Abstract
Expansion of CTG triplet repeats in the 3′ untranslated region of the DMPK gene causes the autosomal dominant disorder myotonic dystrophy. Instability of CTG repeats is thought to arise from their capacity to form hairpin DNA structures. How these structures interact with various aspects of DNA metabolism has been studied intensely for Escherichia coli and Saccharomyces cerevisiae but is relatively uncharacterized in mammalian cells. To examine the stability of (CTG)17, (CTG)98, and (CTG)183 repeats during homologous recombination, we placed them in the second intron of one copy of a tandemly duplicated pair of APRT genes. Cells selected for homologous recombination between the two copies of the APRT gene displayed distinctive patterns of change. Among recombinants from cells with (CTG)98 and (CTG)183, 5% had lost large numbers of repeats and 10% had suffered rearrangements, a frequency more than 50-fold above normal levels. Analysis of individual rearrangements confirmed the involvement of the CTG repeats. Similar changes were not observed in proliferating (CTG)98 and (CTG)183 cells that were not recombinant at APRT. Instead, they displayed high frequencies of small changes in repeat number. The (CTG)17 repeats were stable in all assays. These studies indicate that homologous recombination strongly destabilizes long tracts of CTG repeats.
Expansions of trinucleotide (triplet) repeats underlie many human neurological diseases, including myotonic dystrophy (DM), Huntington disease, fragile X syndrome, Friedreich ataxia, and several spinocerebellar ataxias (2, 7, 10, 78, 81). Healthy individuals generally have fewer than 30 repeats at a disease locus, whereas affected individuals may have from 40 to several thousand repeats, depending on the specific disease. Triplet repeats associated with disease have a propensity to form stable secondary structures in vitro, and it is thought that these unusual structures interfere with aspects of DNA metabolism in cells, leading to repeat expansion and disease (81). Here, we examine the instability of the CTG · CAG trinucleotide repeats (which we will refer to as CTG repeats) that in their expanded form cause DM.
DM is the most common inherited neuromuscular disorder in adults, affecting 1 in 8,000 people worldwide (21). The DM mutation, which is autosomally dominant, arises due to expansion of a CTG repeat located in the 3′ untranslated region of the gene for DM protein kinase (DMPK) (78). The mechanism by which the expanded CTG repeats cause the characteristic features of the disease is not well defined and is likely to be complicated. It has been shown previously that transcripts from the mutant allele are retained in the nucleus (12), that the distribution of DMPK cytoplasmic mRNA isoforms is altered (76), that expression of the adjacent SIX5 homeodomain gene is decreased (34), and that binding of CUG binding proteins to mutant DMPK mRNAs (75) can alter the splicing of other, unrelated mRNAs (42, 56). The lack of DM patients with point mutations in the DMPK gene suggests that simple haploinsufficiency does not account for the disease phenotype (78), a conclusion supported by the relatively minor phenotype displayed by DMPK-knockout mice (28).
The size distributions of normal and mutant DM alleles, along with extensive analyses of CAG · CTG repeats (hereafter, CAG repeats) in sperm (39, 84), suggest that the instability thresholds for CTG repeats in DM and for CAG repeats in other diseases are around 30 repeats (80). Typically, there is a bias for male germ line transmission to generate the first clinically recognized disorder in a DM pedigree (40) and a bias for female germ line transmission to generate congenital DM, the most severe form of the disease (20). It is unclear whether germ line instability of CTG repeats in DM results from meiotic or from mitotic events. Instability of CAG triplet repeats, which are associated with other diseases, can occur before meiosis (39), during arrest in prophase I (32), and in postmeiotic haploid cells (35). It is clear that CTG instability can occur during mitotic divisions, based on changes in repeat length in different tissues from DM patients (22, 43, 83), in patient-derived cell lines upon serial passage (1, 82), and in tissues and cell lines from transgenic mouse models (19, 79).
The ability of long CTG repeats to form unusual secondary structures is the likely basis for expansion at the DMPK locus (81). Several in vitro studies have shown that CTG and CAG repeats can form hairpins (18, 73, 74) and slipped-strand DNA duplexes (55). In addition, studies of Escherichia coli (11, 30, 67) and Saccharomyces cerevisiae (48) suggest that CTG and CAG repeats form unusual DNA structures in cells. One of the strongest links between unusual secondary structures and disease progression is the effect of nucleotide changes within the repeat sequence. Interruptions that decrease the propensity of the repeat to form secondary structures also reduce the frequency of expansions and the potential for disease (7, 37, 73).
Both E. coli and S. cerevisiae provide useful model systems to probe genetic influences on triplet repeat instability, especially of CTG and CAG repeats. Virtually every process that exposes single strands of DNA destabilizes triplet repeats, including transcription (6, 70), nucleotide excision repair (50, 53), mismatch repair (29, 61, 68, 71), replication (23, 30, 44, 67), and recombination (17, 24, 25, 26, 27, 58, 59). CTG and CAG triplet repeats also cause double-strand DNA breaks in yeast (17, 26). Differences in assays and differences in frequencies of contractions and expansions make it difficult to rank order these processes in terms of their effects on repeat instability. Nevertheless, it is thought that errors in mismatch repair correspond to small changes in repeat length, errors in replication correspond to small and intermediate changes, and errors during recombination correspond to intermediate and large changes (7, 78). Studies in these systems have shaped present models for replication-based and recombination-based instability of triplet repeats (7, 60, 78).
The causes of triplet repeat instability in mammalian cells are difficult to investigate directly. Changes in repeat length upon serial passage of cell lines derived from patients (1, 82) or transgenic mice (16, 19) have been characterized, and the influences of mismatch repair (36, 79) and replication (9, 51) have been examined. Present approaches in mammalian cells are limited, however, because different sizes of repeats cannot be readily tested in the same genetic background, different genomic contexts and orientations of the repeats are difficult to compare, and different genetic influences cannot be easily examined. An additional significant restriction is a lack of sensitivity, which is limited to a frequency of 10−2 to 10−3 for changes in repeat length that must be screened for in the cell population.
In this paper we describe a model system in CHO cells that is designed to overcome some of these limitations. We have deposited CTG repeats from the DM gene into an intron in one copy of a tandemly duplicated pair of APRT genes at their endogenous locus in CHO cells. By selecting for homologous recombination between the duplicated copies of the APRT gene, we can examine changes to the inserted CTG repeats in a population of cells that have experienced a nearby recombination event. Within this selected population we show that long CTG repeats experience large contractions and generate a high frequency of rearrangements that extend beyond the repeat tract. These distinctive changes were not observed for long tracts of CTG repeats in replicating cells that were not recombinant at APRT. Instead, replicating cells displayed a high frequency of expansions and contractions that usually involved just one to three triplets. These results indicate that homologous recombination dramatically destabilizes long CTG repeats in CHO cells.
MATERIALS AND METHODS
Construction of vectors.
