpt3, dAda2a, and dAda2b (Fig. 2B). We found that the concentrations of dGcn5, dSpt3, and dAda2b are slightly higher during the blastoderm stage than during the following developmental stages (Fig. 2B, lane 0-3). Since zygotic gene expression starts at the end of the blastoderm stage, these signals most likely represent maternally loaded protein. Immunodetection assays in whole-mount embryos confirmed that these proteins indeed are maternally loaded (not shown). During the following embryonic developmental stages, the protein levels of dGcn5, dSpt3, and dAda2b do not significantly change. Also, during larval and pupal development, the levels of these proteins are relatively constant. A remarkable difference was observed between adult male and female flies. While the levels of dGcn5, dSpt3, and dAda2b proteins in adult females are comparable to those of developing animals, significantly lower levels of these factors were detected in adult males. Although the reason for this difference remains unclear, it is tempting to speculate that higher concentrations of these factors might be required during oogenesis. Changes in the concentration of dAda2a are slightly different from those for dGcn5, dSpt3, and dAda2b. For instance, dAda2a levels increase slightly from 13 to 24 h of development. During this phase, most of the embryonic differentiation processes occur. During third-instar larval development, the dAda2a levels are lower than those in earlier larval stages. Finally, the differences between the dAda2a protein levels in males and females are rather minute. Taken together, dGcn5 and other candidate subunits of GNAT complexes appear to be present throughout the entire life cycle of Drosophila.
Gcn5 homologues from yeast and other eukaryotes exhibit similar substrate specificities (for a review, see reference 11). As recombinant proteins, all Gcn5 homologues so far characterized acetylated the free core histone H3. Only when associated with other GNAT subunits was Gcn5 able to utilize nucleosomal histone H3 as a substrate (4). In addition, some GNAT complexes were able to acetylate the histones H2B and H4, although to a lesser extent than H3. Thus, testing immunoprecipitates from nuclear extracts for nucleosomal HAT activity would allow us to determine whether the antisera against Drosophila GNAT subunits were capable of immunoprecipitating GNAT complexes. First, we determined whether the anti-dGcn5 antibody would immunoprecipitate a Gcn5-like enzymatic activity. A bacterially expressed recombinant dGcn5, which can acetylate only free histone H3, was used as a control (Fig. 2C, rdGcn5) (42). In contrast to rdGcn5, the immunoprecipitates from anti-dGcn5 antibodies acetylated nucleosomal H3 and also recognized the histones H2B and H4 as substrates (Fig. 2C). This suggested that the anti-dGcn5 antiserum recognized dGcn5 and was able to immunoprecipitate GNAT complexes with HAT activity from S2 nuclear extract. Consistent with this possibility, the antisera against dAda3, dSpt3, dAda2a, and dAda2b also coimmunoprecipitated a HAT activity with similar histone substrate preference. This suggests that all these proteins are associated with dGcn5 (Fig. 2C). These results confirm the specificities of the antisera and indicate that these antisera do not significantly disrupt or inhibit the immunoprecipitated GNAT complexes. The antiserum against dTra1, however, did not immunoprecipitate any HAT activities (not shown). We assume that this antiserum might fail to recognize native dTra1 or that the epitope which was used for the generation of the antibodies might be masked within the context of HAT complexes.
We further confirmed the association of dSpt3, dAda2a, and dAda2b with dGcn5 by probing Western blots of immunoprecipitates with antibodies against dGcn5. All GNAT complexes so far characterized contained Ada3 (for a review, see reference 11). Thus, by simultaneously probing the blotting membranes for dAda3 and dGcn5, it would be possible to monitor any dissociation of these proteins during the immunoprecipitation experiments. As shown in Fig. 2D, antibodies against dAda2b (lane 1), dAda2a (lane 2), and dSpt3 (lane 3) immunoprecipitated dAda3 and dGcn5 from S2 nuclear extracts.
Thus, all three factors were stably associated with dGcn5 and dAda3. However, antibodies against dSpt3 precipitate much higher levels of dGcn5 from nuclear extracts, whereas the antisera against the two Ada2 proteins, in particular anti-dAda2a antibodies, appear to partially disrupt the interactions of dGcn5 and dAda3, respectively. In addition, antibodies against the two Ada2 homologues immunoprecipitated weaker HAT activities than antisera against dSpt3 or dAda3 did (see Fig. 2C). It has been suggested that the yeast Ada2 protein interacts with both Ada3 and Gcn5 (13), making it likely that a fraction of the polyclonal antibodies reduce the interactions between the fly Ada2 homologues and dAda3 or dGcn5.
dAda2b, but not dAda2a, associates with proteins that are characteristic of dSAGA-type complexes.
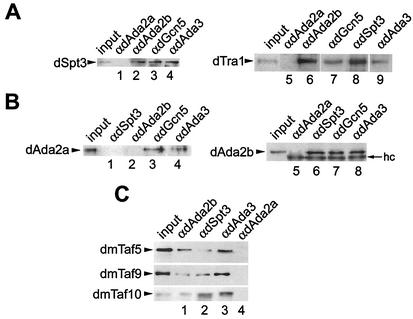
We next wished to address any functional differences between the two fly Ada2 variants. In a first approach, we tested in coimmunoprecipitation assays whether each of the fly Ada2 homologues associates with the SAGA-specific subunits dSpt3 and dTra1. To our surprise, only dAda2b, dGcn5, and dAda3 coimmunoprecipitated with these proteins (Fig. 3A, lanes 2 to 4 and 6 to 9); antibodies against dAda2a failed to immunoprecipitate dSpt3 and dTra1 (Fig. 3A, lanes 1 and 5). Also, in reciprocal immunoprecipitation experiments, the dAda2a variant did not interact with dSpt3 (Fig. 3B, lane 1). We also wished to address whether dAda2b and dAda2a might coexist in non-SAGA-type GNAT complexes. However, the two fly Ada2 variants did not interact in coimmunoprecipitation assays (Fig. 3B, lanes 2 and 5).