CTG triplet repeats that included 19 nucleotides of 5′- and 43 nucleotides of 3′-flanking sequences from the DM locus were inserted into the polylinker in intron 2 of the APRT gene in pGS100, as described previously (23). Plasmids pRW3502, pRW3504, and pRW3506 carry (CTG)17, (CTG)98, and (CTG)175, respectively, with the repeats oriented so that the CTG sequences are in the noncoding strand of the DNA (23). This is the same orientation as at the DM locus. The (CTG)17 and (CTG)98 tracts of repeats are pure, whereas the (CTG)175 repeat is interrupted by two G-to-A changes, which in the CTG orientation are located in repeats 27 and 67 (30). The APRT genes in plasmids pRW3502, pRW3504, and pRW3506, like the one in their parent plasmid pGS100, are truncated in exon 5 and are nonfunctional as a result.
Construction of cell lines.
Site-specific recombination involving the FLP recombinase and the FLP recombinase recombination target (FRT) site was used to generate tandemly duplicated APRT genes, as described previously (46). The APRT− gene in the RMP41 cell line carries a point mutation in exon 2 that eliminates APRT function and removes an EcoRV recognition sequence (63). The RMP41 cell line also carries an FRT site in intron 2, which does not affect the function of the APRT gene (46). Plasmids pRW3502, pRW3504, and pRW3506 each carry an FRT site adjacent to the tracts of inserted CTG repeats. FLP recombinase-mediated site-specific recombination between the FRT sites in these plasmids and the FRT site in RMP41 cells was used to generate APRT+ colonies with the tandemly duplicated gene structures shown in Fig. 1. Sequencing of the triplet repeats in each cell line showed that GS3502 carried (CTG)17 and GS3504 carried (CTG)98, as expected. Cell line GS3506, however, carried (CTG)183 instead of the expected (CTG)175. The presence of 183 CTG repeats, instead of the expected 175, presumably reflects some instability that occurred in the growth of the plasmid in E. coli or in the targeting and establishment of the cell line. In GS3506 the G-to-A changes are located in CTG repeats 27 and 70 (instead of CTG repeats 27 and 67). In each of these cell lines, the upstream copy of APRT is inactive due to the point mutation in exon 2 and the exon 5 truncation, whereas the downstream APRT gene, which carries the CTG triplet repeats in intron 2, is functional. Structures of all cell lines were verified by Southern blotting after digestion with restriction enzymes diagnostic for the predicted structure.
FIG. 1.
Structures of the APRT locus in parental CHO cells and in colonies isolated under various selections. (A) Molecular structures of the tandem duplication of APRT sequences at the endogenous locus. The locations of the CTG triplet repeats in the parental cell lines are shown above their common site of insertion in the second intron of the downstream, functional APRT gene (the five exons of APRT are shown as boxes). The sequences immediately surrounding the upstream FRT site (black triangle) and downstream FRT site (open triangle) are different and therefore distinguishable. The upstream copy of APRT is nonfunctional by virtue of a truncated fifth exon and a point mutation in exon 2 that eliminates an EcoRV site (filled box). The upstream and downstream copies share 6.8 kb of homology indicated by brackets: 4.5 kb upstream of the APRT gene (thick line) and 2.3 kb of homology within the gene itself. (B) Molecular structures of the APRT locus in APRT− and TK− APRT− colonies. Products were distinguished by a combination of Southern blotting and PCR analysis. Conversions have a structure like the parental tandem duplication, except that some lose the insert as part of the conversion process (status of the insert is indicated by +/−). CTG+ conversions were distinguished from mutations by PCR analysis. Mutations are assumed to carry point mutations or small deletions elsewhere in the APRT gene; however, they were not further characterized. Crossovers have a single copy of the APRT gene whose digestion pattern depends on whether the insert (+) was retained or lost (−). Rearrangements yield a Southern blot pattern that does not correspond to conversions, crossovers, or mutations.
Cell culture and fluctuation analysis.
Cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with amino acids and 10% fetal calf serum. Selections were carried out as previously described (63). APRT+ cells were selected by growth in ALASA medium (25 μM alanosine, 50 μM azaserine, 100 μM adenine). APRT− cells were selected by growth in medium made with 10% dialyzed fetal calf serum and supplemented with 400 μM 8-azaadenine. TK− APRT− cells were selected by growth in APRT− selection medium supplemented with 0.3 μM fluoroiodoarabinosyluridine (FIAU).
Fluctuation analysis (38, 41) was carried out by using 12 parallel cultures grown from initial populations of 50 to 100 cells for each rate determination, as described previously (63). The numbers of APRT− or TK− APRT− colonies in parallel cultures were used to calculate rates by the method of the median (38). A single colony was picked from each parallel culture to ensure that all analyzed colonies arose independently.
Southern analyses, PCR analysis, and DNA sequencing.
Southern analyses were carried out according to standard protocols (62). The probe for Southern analysis was the 3.9-kb BamHI fragment containing the entire APRT gene, labeled by random priming with [32P]dCTP. PCR analysis of the recombination products was carried out as previously described (63). The distinction between conversions and point mutations was based on restriction digestion of a PCR product that includes exon 2. The copy of exon 2 in the downstream APRT gene was specifically amplified by using one primer that was located in unique sequences adjacent to the CTG repeat in intron 2. PCR products from convertants give rise to PCR products that are not cleaved by EcoRV because they have picked up the EcoRV mutation from the upstream copy of the APRT gene. By contrast, colonies that have acquired point mutations elsewhere in the APRT gene give rise to a PCR fragment that is cleaved by EcoRV.
PCR amplification of triplet repeat fragments for sequencing used one primer in intron 2 and a second primer at the 3′ end of the gene, which is unique to the downstream copy. This choice of primers allowed the downstream insertion site to be specifically amplified. In addition, it was found that embedding the CTG repeats in a larger PCR fragment gave more reliable sequencing results. The locations of PCR primers used for analysis of rearrangements and their sequences are available on request. DNA sequencing was carried out on amplified PCR fragments to determine the numbers of triplet repeats and to decipher the structures of the rearrangement junctions.
Statistical analysis.
Means were compared by the two-tailed t test. Standard errors of the means are reported in Table 2 and were used to determine the propagated errors reported in Table 4. For t test comparisons of the means in Table 4, however, the propagated error was recalculated by using standard deviations (which is the standard error × the square root of the sample size). Distributions were compared by the chi-square test. For two-by-two comparisons with 1 df, the Yates adjustment was used to compensate for the tendency of such comparisons to exaggerate significance. This adjustment consists of changing the observed frequencies by half a unit to give smaller deviations from the expected values, thereby giving a larger, more realistic P value (8). For all comparisons a P value of 0.05 was used to accept or reject the null hypothesis, which was that the means or distributions were the same. All calculations for the statistical tests were performed by using the PHStat add-in for Excel.
TABLE 2.
Rates of APRT− and TK− APRT− colony formation
| Cell line(s) | Insert | Insert length (bp) | Rate of colony formatione (10−7)
|
|
|---|---|---|---|---|
| APRT− | TK−APRT− | |||
| GS3502 | (CTG)17 | 270 | 7.1 ± 2.5 | 0.90 ± 0.10 |
| GS3504 | (CTG)98 | 513 | 6.9 ± 1.2 | 2.0 ± 1.4 |
| GS3506 | (CTG)183 | 768 | 9.0 ± 2.7 | 3.9 ± 1.7 |
| GSC4, GSC6a | None | 0 | 15.5 ± 2.5 | 0.75 ± 0.15 |
| GS21-15b | FRT | 67 | 14.0 ± 1.9 | 1.40 ± 0.27 |
| GSE1, GSE3a | HOFRTf | 114 | 12.3 ± 2.8 | 1.33 ± 0.53 |
| GSB2, GSB5a | (GT)29 | 200 | 16.3 ± 1.8 | 1.55 ± 0.24 |
| AK209c | HPRTg intron | 810 | 14.1 ± 3.8 | 1.20 ± 0.40 |
| AK784d | HPRT intron plus I-SceI | 835 | 17.0 | 1.10 |
| Mean | Non-CTG containing | 14.4 ± 1.1 | 1.29 ± 0.14 | |
Results for GS21-15 were previously published (65).