FIG. 3.
dAda2b, but not dAda2a, associates with dmTaf5, dmTaf9, dmTaf10, dSpt3, and dTra1. Affinity-purified antibodies were conjugated to protein A-Sepharose beads and were used in immunoprecipitation assays of 300 μg of nuclear extract from S2 cells. Input, 30 μg of nuclear extract. (A) Western blots immunoprobed for dSpt3 (left) and dTra1 (right). Lane 1, immunoprecipitates from anti-dAda2a (αdAda2a); lane 2, immunoprecipitates from anti-dAda2b; lane 3, immunoprecipitates from anti-dGcn5; lane 4, immunoprecipitates from anti-dAda3; lane 5, immunoprecipitates from anti-dAda2a; lane 6, immunoprecipitates from anti-dAda2b. (B) Western blots probed for dAda2a (left) and dAda2b (right). Lane 1, immunoprecipitates from anti-dSpt3; lane 2, immunoprecipitates from anti-dAda2b; lane 3, immunoprecipitates from anti-dGcn5; lane 4, immunoprecipitates from anti-dAda3; lane 5, immunoprecipitates from anti-dAda2a; lane 6, immunoprecipitates from anti-dSpt3; lane 7, immunoprecipitates from anti-dGcn5; lane 8, immunoprecipitates from anti-dAda3. hc, IgG heavy chain. (C) Western blots immunolabeled with antibodies against dmTaf5, dmTaf9, and dmTaf10. Lane 1, immunoprecipitates from anti-dAda2b; lane 2, immunoprecipitates from anti-dSpt3; lane 3, immunoprecipitates from anti-dAda3; lane 4, immunoprecipitates from anti-dAda2a.
A number of Tafs have been identified as subunits of SAGA-type GNAT complexes from yeast and humans (for a review, see references 11, 35, and 46). In Drosophila, several Tafs show sequence homology with typical SAGA Tafs from yeast and humans, and it is possible that these Tafs are also components of fly SAGA complexes. Three of these candidate dSAGA-associated Tafs are dmTaf5, dmTaf9, and dmTaf10 (for a review, see reference 48), and we tested whether these Tafs coimmunoprecipitate from nuclear extracts with dAda3, dSpt3, and the two dAda2s. Indeed, we were able to detect all three fly Tafs in the immunoprecipitates from antibodies against dAda2b (Fig. 3C, lane 1), dSpt3 (Fig. 3C, lane 2), and dAda3 (Fig. 3C, lane 3), whereas antibodies against dAda2a failed to coimmunoprecipitate any of these Tafs (Fig. 3C, lane 4). We conclude that dmTaf5, dmTaf9, and dmTaf10 are components of SAGA-type complexes in Drosophila, as has been reported for their yeast and human counterparts. Taken together, these results strongly suggest that only the Ada2b variant is a component of dSAGA-type complexes, whereas dAda2a did not associate with proteins that are characteristic of SAGA-type complexes, such as dSpt3, Tafs, and dTra1. Ada2a interacted with dAda3 and dGcn5 in coimmunoprecipitation assays (Fig. 2D, lane 2; Fig. 3B, lanes 3 and 4), and antibodies against dAda2a immunoprecipitated a dGcn5-specific HAT activity (Fig. 2B). We conclude that dAda2a is a component of dGcn5-containing complexes other than SAGA-type GNATs. Possible candidates for such complexes might be the Drosophila relatives of the yeast ADA or HAT A2 complex (8, 19, 22, 39).
dAda2a is not solely associated with dGcn5-containing multiprotein complexes.
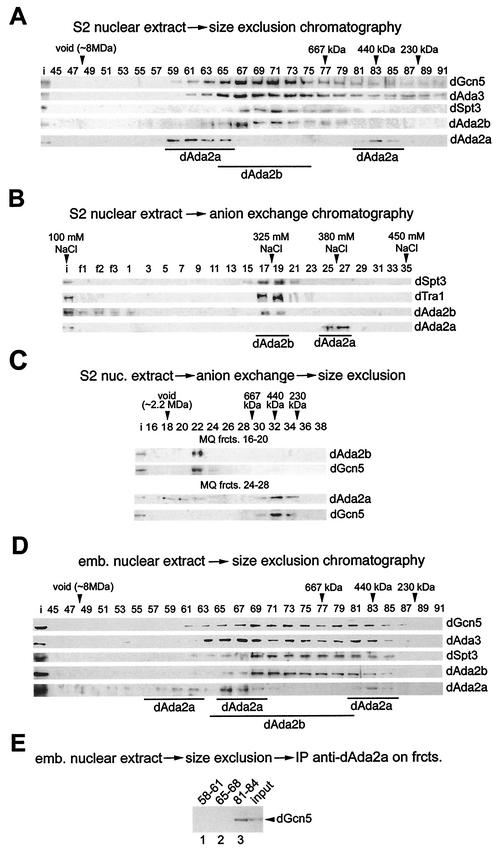
SAGA-type HAT complexes from yeast and humans are ∼1.8 to 2 MDa in mass, whereas the non-SAGA-type ADA and A2 complexes from yeast are significantly smaller and have estimated molecular masses of 800 and 200 kDa, respectively (11, 33). To better understand the sizes and complexity of distinct Drosophila GNAT complexes, we fractionated nuclear extracts from S2 cells on a Sephacryl S400 size exclusion column. The resulting fractions were then immunoprobed for dGcn5, dAda3, dSpt3, dAda2b, and dAda2a (Fig. 4A). dGcn5 and dAda3 coeluted in a rather broad profile from ∼2 MDa to 200 kDa. Ada3 is contained within all GNAT-type complexes so far characterized, and the elution profiles of the fly dGcn5 and dAda3 proteins suggest that flies might possess several GNAT complexes with different functions and compositions. dAda2b and dSpt3 coeluted in a molecular mass range between 2 and 1 MDa, supporting the notion that these two proteins might be components of larger GNAT complexes, such as SAGA-type complexes. dAda2a, however, eluted in two peaks with the approximate molecular masses of 2.5 MDa and 440 kDa. This suggests that dAda2a is part of at least two molecular mass complexes. Note that the larger dAda2a-containing complexes eluted in a much broader peak, which indicates that these complexes might be more abundant than the 440-kDa dAda2a complexes.