Results for AK209 were previously published (33).
Results for AK784 were previously published (66).
Rates were determined by fluctuation analysis. For GS3502 the rates for formation of TK− APRT− and APRT− colonies were determined from two fluctuation analyses. For GS3504 the rates were determined from four fluctuation analyses. For GS3506 the rates were determined from three fluctuation analyses. The overall mean rates of APRT− and TK− APRT− colony formation for cell lines that did not contain a CTG insert are the averages of 17 and 16 fluctuation analyses, respectively. The standard error of the mean is shown for all rates.
HOFRT, the recognition sequence for HO endonuclease is located adjacent to the FRT site.
HPRT, hypoxanthine phosphoribosyltransferase.
TABLE 4.
Individual rates of formation of different types of colonies
| Colony type and cell line | Rate (10−7)a
|
|||
|---|---|---|---|---|
| Conversions | Cross- overs | Mutations | Rearrange- ments | |
| APRT− | ||||
| GS3502 (CTG)17 | 4.9 ± 1.9 | 1.3 ± 0.6 | 0.9 ± 0.5 | <0.13 |
| GS3504 (CTG)98 | 3.5 ± 0.9 | 1.4 ± 0.5 | 1.3 ± 0.5 | 0.7 ± 0.3 |
| GS3506 (CTG)183 | 3.5 ± 1.2 | 3.8 ± 1.3 | 0.9 ± 0.4 | 0.8 ± 0.4 |
| Otherb | 11.8 ± 1.3 | 1.9 ± 0.4 | 0.7 ± 0.2 | <0.07 |
| TK−APRT− | ||||
| GS3502 (CTG)17 | 0.9 ± 0.3 | <0.06 | ||
| GS3504 (CTG)98 | 1.7 ± 1.3 | 0.3 ± 0.3 | ||
| GS3506 (CTG)183 | 3.5 ± 1.7 | 0.4 ± 0.3 | ||
| Otherb | 1.3 ± 0.2 | <0.006 | ||
Rates were obtained by multiplying the overall rate (Table 1) by the proportion of all colonies represented by the specific type (Table 3). For example, for GS3502 APRT− conversions the rate is 7.1 × (38/55) = 4.9. Standard rules for propagation of error were used to combine the standard errors associated with the rates in Table 2 with the sampling errors associated with the numbers of events in Table 3, where the sampling error is plus or minus the square root of the number of events. Specifically, the standard errors and sampling errors were expressed as fractional errors, and the fractional error of the product was calculated as the square root of the sum of the squares of the fractional errors. For GS3502 APRT− conversions, the propagated error is 4.9 × [(2.5/7.1)2 + (6.2/38)2]0.5 = 1.9.
Other cell lines are those that do not contain a CTG repeat. The mean rates shown in Table 1 were used for these calculations.
RESULTS
Construction of cell lines and experimental rationale.
To test the effects of homologous recombination on the stability of CTG triplet repeats, we used site-specific recombination to construct cell lines that carried tandem duplications of the APRT gene (Fig. 1A; see Materials and Methods). In each cell line, the upstream, APRT− copy of the gene carries an inactivating point mutation that eliminates an EcoRV site in exon 2, a deletion of its 3′ end beginning in exon 5, and the site-specific recombination target—the FRT site—in intron 2. The downstream, APRT+ copy is wild type, except that it carries a tract of CTG repeats located in a polylinker adjacent to the FRT site in intron 2. Homologous recombination between the tandem copies of APRT is detected by the appearance of APRT− cells that have incorporated the upstream point mutation into the downstream copy of the gene by gene conversion or crossover recombination (Fig. 1B).
We chose to test CTG repeats in the same orientation in which they occur in the DM gene, that is, so that the RNA transcript carries CUG repeats. In contrast to the DMPK mRNA, which retains the CUG sequences, the CUG repeats are absent from the APRT mRNA because they are removed by splicing, along with the rest of the sequences in intron 2. Because cells that carry the CTG repeats in the otherwise wild-type copy of the gene are phenotypically APRT+, the CUG repeats in the RNA evidently do not interfere with normal splicing, nor do they have any observable effects on cell growth. We tested three lengths of CTG repeats. (CTG)17 is within the range of repeats in healthy individuals and was anticipated to be unaffected by DNA replication and homologous recombination. By contrast, (CTG)98 and (CTG)183 are in the range found in affected individuals and have been demonstrated previously to cause instability in E. coli (30).
CTG repeat stability in replicating cells.
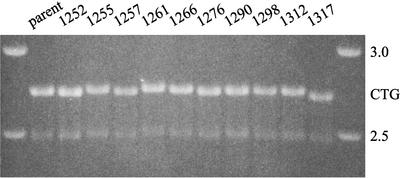
To measure the influence of replication on CTG repeat stability, we grew the (CTG)17, (CTG)98, and (CTG)183 cell lines (GS3502, GS3504, and GS3506, respectively) through about 30 cell doublings and then isolated individual colonies from each cell line. Individual colonies were grown to about 106 cells and analyzed by PCR amplification with flanking primers. Out of 43 colonies from the (CTG)17 cell line, 118 colonies from the (CTG)98 cell line, and 134 colonies from the (CTG)183 cell line, no colonies gave rise to a PCR fragment that differed substantially from its siblings, suggesting that these repeat lengths are reasonably stable during cell proliferation (data not shown). However, a slight unevenness in the alignment of bands was evident in gels from (CTG)183 colonies, as is apparent in the long gel run shown in Fig. 2.
FIG. 2.
Agarose gel electrophoresis of PCR fragments generated by amplification across the CTG repeats in individual colonies isolated from a population of proliferating GS3506 cells. The individual colonies correspond to those in Table 1, and the PCR products shown here (marked by CTG) were the ones that were isolated and subjected to DNA sequence analysis. The bands in the outside lanes show standard 2.5- and 3.0-kb markers from a commercial ladder. To accentuate the slight differences between bands, the fragments were electrophoresed through about 20 cm of gel. The source of the faint band at around 2.5 kb in all lanes is unknown.