FIG. 4.
dAda2b associates with dSAGA-specific subunits in biochemical fractionation assays. (A) Elution profiles of dGcn5, dAda3, dSpt3, dAda2b, and dAda2a from a Sephacryl S400 gel filtration column; 5 mg of S2 nuclear extract was separated on the column, and 15 μl of each fraction was subjected to SDS-PAGE, followed by Western blotting (fraction numbers indicated at the top). dGcn5 and dAda3 coelute in a range from ∼2.2 MDa to 300 kDa. dAda2b coelutes with dSpt3 in a range of ∼2 MDa to 800 kDa. dAda2a elutes in two peaks around 2.5 MDa and 440 kDa. i, input (30 μg of embryonic nuclear extract). (B) dSpt3, and dTra1 cofractionate with dAda2b, but not dAda2a, in anion-exchange chromatography. Bound proteins were eluted in a linear gradient of 100 to 450 mM sodium chloride from a 1-ml MonoQ anion-exchange column. The salt concentrations of the peak fractions are indicated above the blot. Fifteen microliters of each fraction was subjected to SDS-PAGE, followed by Western blotting (fraction numbers indicated at the top; f1 to f3, flowthrough). (C) MonoQ (MQ) fractions (frcts.) 16 to 20 (top) and 24 to 28 (bottom) of the anion-exchange chromatography were concentrated and subjected to size exclusion chromatography. dAda2b and dGcn5 coelute in a range of ∼1.8 MDa. dAda2a elutes in fractions of >2 MDa and ∼450 kDa. dGcn5 is detectable only in fractions containing the smaller dAda2a complex. nuc., nuclear. (D) Gel filtration analysis of nuclear extract from 0- to 12-h-old embryos as described for panel A. A third dAda2a peak is observed in a molecular-mass range of ∼1.8 MDa. (E) Pooled dAda2a peak fractions from embryonic extracts (panel D) were subjected to immunoprecipitation assays with affinity-purified anti-dAda2a antibodies. The immunoprecipitates (IP) were separated by SDS-PAGE and were probed for dGcn5. Lane 1, fractions 58 to 61; lane 2, fractions 65 to 68; lane 3, fractions 81 to 84.
We next subjected S2 nuclear extracts to a 100- to 450-mM salt gradient on a MonoQ anion-exchange column. As shown in Fig. 4B, dAda2b coeluted with both dSpt3 and dTra1 at salt concentrations of ∼325 mM sodium chloride, whereas dAda2a-containing complexes eluted at higher salt concentrations of ∼380 mM. Consistent with the coimmunoprecipitation assays, none of the dAda2a peak fractions contained detectable amounts of dSpt3, dTra1, or dAda2b (Fig. 3). These findings further illustrate that dAda2a is not associated with dSAGA-type complexes.
The Drosophila S2 cell lines have been reported to derive from 16- to 24-h-old male embryos (40, 43), and it is possible that these cells contain only a portion of chromatin-remodeling complexes. To test this, we also analyzed nuclear extracts prepared from 0- to 12-h-old wild-type embryos (Fig. 4D). The elution profiles of dGcn5, dAda3, dSpt3, and dAda2b were comparable to those from S2 cells, although higher levels of dAda2b and dSpt3 were also found in the lower-molecular-mass range. For dAda2a, however, we detected an additional peak around 1.8 MDa, which we could not detect in S2 nuclear extracts. It remains unclear whether these fractions represent partially degraded 2.5-MDa complexes or a third cell-type- or stage-specific complex.
We next wished to test which of the dAda2a complexes from embryos contain dGcn5, particularly since the second-largest dAda2a peak from the size exclusion chromatography of embryonic extracts coeluted with dSpt3, dAda2S, dAda3, and dGcn5 (Fig. 4D). For this purpose, we performed coimmunoprecipitation assays with the pooled dAda2a peak fractions from the S400 size exclusion chromatography (Fig. 4D, fractions 58 to 61, 65 to 68, and 81 to 84). When immunoprecipitates from anti-dAda2a antibodies against dGcn5 were probed, the protein was detectable only in the low-molecular-mass dAda2a peak fractions (Fig. 4E, lane 3) and not in the two high-molecular-mass peak fractions (Fig. 4E, lanes 1 and 2).
Thus, dAda2a is a component of at least two complexes, a 440-kDa complex containing dAda3 and dGcn5 and a complex of ∼2.5 MDa lacking these two proteins; 0- to 12-h-old embryos possess a third 1.8-MDa complex. It remains unclear whether this third ∼1.8-MDa dAda2a complex is specific for this developmental stage or is missing in S2 cells or whether these fractions contain a partially degraded 2.5-MDa dAda2a-containing complex.
dAda2b-containing GNAT complexes interact with the transcription activators VP16 and Dmp53.
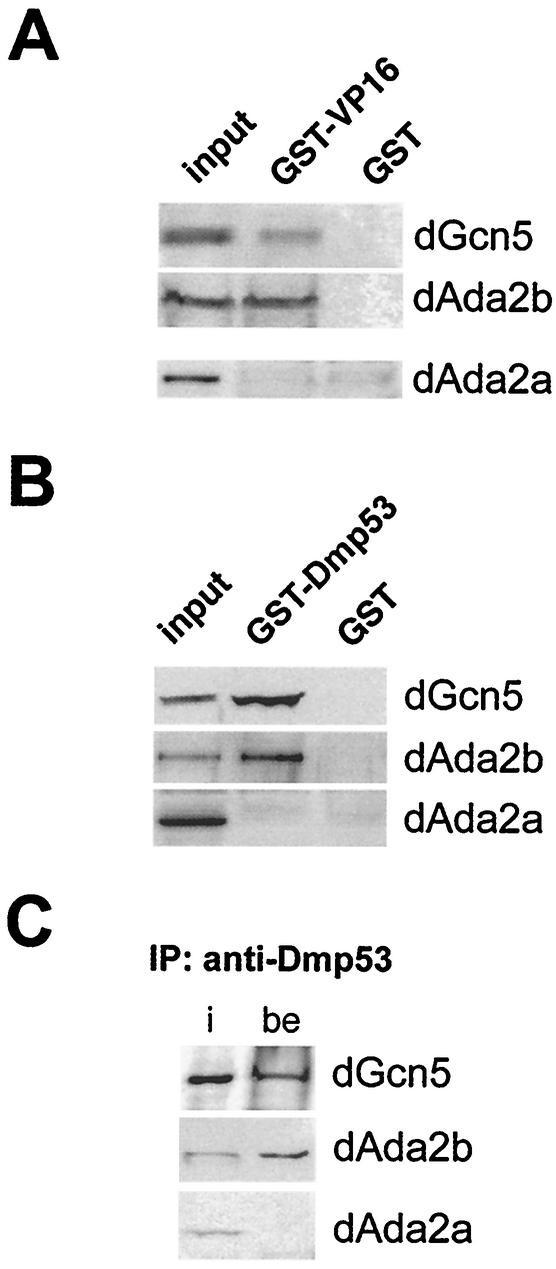
It has been shown that strong transcription activators containing acidic activation domains, such as the herpes simplex virus activator VP16, interact with the yeast SAGA complex but not with the ADA complex (27, 49). It has recently been demonstrated that the yeast SAGA subunit Tra1 contributes to this interaction (10). Tra1 and Spt3 are subunits of yeast SAGA-like complexes. Accordingly, one would expect that Drosophila dSAGA-type complexes, which should contain the dSAGA-specific subunit dAda2b, exhibit similar functional properties. We therefore tested whether dAda2b-associated complexes interact with transcription activators such as VP16 in pull-down experiments. Moreover, since we did not observe any association of dAda2a with dTra1 (Fig. 5A), one would expect that dAda2a-containing complexes would fail to interact with acidic activators.
FIG. 5.
VP16 and Dmp53 interact with dAda2b-containing GNAT complexes. (A) GST pull-down assays were performed with GST-VP16 or GST alone bound to GSH-Sepharose beads and 300 μg of S2 nuclear extract. The precipitates were resolved by SDS-PAGE and transferred onto nitrocellulose membranes for immunodetection assays with antisera against dGcn5, dAda2b, and dAda2a. Input, 30 μg of nuclear extract. (B) GST pull-down assays using a GST-Dmp53 fusion protein. (C) Coimmunoprecipitation assays were performed with anti-Dmp53 antibodies conjugated to protein A-Sepharose and 300 μg of S2 nuclear extract. The immunoprecipitated (IP) proteins were separated by SDS-PAGE prior to Western blotting analyses using antisera against dGcn5, dAda2b, and dAda2a. i, input; be, beads.
A bacterially expressed and purified GST-VP16 fusion protein coprecipitated dAda2b, but not dAda2a, from S2 nuclear extracts (Fig. 5A). This suggests that SAGA-type complexes from yeast, humans, and flies have similar properties and further supports the notion that dAda2b is associated with this type of GNAT complex.
We wished to extend these observations and to test whether dAda2b is capable of interacting with a putative acidic transcription activator from Drosophila. One good candidate for such an activator is the Drosophila p53 homologue Dmp53 (9, 29, 37). The human p53 protein possesses two acidic regions that were shown to interact with GNAT complexes (15). Although these acidic activation domains are rather poorly conserved between the fly and human p53 homologues, both proteins control conserved apoptotic pathways and therefore might have similar regulatory properties (9).
As shown in Fig. 5B, a GST-Dmp53 fusion protein coprecipitated with dAda2b and dGcn5 but not with dAda2a. Moreover, antibodies against Dmp53 immunoprecipitated dAda2b and dGcn5 but not dAda2a from nuclear extracts (Fig. 5C). Thus, these endogenous proteins interact at native concentrations, suggesting that dSAGA-type complexes interact with Dmp53 in vivo.
Taken together, these results further support the notion that dAda2b is a component of dSAGA-type complexes, which might have regulatory capacities similar to those of their yeast and mammalian counterparts.
Distribution of Drosophila dGcn5, dAda3, dAda2a, and dAda2b on polytene chromosomes.
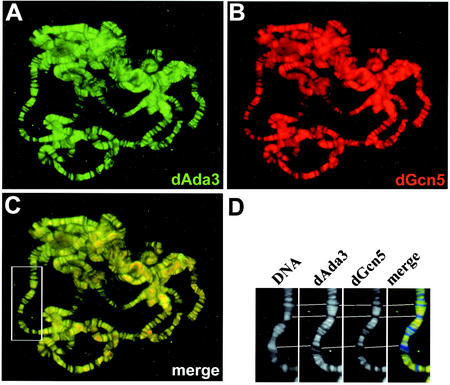
Several lines of evidence suggest that dAda2b is specific for Drosophila dSAGA-like HAT complexes, whereas dAda2a appears to be a component of at least two different multiprotein complexes (Fig. 3 and 4). The larger complexes seemed to lack stoichiometric amounts of dGcn5 and dAda3, whereas a smaller 440-kDa complex contained both dGcn5 and dAda3. To further investigate the relationships among these proteins and complexes, we performed immunocolocalization studies of native chromatin using Drosophila polytene chromosomes from third-instar larval salivary glands. According to our previous findings, one would expect a tight association of dAda3 and dGcn5. Indeed, when polytene chromosomes were immunoprobed for dAda3 (Fig. 6A) and dGcn5 (Fig. 6B), both proteins displayed full colocalization (Fig. 6C). A control staining of the DNA with DAPI, which stains condensed heterochromatic regions more intensely, revealed that dAda3 and dGcn5 predominantly localize to euchromatic interband regions (Fig. 6D). Our findings suggest that all Drosophila GNAT complexes contain dAda3 and that dAda3 is generally associated with dGcn5. The localization of dGcn5 and dAda3 to euchromatin supports their potential roles in transcription. Nevertheless, we observed that certain heterochromatic regions, such as the pericentric chromatin, stain weakly for both proteins and might contain low levels of dAda3 and dGcn5 (Fig. 6 and data not shown).
FIG. 6.
dAda3 and dGcn5 colocalize on polytene chromosomes from third-instar larvae. Polytene chromosome spreads were double labeled with affinity-purified antibodies against dAda3 (green) and dGcn5 (red). (A) Staining with rabbit anti-dAda3 antibodies. (B) Staining with rat anti-dGcn5 antibodies. (C) Overlap of green and red stains appears in the merge of panels A and B as yellow, yellow-green, and orange. dGcn5 and dAda3 show almost complete overlap. (D) Magnification of the boxed region in panel C. DNA was counterstained with DAPI, which preferentially labels heterochromatin. Note that dAda3 and dGcn5 mainly localize to DAPI-negative euchromatic regions.