As a more sensitive measure, we sequenced the PCR products from several individual colonies for each of the three cell lines. Sequencing of longer CTG repeats typically gave a clear pattern of repeats followed abruptly by unreadable sequence. If the population comprises PCR fragments with different numbers of repeats—either due to slippage during PCR or due to a mixture of lengths in the starting population—the repeat pattern will remain evident through the longest major species. Beyond the repeats, however, the unique sequences will be out of phase, giving rise to unreadable sequences. One manifestation of this effect was evident in the sequences of the (CTG)183 PCR products. Each of the G-to-A changes in triplets 27 and 70 was represented as a major peak at the expected position, preceded by two to three smaller A peaks. Despite these problems, numbers of CTG repeats could be determined reproducibly from the sequencing runs. As shown in Table 1, no changes were detected in (CTG)17 repeats, but small changes in numbers of repeats occurred in 5 of 11 colonies isolated from the (CTG)98 cell line and in 7 of 10 colonies from the (CTG)183 cell line. Of the 12 colonies with altered repeat length, seven had longer repeats and five carried shorter ones. These frequent, minor changes serve as a baseline against which to evaluate the changes found among homologous recombinants.
TABLE 1.
Lengths of CTG triplet repeats in colonies from a population of proliferating cells
| Colonya | CTGb |
|---|---|
| GS3502 (parent) | 17 |
| GS3502-234 | 17 |
| GS3502-238 | 17 |
| GS3502-239 | 17 |
| GS3502-244 | 17 |
| GS3502-253 | 17 |
| GS3502-257 | 17 |
| GS3502-262 | 17 |
| GS3502-265 | 17 |
| GS3502-312 | 17 |
| GS3502-314 | 17 |
| GS3504 (parent) | 98 |
| GS3504-954 | 99 |
| GS3504-978 | 98 |
| GS3504-1051 | 98 |
| GS3504-1052 | 98 |
| GS3504-1059 | 98 (2)c |
| GS3504-1061 | 99 |
| GS3504-1063 | 99 |
| GS3504-1064 | 99 |
| GS3504-1068 | 99 (2)c |
| GS3504-1069 | 98 (2)c |
| GS3504-1072 | 98 (3)c |
| GS3506 (parent) | 183 |
| GS3506-1252 | 180 |
| GS3506-1255 | 184 |
| GS3506-1257 | 180 |
| GS3506-1261 | 185 |
| GS3506-1266 | 183 |
| GS3506-1276 | 181 |
| GS3506-1290 | 183 |
| GS3506-1298 | 182 |
| GS3506-1312 | 183 |
| GS3506-1317 | 174 |
Colonies from which the cell population was grown are indicated as “parent.” After about 30 cell doublings, individual colonies (numbered) were isolated and analyzed.
The length of the repeat is indicated as the number of CTG triplets.
Numbers in parentheses indicate the number of times that a sample was independently PCR amplified and sequenced. For each colony the results of independent analyses were the same.
Effects of CTG repeats on homologous recombination.
To assess whether CTG repeats affected homologous recombination at APRT, we selected for APRT− colonies from the APRT+ parent cell lines, which carry tandemly duplicated copies of APRT genes (Fig. 1B). Although these tandem duplications can give rise to APRT− cells in several ways, previous analyses of spontaneous events at the APRT locus in CHO cells indicated that homologous events were predominant, accounting for about 95% of the APRT− products, compared to 5% for mutations and <0.5% for rearrangements (63, 65). Homologous recombination can generate APRT− cells by crossover recombination, which eliminates one copy of the APRT gene, and by gene conversion, in which the exon 2 mutation in the upstream copy is transferred to the downstream copy (5) (Fig. 1B). In those studies we also showed that TK− APRT− cells were generated entirely by crossover recombination.
Initially, we measured the rates of production of APRT− and TK− APRT− cells to determine whether CTG repeats might stimulate homologous recombination, as long CTG repeats have been shown elsewhere to do in E. coli and yeast (17, 24, 25, 26, 27, 58, 59). As shown in Table 2, the (CTG)17, (CTG)98, and (CTG)183 cell lines yielded APRT− and TK− APRT− colonies at rates that were not substantially different from the mean rates for the other cell lines, which each carried the identical tandemly duplicated APRT locus, but with different DNA inserts in the downstream copy of the gene. Thus, CTG repeats of these lengths do not dramatically stimulate homologous recombination in mammalian cells.
To measure the contributions of homologous recombination, mutation, and rearrangement to the measured rates, independent APRT− and TK− APRT− colonies were isolated and examined by Southern blotting and PCR analysis. A combination of these two approaches was used to identify the molecular structure of the APRT locus and to assign each colony to a specific category of event. An example of a Southern blot analysis of BamHI-cleaved DNA from several APRT− colonies derived from the (CTG)98 cell line is shown in Fig. 3B. Colonies 194, 198, 207, 208, 210, and 218 (lanes 1, 5, 10, 11, 12, and 16, respectively) have Southern blot patterns that are consistent either with a conversion event that has retained the CTG sequence or with a point mutation elsewhere in the APRT gene. Subsequent PCR analyses showed that colonies 198, 207, and 218 (lanes 5, 10, and 16, respectively) are conversion events and that colonies 194, 208, and 210 (lanes 1, 11, and 12, respectively) arose by point mutation. Colonies 195 and 196 (lanes 2 and 3) are conversion events that have lost the repeats. Colonies 215 and 217 (lanes 13 and 15) are crossover events that have retained the CTG repeat, and colony 199 (lane 6) is a crossover event that has lost the repeat. Colonies 200 and 203 (lanes 7 and 8) are rearrangements. The results of all such analyses are listed in Table 3.
FIG. 3.
Southern analysis of APRT− colonies isolated from the (CTG)98 cell line, GS3504. (A) Restriction map for conversions and crossovers. Arrows indicate the sites at which BamHI cleaves, and the sizes of the resulting fragments are indicated in kilobases. Because a BamHI site is located adjacent to the CTG sequence in the insert, the Southern blot pattern depends on whether the insert is retained or lost. A conversion that has lost the insert yields fragments of 12.3 and 4.0 kb. If the insert is retained, the 4.0-kb fragment is replaced by a pair of fragments at 1.3 and 3.1 kb. Similarly, a crossover that has lost the insert yields a single band at 4.0 kb, whereas a crossover that has retained the repeat yields bands at 1.3 and 3.1 kb. (B) Southern blot of BamHI-digested DNA from APRT− colonies isolated from the (CTG)98 cell line, GS3504. DNAs from individual colonies were digested with BamHI, and the fragments were resolved by gel electrophoresis and made visible by Southern blotting. Numbers at the side indicate the lengths of fragments in kilobases. Numbers at the top identify the individual colonies. The structures of GS3504-200 and GS3504-203, which are rearrangements, are shown in more detail in Fig. 4.
TABLE 3.
Distribution of types of products among APRT− and TK− APRT− colonies
| Colony type and cell line | No. of products (% of total)
|
||||
|---|---|---|---|---|---|
| Conversions | Cross- overs | Mutations | Rearrange- ments | Total | |
| APRT− | |||||
| GS3502 (CTG)17 | 38 (69) | 10 (18) | 7 (13) | 0 | 55 |
| GS3504 (CTG)98 | 27 (51) | 11 (21) | 10 (19) | 5 (9) | 53 |
| GS3506 (CTG)183 | 31 (39) | 34 (43) | 8 (10) | 7 (9) | 80 |
| Othera | 172 (82) | 28 (13) | 10 (5) | 0 | 210 |
| TK−APRT− | |||||
| GS3502 (CTG)17 | 0 | 14 (100) | 0 | 0 | 14 |
| GS3504 (CTG)98 | 0 | 11 (85) | 0 | 2 (15) | 13 |
| GS3506 (CTG)183 | 0 | 27 (90) | 0 | 3 (10) | 30 |
| Otherb | 0 | 219 (100) | 0 | 0 | 219 |
Analysis of the distributions in Table 3 gives the first indication of a length-dependent effect of CTG repeats during recombination at the APRT locus. The distribution of APRT− products from the (CTG)17 cell line is not significantly different from the pooled distribution of products from the other cell lines (P = 0.06); however, the distributions for the (CTG)98 and (CTG)183 cell lines are dramatically different (P = 3 × 10−5 and P = 8 × 10−13, respectively). Similarly, the distributions of TK− APRT− products from the (CTG)98 and (CTG)183 cell lines are significantly different from the pooled distribution (P = 2 × 10−5 and P = 1 × 10−4, respectively).