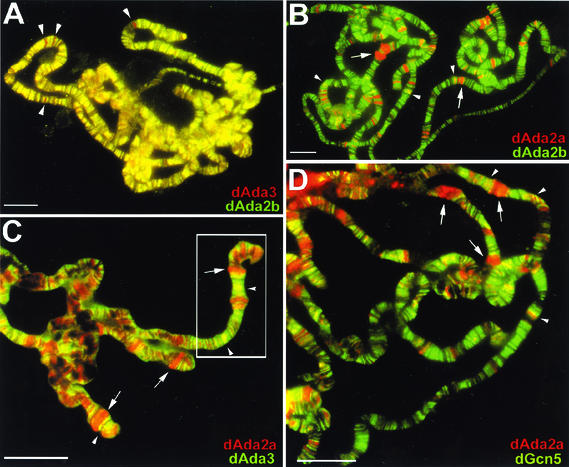
To look more specifically at dSAGA-related complexes, we compared the distributions of dAda2b and dAda3 on polytene chromosomes (Fig. 7A). Most of the loci that stained for dAda3 were also positive for dAda2b (Fig. 7A), suggesting that dSAGA-related complexes are relatively abundant. However, a few chromosome bands stained only for dAda3 (Fig. 7A). We assume that GNAT complexes other than dSAGA relatives might occupy these regions. We next determined the abundance of dAda2a-containing complexes and compared their distribution to that of dAda2b-containing complexes. As shown in Fig. 7B, dAda2a and dAda2b colocalized to only a few bands on polytene chromosomes, whereas most of the signals were not overlapping. Remarkably, a significant portion of dAda2a-containing complexes localized to decondensed chromosome puffs (Fig. 7B). In chromosome preparations double labeled for dAda2a and dAda3 (Fig. 7C), dAda3 was not concentrated in puffed regions that stained for dAda2a (Fig. 7C). However, both factors colocalize in a few regions, predominantly in condensed regions and in regions flanking dAda2a-positive puffs (Fig. 7C). In agreement with these findings, dAda2a and dGcn5 also colocalize to only a few regions (Fig. 7D), and puffed dAda2a-positive chromosomal regions did not show intense dGcn5 signals. This became even more obvious when the distribution of dAda2a and dAda3 were compared in separate panels at higher magnification (compare Fig. 7E to the boxed region in Fig. 7C). dAda2a is highly enriched in decondensed puffs, and these regions stain only weakly for dAda3 (Fig. 7E). As already shown for dAda3 and dGcn5 (Fig. 6D), dAda2a-containing complexes preferentially localize to DAPI-negative euchromatic regions. It was obvious that the regions positive for dAda2a, dGcn5, and dAda3 are less abundant. This suggests that dAda2a-containing GNAT complexes represent only a minor portion of the GNAT complexes present in salivary glands from third-instar larvae, whereas the majority of GNAT complexes in these cells appear to be dSAGA related. This was also observed in embryonic and S2 nuclear extracts, although the ratio of dGcn5-dependent dAda2a complexes appeared to be higher during these developmental stages (Fig. 4).
FIG. 7.
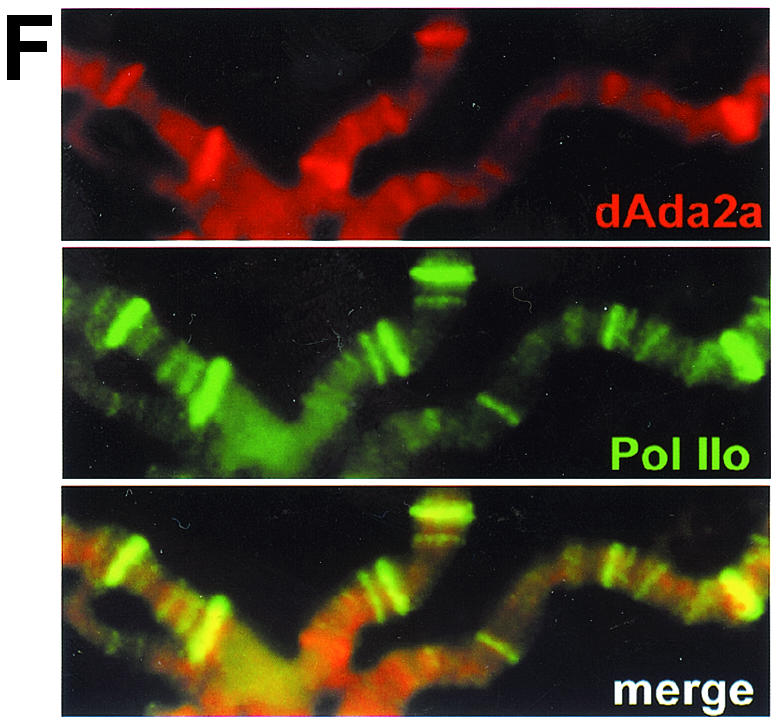
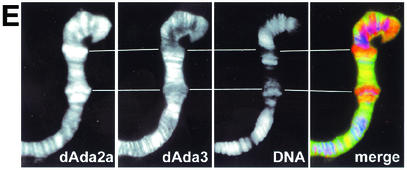
Localization studies of dAda2b and dAda2a on polytene chromosomes from third-instar larvae. Polytene chromosome spreads were double-labeled with the indicated affinity-purified antibodies. The pictures were taken at different magnifications; the bars correspond to 20 μm. (A to D) Merged red and green channels. (A) Costaining using rabbit anti-dAda3 antibodies (red) and rat anti-dAda2b antibodies (green). The arrowheads indicate selected regions exclusively staining for dAda3. (B) Costaining with rabbit anti-dAda2a (red) and rat anti-dAda2b (green). The arrowheads indicate selected regions of signal overlap. The arrows indicate puffs strongly staining for dAda2a. (C) Costaining with rat anti-dAda2a (red) and rabbit anti-dAda3 (green). The arrowheads indicate selected regions of signal overlap. The arrows indicate puffs strongly staining for dAda2a. (D) Costaining with rabbit anti-dAda2a (red) and rat anti-dGcn5 (green). The arrowheads indicate selected regions of overlap. The arrows indicate puffs strongly staining for dAda2a. (E) Magnification of the boxed region in panel C. DNA was stained with DAPI. dAda2a strongly stains decondensed euchromatic regions (compare the white bars). dAda3 and dAda2a colocalize in less condensed euchromatic regions. (F) dAda2a and phosphorylated RNA polymerase II colocalize on polytene chromosomes from third-instar larvae. Top, staining with rabbit anti-dAda2a antibodies; middle, staining with mouse anti-phosphorylated C-terminal domain of RNA polymerase II antibodies; bottom, overlap of green and red stains appears in the merge of panels A and B as yellow, yellow-green, and orange.
dAda2a is highly enriched in decondensed chromosome puffs, which usually contain transcriptionally very active loci. We therefore wished to test whether dAda2a colocalizes with transcribing RNA polymerase II, which can be detected with the help of antibodies against its phosphorylated C-terminal domain (50). As shown in Fig. 7E, high levels of dAda2a are detectable in puffs that are also enriched for phosphorylated RNA polymerase II. This suggests that the dGcn5-independent dAda2a complexes might play a role in RNA polymerase II-dependent transcription.
DISCUSSION
Drosophila has SAGA-type and other Gcn5-related N-acetyltransferase complexes.
We have characterized the Drosophila homologues of the Gcn5-associated factors Ada3, Ada2, Spt3, and Tra1/TRRAP (Fig. 1) and have demonstrated that these proteins are components of large multiprotein complexes and that they associate with Gcn5 HAT activities (Fig. 2). The human and yeast homologues of these factors were shown to be components of the SAGA-related GNAT complexes SAGA, PCAF, STAGA, and TFTC (8, 22, 33, 36). SAGA-related complexes share a significant number of conserved subunits, and the SAGA, PCAF, and STAGA complexes have similar molecular masses of ∼1.8 to 2 MDa. The TFTC complex is ∼1.35 MDa, suggesting that higher eukaryotes might have more SAGA-related subcomplexes than yeast. We observed that Drosophila SAGA-related complexes elute from size exclusion columns in a comparable molecular-mass range (Fig. 4A and D). Moreover, size exclusion chromatography using a S400 column suggests that dGcn5, dAda3, dAda2b, and dSpt3 eluted in two peaks of ∼2 and 1.6 MDa (Fig. 4A and D). It therefore appears likely that Drosophila also might have different subtypes of SAGA-related complexes, as has been described for humans. The striking similarity in the sizes of Drosophila and human SAGA-type complexes suggests that their compositions might be very similar. This is supported by our findings that Drosophila homologues of SAGA-specific human Tafs, such as dmTaf5, dmTaf9, and dmTaf10, associate with the dSAGA subunits dAda3, dAda2b, and dSpt3 (Fig. 3C). In addition, we were able to demonstrate that dSAGA-type complexes interact with the acidic activator VP16 and the p53 homologue Dmp53 (Fig. 5). This suggests that these chromatin-remodeling complexes might have similar functions in the regulation of gene expression in flies and humans.
The existence of a 440-kDa GNAT complex containing dAda2a, dGcn5, and dAda3 indicates that Drosophila has smaller GNAT complexes distinct from dSAGA. This novel finding suggests that higher eukaryotes also might possess non-SAGA-type GNAT complexes, as found in yeast. So far, only two yeast non-SAGA-type GNAT complexes, namely, the 800-kDa ADA and the 200- to 500-kDa HAT A2, have been identified (19, 22, 39). However, the functions of these complexes are not yet understood. Unfortunately, we were not able to find specific subunits of non-SAGA complexes, such as a fly homologue of the yeast Ahc1 protein, but the possibility that a functionally equivalent protein exists in ADA-type complexes from higher eukaryotes cannot be excluded. We are pursuing studies addressing the structure and function of the distinct dAda2-containing complexes.
Drosophila and mammalian Ada2 homologues form two distinct subgroups.
In an attempt to address the functions of GNAT complexes in multicellular organisms, we sought to exploit the genetically well-studied protostome model organisms Caenorhabditis elegans and D. melanogaster. To our surprise, we were not able to detect any sequences with homology to GNAT complex subunits in the genome of C. elegans, whereas D. melanogaster possesses homologues of Gcn5, Ada2, Ada3, and Spt3 (Fig. 1). The possibility that these factors are less conserved in nematodes cannot be excluded; however, the Tra1/TRRAP homologue from C. elegans is highly similar to Tra1/TRRAP from other organisms. Tra1/TRRAP is a component of the histone H4-specific NuA4 and Tip60 complexes (2, 28), suggesting that C. elegans might have H4-specific HAT complexes. This is supported by the fact that C. elegans has homologues of Tip60 and Esa1, the catalytic subunit of NuA4. It remains unclear at this point why the protostome C. elegans may lack GNAT-type HAT complexes whereas the protostome Drosophila possesses GNAT complexes with significant homologies to their mammalian counterparts. Studies of mice indicate that GNAT complexes might play essential roles during development (54). The maternal load of GNAT components and their higher concentration in female flies suggest that they might play important roles during oogenesis and in the regulation of early embryonic gene regulation (Fig. 2B). We are investigating the developmental functions of the distinct GNAT subunits in D. melanogaster.
It is interesting that both flies and mammals contain two subgroups of Ada2 proteins, namely, the Ada2a-like and Ada2b-like adapter proteins (Fig. 1B). Thus far, we have not found a sequence motif that is specific for either of the subgroups; however, phylogenetic comparisons strongly suggest that both dAda2a and dAda2b from flies and mammals form distinct subgroups. This is supported by our findings that the Drosophila members of these subgroups are in distinct multiprotein complexes. It remains to be demonstrated that the human or mouse homologues have similar features and associate with distinct GNAT complexes. At present, only the human PCAF complex has been shown to contain a homologue of Ada2a (Fig. 1B, H.s. Ada2). The PCAF complex contains subunits that are characteristic of SAGA-type complexes, suggesting that mammalian Ada2a homologues can associate with SAGA-type GNAT complexes. Nevertheless, phenotypic studies of mice have demonstrated that PCAF is dispensable for viability whereas the ubiquitously expressed Gcn5L protein is essential for embryogenesis (54). Thus, Gcn5L-containing complexes appear to be true functional homologues of SAGA-type complexes from flies and yeast. Intriguingly, the human Ada2a homologue was not detected in the Gcn5L-containing STAGA and TFTC complexes. Although an uncharacterized protein of the apparent molecular mass of the human Ada2b-like protein copurified with STAGA (33), it remains to be demonstrated that these complexes contain Ada2b-like adapter proteins as their fly homologues do.
Novel GNAT-independent function of Ada2 in transcription.