As expected, most of the APRT− colonies for all CTG cell lines arose by homologous recombination. The proportion of homologous recombinants for the (CTG)17 cell lines (87%) is not significantly less than the 95% for the pooled data from the other cell lines (P = 0.07); however, the proportions of homologous recombinants for the (CTG)98 and (CTG)183 cell lines (72 and 82%, respectively) are significantly less (P = 7 × 10−7 and P = 4 × 10−4, respectively). This suggests that these two cell lines generate homologous recombinants at lower rates and/or generate mutations and rearrangements at higher rates.
To determine the rates for specific types of events for each cell line, the percentage of each event (Table 3) was multiplied by the overall rate of APRT− or TK− APRT− colony formation (Table 2). The individual rates are presented in Table 4. All three CTG cell lines have significantly lower rates of conversion relative to the pooled data from the other cell lines [(CTG)17, P = 0.02; (CTG)98, P = 0.0004; (CTG)183, P = 0.001]. In addition, the rates of crossover recombination in the APRT− population and in the TK− APRT− population are significantly elevated for the (CTG)183 cell line (P = 0.005 for both). These changes are evident in the ratios of conversions to crossovers, which decrease from 6.1 in cell lines that do not contain CTG repeats to 3.8 for the (CTG)17 cell line, 2.5 for the (CTG)98 cell line, and 0.9 for the (CTG)183 cell line.
Effects of homologous recombination on CTG repeats.
These results indicate that CTG repeats affect homologous recombination processes in their vicinity; however, they do not address the issue of CTG repeat stability during homologous recombination. One way that instability might manifest itself would be as changes in repeat length in APRT− and TK− APRT− recombinants. To detect such changes, conversion and crossover recombinants that retained their CTG repeats were screened by PCR. No changes were found in recombinants isolated from the (CTG)17 cell line. However, in addition to the expected small variations detected in replicating populations of (CTG)98 and (CTG)183 cell lines (data not shown), five large contractions were found among the 95 CTG-positive recombinants from the (CTG)98 and (CTG)183 cell lines. Contractions to 58, 76, and 81 repeats were isolated from the (CTG)98 cell line; contractions to 96 and 157 repeats were isolated from the (CTG)183 cell line. These contractions were all associated with homologous recombination events: three with conversions and two with crossovers. The contraction to 76 repeats, which arose in a conversion event, is shown in lane 10 in Fig. 3B. Five large contractions among 95 recombinants (5%) represent a significant increase (P = 0.002) over replicating cell populations, which yielded no large changes out of 252 colonies (<0.4%). These results indicate that long CTG repeats yield large contractions at a >10-fold-higher rate during homologous recombination than during replication.
A second way that instability might manifest itself is as rearrangements that disrupt the function of the APRT gene, or of both the TK and APRT genes. Among 176 APRT− and TK− APRT− colonies isolated from the (CTG)98 and (CTG)183 cell lines, 17 were rearrangements, accounting for nearly 10% of all colonies (Table 3). In contrast, the (CTG)17 cell line yielded no rearrangements out of 69 colonies, and the other cell lines (33, 63, 65, 66) have yielded no rearrangements out of 429 APRT− and TK− APRT− colonies (Table 3). Seventeen rearrangements out of 176 colonies (10%) from the (CTG)98 and (CTG)183 cell lines is significantly different (P = 2 × 10−12) from 0 rearrangements out of 498 colonies (<0.2%) from the (CTG)17 cell line and the pooled data from the other cell lines. Thus, in a population that has been selected for a nearby homologous recombination event, long CTG repeats are associated with a >50-fold-higher frequency of rearrangements (10% compared with <0.2%).
To determine whether there might be a direct link between recombination and the formation of rearrangements, 12 of the 17 rearrangements were examined by more extensive Southern blotting and PCR analyses (Fig. 4). These rearrangements include a variety of deletions and insertions, but their common feature is that they all involve the CTG sequence. In 7 of the 12 rearrangements, one or both of the restriction sites that flank the triplet repeat were deleted, indicating that the rearrangement encompasses the CTG repeat or ends within or adjacent to it. In the other five rearrangements, extra DNA was present at the site of the repeat.
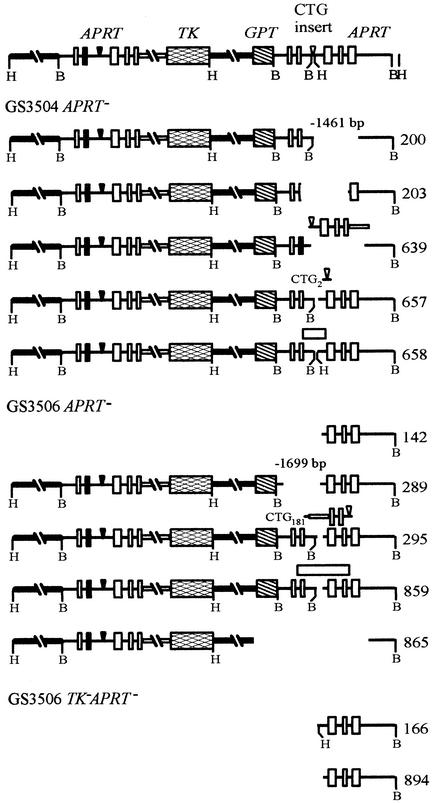
FIG. 4.
Molecular structures of rearrangements. The structure of the APRT locus in the parental (CTG)98 and (CTG)183 cell lines, GS3504 and GS3506, respectively, is indicated at the top. B and H indicate sites of BamHI and HindIII cleavage, respectively. The CTG tracts in these cell lines carry a BamHI site to the left of the CTG sequence and a HindIII site to the right. Numbers to the right identify individual colonies. Southern blotting was carried out on BamHI-digested DNA and on HindIII-digested DNA for each colony. PCR analyses were used to refine estimates of the positions of the ends of the rearrangements. Boxes above the site of the original CTG sequence indicate various inserted sequences. Open boxes in GS3504-658 and GS3506-859 indicate insertions that have not been further characterized. For GS3504-200, GS3504-639, GS3504-657, GS3506-289, and GS3506-295 the rearrangement junction fragment was isolated and sequenced.
In five cases the junction fragments were successfully amplified by PCR and sequenced. GS3504-200 from the (CTG)98 cell line and GS3506-289 from the (CTG)183 cell line were shown to be simple deletions in which the entire CTG repeat, along with adjacent sequences, was removed (Fig. 4). In the other three sequenced rearrangements, a segment of DNA including or immediately adjacent to the CTG repeat was deleted and other sequences were inserted in their place. Surprisingly, in each case the inserted sequences were derived from the upstream copy of the APRT gene.