All Ada2 proteins so far characterized show homologies within three conserved regions that mediate important functions of these proteins (Fig. 1A). The most N-terminal ZZ finger motif (38) has been shown to be essential for the interactions of Ada2 and Gcn5 in vitro (13). The second conserved region is the Myb/SANT domain (1, 32). Myb/SANT domains have been found in the Myb protein family (32) as well as in several other chromatin-associated proteins, such as Swi3, TFIIIB, and the nuclear receptor corepressor N-CoR (1). Deletions of this domain cripple the ability of Ada2 to support transcriptional activation in vivo (13). Recent studies have suggested that this domain is important for the proper acetylation of nucleosomal histones (45). This is consistent with the finding that Ada2 potentiated the catalytic function of Gcn5 (4). A third central region of high homology was shown to mediate interaction with Ada3 in vitro (5, 13, 14). These three regions are conserved in virtually all Ada2 proteins so far characterized (Fig. 1A), supporting a tight functional link between Ada2- and Gcn5-containing complexes. To our surprise, the association of either of the two Drosophila Ada2 homologues was unexpectedly specific for distinct GNAT complexes. Only dAda2b associated with the dSAGA-specific subunits dmTaf5, dmTaf9, dmTaf10, dSpt3, and dTra1 (Fig. 3 and 4), and dAda2b-containing GNAT complexes have features that are typical of SAGA-related complexes from yeast and humans (Fig. 5). dAda2a, however, exhibits an even more unexpected promiscuity and is incorporated into a large multiprotein complex lacking dGcn5. We cannot exclude the possibility that a second copy of Gcn5 exists in flies as it does in mammals and that this second Gcn5 homologue is a catalytic subunit of the larger dAda2a-containing complexes. However, several lines of evidence do not support this. Ada3 has been found in all GNAT complexes so far characterized (11, 33) and is essential for the acetyltransferase activities and specificities of these complexes (4). One would therefore expect dAda3 to be associated with the 2.5-MDa dAda2a-containing complexes. However, dAda3 did not coelute with these complexes in chromatography assays (Fig. 4A and D). Immunocolocalization studies of polytene chromosomes revealed that dAda2a was enriched at sites with high levels of RNA polymerase II-dependent transcription (Fig. 7F), but these puffed regions do not stain strongly for dAda3 or dGcn5 (Fig. 7C to E). It therefore appears likely that dAda2a functions within large complexes independently of dAda3 and Gcn5, proteins with which Ada2 functions had previously been associated. Intriguingly, the large dAda2a-containing complex localizes to chromosomal regions with high RNA polymerase II activities, pointing to a novel transcription-linked function of this Ada2 variant (Fig. 7F).
Although it will be necessary to identify subunits which are specific for these non-GNAT dAda2a complexes, several lines of evidence suggest that the ratio between distinct dAda2a-containing complexes might vary during the Drosophila life cycle. First, we found three different types of dAda2a-containing complexes in nuclear extracts from 0- to 12-h-old embryos (Fig. 4D). In S2 cells, which derive from 16- to 24-h-old male embryos (40, 43), one of these complexes was not detectable (Fig. 4A). Second, antibodies against dAda2a immunoprecipitated relatively large amounts of dGcn5 from S2 nuclear extracts; however, only a little overlap between dAda2a and dGcn5 or dAda3 was observed on polytene chromosomes from third-instar larvae (Fig. 7C to E). Although the larger dAda2a complexes in general seem to be more abundant than the 440-kDa complex (Fig. 4A and D), these findings were rather unexpected. Nevertheless, developmental-expression studies indicate that dAda2a concentrations are significantly higher in 13- to 24-h-old embryos (Fig. 2B, lane 13-24), from which S2 cells derive. In a third-instar larva, however, dAda2a levels are decreased relative to those in embryos or earlier larval stages. In addition, while the concentrations of dGcn5 and other dSAGA-specific subunits, such as dSpt3 or dAda2b, are relatively low in adult male flies (Fig. 2B), the amounts of dAda2a are equally large in male and female flies. This indicates that the amount of dAda2a is not tightly linked to the levels of dGcn5 expression and suggests that the ratio between distinct dAda2a-containing complexes might be variable. We are pursuing the biochemical characterization of non-GNAT-type dAda2a complexes in order to better understand their compositions and functions and to further address this hypothesis.
ADDENDUM IN PROOF
During the revision of the manuscript, a paper presenting similar findings was published (33a).
Acknowledgments
We are indebted to Celera Genomics, Inc., and members of the Allis, Gilmour, Lucchesi, Nakatani, Reese, Simpson, Tan, and Tora laboratories for reagents and helpful discussions. Special thanks go to C.-H. Wu for providing embryonic nuclear extracts and for technical advice.
REFERENCES
- 1.Aasland, R., A. F. Stewart, and T. Gibson. 1996. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 21:87-88. [PubMed] [Google Scholar]
- 2.Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant, C. J. Brandl, L. Pillus, J. L. Workman, and J. Cote. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18:5108-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Balasubramanian, R., M. G. Pray-Grant, W. Selleck, P. A. Grant, and S. Tan. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277:7989-7995. [DOI] [PubMed] [Google Scholar]
- 5.Berger, S. L., B. Pina, N. Silverman, G. A. Marcus, J. Agapite, J. L. Regier, S. J. Triezenberg, and L. Guarente. 1992. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70:251-265. [DOI] [PubMed] [Google Scholar]
- 6.Biggin, M. D., and R. Tjian. 1988. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell 53:699-711. [DOI] [PubMed] [Google Scholar]
- 7.Brand, M., J. G. Moggs, M. Oulad-Abdelghani, F. Lejeune, F. J. Dilworth, J. Stevenin, G. Almouzni, and L. Tora. 2001. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 20:3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brand, M., K. Yamamoto, A. Staub, and L. Tora. 1999. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 274:18285-18289. [DOI] [PubMed] [Google Scholar]
- 9.Brodsky, M. H., W. Nordstrom, G. Tsang, E. Kwan, G. M. Rubin, and J. M. Abrams. 2000. Drosophila p53 binds a damage response element at the reaper locus. Cell 101:103-113. [DOI] [PubMed] [Google Scholar]
- 10.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333-2337. [DOI] [PubMed] [Google Scholar]
- 11.Brown, C. E., T. Lechner, L. Howe, and J. L. Workman. 2000. The many HATs of transcription coactivators. Trends Biochem. Sci. 25:15-19. [DOI] [PubMed] [Google Scholar]
- 12.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 13.Candau, R., and S. L. Berger. 1996. Structural and functional analysis of yeast putative adaptors. Evidence for an adaptor complex in vivo. J. Biol. Chem. 271:5237-5245. [DOI] [PubMed] [Google Scholar]
- 14.Candau, R., P. A. Moore, L. Wang, N. Barlev, C. Y. Ying, C. A. Rosen, and S. L. Berger. 1996. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol. Cell. Biol. 16:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Candau, R., D. M. Scolnick, P. Darpino, C. Y. Ying, T. D. Halazonetis, and S. L. Berger. 1997. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene 15:807-816. [DOI] [PubMed] [Google Scholar]
- 16.Carter, K. C., L. Wang, B. K. Shell, I. Zamir, S. L. Berger, and P. A. Moore. 1997. The human transcriptional adaptor genes TADA2L and GCN5L2 colocalize to chromosome 17q12-q21 and display a similar tissue expression pattern. Genomics 40:497-500. [DOI] [PubMed] [Google Scholar]
- 17.Dudley, A. M., L. J. Gansheroff, and F. Winston. 1999. Specific components of the SAGA complex are required for Gcn4- and Gcr1-mediated activation of the his4-912delta promoter in Saccharomyces cerevisiae. Genetics 151:1365-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eberharter, A., S. John, P. A. Grant, R. T. Utley, and J. L. Workman. 1998. Identification and analysis of yeast nucleosomal histone acetyltransferase complexes. Methods 15:315-321. [DOI] [PubMed] [Google Scholar]
- 19.Eberharter, A., D. E. Sterner, D. Schieltz, A. Hassan, J. R. Yates III, S. L. Berger, and J. L. Workman. 1999. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6621-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elgin, S. C. R., and J. L. Workman. 2000. Chromatin structure and gene expression, 2nd ed. Oxford University Press, Oxford, United Kingdom.