GS3504-639 from the (CTG)98 cell line appears to have been derived from a conversion event that went awry. The downstream copy of the APRT gene now resembles the upstream copy: it includes the mutation in exon 2, the FRT site from the upstream sequence, the 3′ truncation of exon 5, and the contiguous plasmid sequences (Fig. 4). This homologous copy of the upstream sequence is joined nonhomologously through its plasmid sequences to the downstream APRT flanking sequences. GS3504-657 from the (CTG)98 cell line has picked up 34 bp from the upstream FRT sequence, which is joined nonhomologously to the remaining two CTG repeats and homologously to sequences on the 3′ side of the downstream FRT site (Fig. 4). GS3506-295 from the (CTG)183 cell line has retained its CTG repeat but carries an insert in place of a 65-bp deletion (Fig. 4). The insert consists of inverted copies of two discontinuous segments of upstream sequences. One segment includes plasmid sequences and contiguous APRT sequences extending from the truncated fifth exon into intron 3. The other segment is an inverted 47-bp copy of sequences around the upstream FRT site. These two segments are linked by a 6-bp sequence whose source is unclear.
The five sequenced rearrangements contain a total of eight nonhomologous junctions. For six of those junctions the sequences of the DNAs that formed the junction are known, allowing the homology at each of these junctions to be determined. These junctions display the microhomology that is typical of nonhomologous junctions in mammalian cells (64). One junction had one nucleotide of homology, one junction had two nucleotides of homology, and four junctions had three nucleotides of homology. Thus, the rearrangements generated during homologous recombination are complex, with elements of both homologous and nonhomologous recombination in evidence.
DISCUSSION
In this study we show that homologous recombination and DNA replication destabilize CTG triplet repeats in distinct ways, indicating that CTG repeats in CHO cells respond differently to these two processes. During replication, the repeats in the (CTG)17 cell line were stable, but those in the (CTG)98 and (CTG)183 cell lines were very unstable, giving rise to colonies with altered repeat lengths at very high frequencies: 45% for (CTG)98 and 70% for (CTG)183. These changes included expansions and contractions, usually less than three repeats in length.
In contrast to the small changes observed in replicating cells, large changes were common among homologous recombinants. Once again, the changes were confined to the longer CTG repeats, with (CTG)17 repeats being stable. The (CTG)98 and (CTG)183 cell lines showed two kinds of instability in response to recombination. Large contractions of these CTG repeats (greater than 15 repeats) were stimulated more than 10-fold by nearby homologous recombination events. High frequencies of recombination-stimulated repeat instability have also been reported for E. coli and yeast. In experiments where both contractions and expansions could be assayed, some studies have reported a two- to eightfold preponderance of contractions (26, 58, 59), whereas others have shown mainly expansions (17, 24, 25). Given the small number of characterized examples in the present study, we can conclude only that recombination-associated instability in our system is biased in favor of contractions.
The most striking manifestation of recombination-associated repeat instability, however, was a novel class of rearrangements that extended outside the CTG repeats. These occurred at a frequency more than 50-fold above the normal frequency of rearrangements (33, 63, 65, 66). Three of the five sequenced rearrangements had inserted DNA from the upstream copy of the APRT gene at the site of the CTG repeat. In two of these cases, one end of the rearrangement was clearly formed by a homologous recombination event, while the other end was formed by a nonhomologous event. These footprints of recombination at the sites of rearrangements involving the CTG repeats argue that homologous recombination is responsible for these events.
Rearrangements associated with triplet repeats have been noted previously for E. coli (24, 25, 57) and in patients with fragile X syndrome (13, 15, 45, 47, 69). The characterized fragile X rearrangements include deletions that encompass the CGG repeat (13, 69) and deletions that have one end within the repeat (45, 47), which are similar to the types of rearrangements that we have identified for CTG repeats. Like the more common CGG repeat expansion, deletions are observed at the fragile X MR gene (FMR1) because they can eliminate the function of FMR1, which is the basis for the disease phenotype. By contrast, deletions of CTG repeats and flanking DNA sequences have not been observed as the cause of DM, presumably because inactivation of the DMPK gene does not lead to the disease (28, 78). Our results suggest that careful examination of somatic tissues from DM patients would uncover cells with rearrangements of the type described here. Analysis of patient samples, which usually focuses on changes in repeat number with flanking PCR primers, may overlook these rearrangements. In general, rearrangements triggered by long tracts of triplet repeats might be expected to contribute to the progression of diseases such as fragile X syndrome and Friedreich ataxia, which are caused by loss-of-function mutations, but not of diseases such as DM and Huntington disease, which are caused by gain-of-function mutations (10, 78).
For both replication and transcription, the effects on triplet repeat stability in E. coli and yeast (6, 17, 23, 44, 53, 67) are accompanied by a reciprocal effect on the process itself. Triplet repeats have been shown elsewhere to interfere, for example, with the progress of DNA polymerase (31, 77) and RNA polymerase (54). These reciprocal effects are presumed to have the same root cause: the unusual structure adopted by the repeat. In this study we have shown not only that CTG triplet repeats are significantly destabilized by homologous recombination but also that CTG repeats have a reciprocal effect on homologous recombination. The most dramatic effect was observed for the (CTG)183 cell line, which displayed a three- to fourfold-lower rate of conversion and a two- to threefold-higher rate of crossover. In this cell line, an additional difference was evident among the crossover class of recombinants. In previous studies with other inserted sequences (63), the insert was retained (25 examples) about as often as it was lost (29 examples). Equal retention versus loss was also observed for (CTG)17 (12 versus 12) and for (CTG)98 (9 versus 13); however, for (CTG)183 there was a significant bias (P = 0.002) toward loss of the repeat (10 retained versus 41 lost).
In yeast, long triplet repeat sequences have been shown previously to cause frequent double-strand DNA breaks, which stimulate homologous recombination (3, 17, 26). In our studies, no substantial increase in homologous recombination was observed, suggesting that CTG repeats do not induce frequent DNA breaks at the APRT locus in CHO cells. Unlike E. coli and S. cerevisiae, however, which repair breaks almost exclusively by homologous recombination, mammalian cells repair breaks by both homologous and nonhomologous processes. It was shown previously that I-SceI-induced double-strand breaks at the site where the CTG repeat sequences were inserted lead to a 100-fold increase in homologous recombinants and to a 1,000-fold increase in rearrangements (64). In the present studies we observed more than a 50-fold increase in rearrangements for long CTG repeats. If these rearrangements had resulted from CTG-induced breaks and the ratio of break resolution by homologous and nonhomologous recombination were maintained, we might have expected a fivefold increase in homologous recombination, which would have been readily detected. This line of reasoning does not rule out a low frequency of CTG-induced breaks in CHO cells, but it does encourage us to think about other ways by which long CTG repeats might influence homologous recombination.
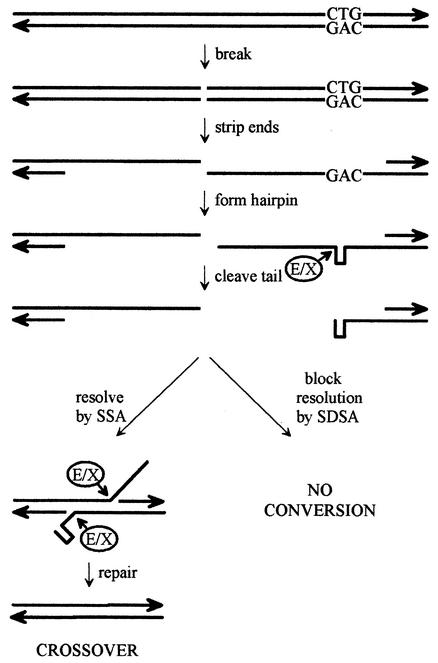
Focusing on the (CTG)183 cell line, where the effects were most evident, and on known recombination activities, we can envision the following pathway. We assume that recombination is initiated at a spontaneous double-strand break (that is, one not induced by the CTG repeat) and the ends are stripped normally, as outlined in Fig. 5. When the repeat becomes single stranded, however, we propose that it forms a hairpin, which triggers removal of the single-stranded tail by Ercc1/Xpf endonuclease (14). This nuclease is known to remove single-strand tails from standard DNA hairpins (14) but has not been tested against triplet repeat hairpins. The resulting hairpin cap might reasonably be expected to alter the ability of this intermediate to participate in conversions and crossovers. For example, if the terminal hairpin blocked strand invasion by preventing the loading of Rad51 (4, 49, 72), then gene conversion, which occurs predominantly by synthesis-dependent strand annealing (SDSA), would be inhibited, as it is in the (CTG)183 cell line. By contrast, formation of crossovers, which occurs mainly by single-strand annealing (SSA) and is not dependent on Rad51 (52), would not be inhibited and might be increased, as it is in the (CTG)183 cell line, if recombination intermediates are diverted from the more common SDSA pathway. This proposed scenario would also account for the preferential loss of repeats from the crossover class of recombinants in the (CTG)183 cell line. Finally, it is possible that the hairpin cap shunts this abnormal intermediate toward resolution by a nonhomologous pathway, leading to rearrangements. This model makes several testable predictions.
FIG. 5.
Speculative model for interaction of CTG repeats with homologous recombination. Although the upstream and downstream copies of the two APRT genes are not shown explicitly, the CTG repeats are present in the top strand of the downstream copy, as illustrated. To generate APRT− crossovers by SSA requires a double-strand break between the mutant and wild-type copies of exon 2, that is, to the left of the CTG repeat, as shown. Conversions by SDSA can be generated in a variety of ways that depend on invasion of a 3′ end, priming of DNA synthesis, release of the extended strand, and pairing with sequences on the other side of the break. The depicted events depart from the standard models for SSA and SDSA (52) at the formation of a hairpin by the triplet repeats and cleavage of the hairpin by the Ercc1/Xpf (E/X) endonuclease. It is hypothesized that a single strand ending in a hairpin is compromised for the strand invasion step in the SDSA pathway. If the initial break is in APRT sequences to the right of the repeat, the right-hand end will not have a hairpin and conversion may proceed normally.
In summary, our studies of CTG repeats in a mammalian cell model system have revealed that replication and recombination each destabilize CTG triplet repeats in a length-dependent manner. For (CTG)98 and (CTG)183, but not (CTG)17, replication caused frequent small changes, while homologous recombination caused frequent contractions and rearrangements. The relative roles of these two processes, as well as other aspects of DNA metabolism, in the normal etiology of triplet repeat diseases are unclear. Additional studies with model systems in mammalian cells and in animals will be required to define their contributions.
Acknowledgments
We thank Vincent Dion, Vera Gorbunova, Kathleen Marburger, Anna Pluciennik, and Andrei Seluanov for their comments on the manuscript.
This work was supported by National Institutes of Health grants GM38219 (J.H.W.), GM52982 (R.D.W.), NS37554 (R.D.W.), and ES11347 (R.D.W.) and by funds from the Robert A. Welch Foundation (R.D.W.) and the Muscular Dystrophy Association (J.H.W.).
REFERENCES
- 1.Ashizawa, T., D. G. Monckton, S. Vaishnav, B. J. Patel, A. Voskova, and C. T. Caskey. 1996. Instability of the expanded (CTG)n repeats in the myotonin protein kinase gene in cultured lymphoblastoid cell lines from patients with myotonic dystrophy. Genomics 36:47-53. [DOI] [PubMed] [Google Scholar]
- 2.Ashley, C. T., and S. T. Warren. 1995. Trinucleotide repeat expansion and human disease. Annu. Rev. Genet. 29:703-728. [DOI] [PubMed] [Google Scholar]
- 3.Balakumaran, B. S., C. H. Freudenreich, and V. A. Zakian. 2000. CGG/CCG repeats exhibit orientation-dependent instability and orientation-independent fragility in Saccharomyces cerevisiae. Hum. Mol. Genet. 9:93-100. [DOI] [PubMed] [Google Scholar]
- 4.Benson, F. E., P. Baumann, and S. C. West. 1998. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature 391:401-404. [DOI] [PubMed] [Google Scholar]
- 5.Bollag, R. J., A. S. Waldman, and R. M. Liskay. 1989. Homologous recombination in mammalian cells. Annu. Rev. Genet. 23:199-225. [DOI] [PubMed] [Google Scholar]
- 6.Bowater, R. P., A. Jaworski, J. E. Larson, P. Parniewski, and R. D. Wells. 1997. Transcription increases the deletion frequency of long CTG/CAG triplet repeats from plasmids in Escherichia coli. Nucleic Acids Res. 25:2861-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowater, R. P., and R. D. Wells. 2001. The intrinsically unstable life of DNA triplet repeats associated with human hereditary disorders. Prog. Nucleic Acid Res. 66:159-202. [DOI] [PubMed] [Google Scholar]
- 8.Box, G. E. P., W. G. Hunter, and J. S. Hunter. 1978. Statistics for experimenters. John Wiley & Sons, Inc., New York, N.Y.