- 21.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 22.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 23.Grant, P. A., A. Eberharter, S. John, R. G. Cook, B. M. Turner, and J. L. Workman. 1999. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274:5895-5900. [DOI] [PubMed] [Google Scholar]
- 24.Grant, P. A., D. Schieltz, M. G. Pray-Grant, D. J. Steger, J. C. Reese, J. R. Yates III, and J. L. Workman. 1998. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94:45-53. [DOI] [PubMed] [Google Scholar]
- 25.Grant, P. A., D. Schieltz, M. G. Pray-Grant, J. R. Yates III, and J. L. Workman. 1998. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell 2:863-867. [DOI] [PubMed] [Google Scholar]
- 26.Howe, L., C. E. Brown, T. Lechner, and J. L. Workman. 1999. Histone acetyltransferase complexes and their link to transcription. Crit. Rev. Eukaryot. Gene Expr. 9:231-243. [DOI] [PubMed] [Google Scholar]
- 27.Ikeda, K., D. J. Steger, A. Eberharter, and J. L. Workman. 1999. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol. Cell. Biol. 19:855-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 29.Jin, S., S. Martinek, W. S. Joo, J. R. Wortman, N. Mirkovic, A. Sali, M. D. Yandell, N. P. Pavletich, M. W. Young, and A. J. Levine. 2000. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 97:7301-7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo, M. H., J. E. Brownell, R. E. Sobel, T. A. Ranalli, R. G. Cook, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383:269-272. [DOI] [PubMed] [Google Scholar]
- 31.Kusch, T., and R. Reuter. 1999. Functions for Drosophila brachyenteron and forkhead in mesoderm specification and cell signalling. Development 126:3991-4003. [DOI] [PubMed] [Google Scholar]
- 32.Lane, T., C. Ibanez, A. Garcia, T. Graf, and J. Lipsick. 1990. Transformation by v-myb correlates with trans-activation of gene expression. Mol. Cell. Biol. 10:2591-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Muratgolu, S., S. Georgieva, G. Pápai, E. Scheer, I. Enünlü, O. Komonyi, I. Cserpán, L. Lebedeva, E. Nabirochkina, A. Udvardy, L. Tora, and I. Boros. 2003. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol. Cell. Biol. 23:306-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 35.Ogryzko, V. 2000. Mammalian histone acetyltransferase complexes. Medicina 60:21-26. [PubMed] [Google Scholar]
- 36.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schiltz, T. Howard, X. J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 37.Ollmann, M., L. M. Young, C. J. Di Como, F. Karim, M. Belvin, S. Robertson, K. Whittaker, M. Demsky, W. W. Fisher, A. Buchman, G. Duyk, L. Friedman, C. Prives, and C. Kopczynski. 2000. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell 101:91-101. [DOI] [PubMed] [Google Scholar]
- 38.Ponting, C. P., D. J. Blake, K. E. Davies, J. Kendrick-Jones, and S. J. Winder. 1996. ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem. Sci. 21:11-13. [PubMed] [Google Scholar]
- 39.Ruiz-Garcia, A. B., R. Sendra, M. Pamblanco, and V. Tordera. 1997. Gcn5p is involved in the acetylation of histone H3 in nucleosomes. FEBS Lett. 403:186-190. [DOI] [PubMed] [Google Scholar]
- 40.Schneider, I. 1972. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27:353-365. [PubMed] [Google Scholar]
- 41.Sendra, R., C. Tse, and J. C. Hansen. 2000. The yeast histone acetyltransferase A2 complex, but not free Gcn5p, binds stably to nucleosomal arrays. J. Biol. Chem. 275:24928-24934. [DOI] [PubMed] [Google Scholar]
- 42.Smith, E. R., J. M. Belote, R. L. Schiltz, X. J. Yang, P. A. Moore, S. L. Berger, Y. Nakatani, and C. D. Allis. 1998. Cloning of Drosophila GCN5: conserved features among metazoan GCN5 family members. Nucleic Acids Res. 26:2948-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, E. R., A. Pannuti, W. Gu, A. Steurnagel, R. G. Cook, C. D. Allis, and J. C. Lucchesi. 2000. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 20:312-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sterner, D. E., X. Wang, M. H. Bloom, G. M. Simon, and S. L. Berger. 2002. The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J. Biol. Chem. 277:8178-8186. [DOI] [PubMed] [Google Scholar]
- 46.Stockinger, E. J., Y. Mao, M. K. Regier, S. J. Triezenberg, and M. F. Thomashow. 2001. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 29:1524-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tora, L. 2002. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 16:673-675. [DOI] [PubMed] [Google Scholar]
- 49.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 50.Weeks, J. R., S. E. Hardin, J. Shen, J. M. Lee, and A. L. Greenleaf. 1993. Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: correlations with gene activity and transcript processing. Genes Dev. 7:2329-2344. [DOI] [PubMed] [Google Scholar]
- 51.Winston, F., and P. Sudarsanam. 1998. The SAGA of Spt proteins and transcriptional analysis in yeast: past, present, and future. Cold Spring Harbor Symp. Quant. Biol. 63:553-561. [DOI] [PubMed] [Google Scholar]
- 52.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 53.Xu, W., D. G. Edmondson, Y. A. Evrard, M. Wakamiya, R. R. Behringer, and S. Y. Roth. 2000. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26:229-232. [DOI] [PubMed] [Google Scholar]
- 54.Yamauchi, T., J. Yamauchi, T. Kuwata, T. Tamura, T. Yamashita, N. Bae, H. Westphal, K. Ozato, and Y. Nakatani. 2000. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. USA 97:11303-11306. [DOI] [PMC free article] [PubMed] [Google Scholar]