- 9.Cleary, J. D., K. Nichol, Y. H. Wang, and C. E. Pearson. 2002. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat. Genet. 31:37-46. [DOI] [PubMed] [Google Scholar]
- 10.Cummings, C. J., and H. Y. Zoghbi. 2000. Trinucleotide repeats: mechanisms and pathology. Annu. Rev. Genomics Hum. Genet. 1:281-328. [DOI] [PubMed] [Google Scholar]
- 11.Darlow, J. M., and D. R. Leach. 1995. The effects of trinucleotide repeats found in human inherited disorders on palindrome inviability in Escherichia coli suggest hairpin folding preferences in vivo. Genetics 141:825-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, B. M., M. E. McCurrach, K. L. Taneja, R. H. Singer, and D. E. Housman. 1997. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl. Acad. Sci. USA 94:7388-7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Graaff, E., P. Rouillard, P. J. Willems, A. P. Smits, F. Rousseau, and B. A. Oostra. 1995. Hotspots for deletions in the CGG repeat region of FMR1 in fragile X patients. Hum. Mol. Genet. 4:45-49. [DOI] [PubMed] [Google Scholar]
- 14.de Laat, W. L., E. Appeldoorn, N. G. J. Jaspers, and J. H. J. Hoeijmakers. 1998. DNA structural elements required for ERCC1-XPF endonuclease activity. J. Biol. Chem. 273:7835-7842. [DOI] [PubMed] [Google Scholar]
- 15.de Vries, B. B., A. M. Wiegers, E. de Graaff, A. J. Verkerk, J. O. Van Hemel, D. J. Halley, J. P. Fryns, L. M. Curfs, M. F. Niermeijer, and B. A. Oostra. 1993. Mental status and fragile X expression in relation to FMR-1 gene mutation. Eur. J. Hum. Genet. 1:72-79. [DOI] [PubMed] [Google Scholar]
- 16.Fortune, M. T., C. Vassilopoulos, M. I. Coolbaugh, M. J. Siciliano, and D. G. Monckton. 2000. Dramatic, expansion-biased, age dependent, tissue-specific somatic mosaicism in a transgenic mouse model of triplet repeat instability. Hum. Mol. Genet. 9:439-445. [DOI] [PubMed] [Google Scholar]
- 17.Freudenreich, C. H., S. M. Kantrow, and V. A. Zakian. 1998. Expansion and length-dependent fragility of CTG repeats in yeast. Science 279:853-856. [DOI] [PubMed] [Google Scholar]
- 18.Gacy, A. M., G. Goellner, N. Juranic, S. Macura, and C. T. McMurray. 1995. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell 81:533-540. [DOI] [PubMed] [Google Scholar]
- 19.Gomes-Pereira, M., M. T. Fortune, and D. G. Monckton. 2001. Mouse tissue culture models of unstable triplet repeats: in vitro selection for larger alleles, mutational expansion bias and tissue specificity, but no association with cell division rates. Hum. Mol. Genet. 10:845-854. [DOI] [PubMed] [Google Scholar]
- 20.Harper, P. S. 1975. Congenital myotonic dystrophy in Britain. 2. Genetic basis. Arch. Dis. Child. 50:514-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harper, P. S. 1998. Myotonic dystrophy as a trinucleotide repeat disorder—a clinical perspective, p. 115-130. In R. D. Wells and S. T. Warren (ed.), Genetic instabilities and hereditary neurological diseases. Academic Press, Inc., San Diego, Calif.
- 22.Ishii, S., T. Nishio, N. Sunohara, T. Yoshihara, K. Takemura, K. Hikiji, S. Tsujino, and N. Sakuragawa. 1996. Small increase in triplet repeat length of cerebellum from patients with myotonic dystrophy. Hum. Genet. 98:138-140. [DOI] [PubMed] [Google Scholar]
- 23.Iyer, R. R., A. Pluciennik, W. A. Rosche, R. R. Sinden, and R. D. Wells. 2000. DNA polymerase III proofreading mutants enhance the expansion and deletion of triplet repeat sequences in Escherichia coli. J. Biol. Chem. 275:2174-2184. [DOI] [PubMed] [Google Scholar]
- 24.Jakupciak, J. P., and R. D. Wells. 1999. Genetic instabilities in CTG/CAG repeats occur by recombination. J. Biol. Chem. 274:23468-23479. [DOI] [PubMed] [Google Scholar]
- 25.Jakupciak, J. P., and R. D. Wells. 2000. Gene conversion (recombination) mediates expansions of CTG/CAG repeats. J. Biol. Chem. 275:40003-40013. [DOI] [PubMed] [Google Scholar]
- 26.Jankowski, C., F. Nasar, and D. K. Nag. 2000. Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proc. Natl. Acad. Sci. USA 97:2134-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jankowski, C., and D. K. Nag. 2002. Most meiotic CAG repeat tract-length alterations in yeast are SPO11 dependent. Mol. Genet. Genomics 267:64-70. [DOI] [PubMed] [Google Scholar]
- 28.Jansen, G., P. J. Groenen, D. Bachner, P. H. Jap, M. Coerwinkel, F. Oerlemans, W. van den Broek, B. Gohlsch, D. Pette, J. J. Plomp, P. C. Molenaar, M. G. Nederhoff, C. J. van Echteld, M. Dekker, A. Berns, H. Hameister, and B. Wieringa. 1996. Abnormal myotonic dystrophy protein kinase levels produce only mild myopathy in mice. Nat. Genet. 13:316-324. [DOI] [PubMed] [Google Scholar]
- 29.Jaworski, A., W. A. Rosche, R. Gellibolian, S. Kang, M. Shimizu, R. P. Bowater, R. R. Sinden, and R. D. Wells. 1995. Mismatch repair in Escherichia coli enhances instability of (CTG)n triplet repeats from human hereditary diseases. Proc. Natl. Acad. Sci. USA 92:11019-11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang, S., A. Jaworski, K. Ohshima, and R. D. Wells. 1995. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat. Genet. 10:213-218. [DOI] [PubMed] [Google Scholar]
- 31.Kang, S. M., K. Ohshima, M. Shimizu, S. Amirhaeri, and R. D. Wells. 1995. Pausing of DNA polymerase in vitro at specific loci in CTG and CGG triplet repeats from human hereditary disease genes. J. Biol. Chem. 270:27014-27021. [DOI] [PubMed] [Google Scholar]
- 32.Kaytor, M. D., E. N. Burright, L. A. Duvick, H. Zoghbi, and H. T. Orr. 1997. Increased trinucleotide repeat instability with advanced maternal age. Hum. Mol. Genet. 6:2135-2139. [DOI] [PubMed] [Google Scholar]
- 33.Kilburn, A. E., M. J. Shea, R. G. Sargent, and J. H. Wilson. 2001. Insertion of telomere repeat sequence into a mammalian gene causes chromosome instability. Mol. Cell. Biol. 21:126-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korade-Mirnics, Z., J. Tarleton, S. Servidei, R. R. Casey, M. Gennarelli, E. Pegoraro, C. Angelini, and E. P. Hoffman. 1999. Myotonic dystrophy: tissue-specific effect of somatic CTG expansions on allele-specific DMAHP/SIX5 expression. Hum. Mol. Genet. 8:1017-1023. [DOI] [PubMed] [Google Scholar]
- 35.Kovtun, I. V., and C. T. McMurray. 2001. Trinucleotide expansion in haploid germ cells by gap repair. Nat. Genet. 27:407-411. [DOI] [PubMed] [Google Scholar]
- 36.Kramer, P. R., C. E. Pearson, and R. R. Sinden. 1996. Stability of triplet repeats of myotonic dystrophy and fragile X loci in human mutator mismatch repair cell lines. Hum. Genet. 98:151-157. [DOI] [PubMed] [Google Scholar]
- 37.Kunst, C. B., E. P. Leeflang, J. C. Iber, N. Arnheim, and S. T. Warren. 1997. The effect of FMR1 CGG repeat interruptions on mutation frequency as measured by sperm typing. J. Med. Genet. 34:627-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lea, D. E., and C. A. Coulson. 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49:264-286. [DOI] [PubMed] [Google Scholar]
- 39.Leeflang, E. P., S. Tavare, P. Marjoram, C. O. S. Neal, J. Srinidhi, H. MacFarlane, M. E. MacDonald, J. F. Gusella, M. de Young, N. S. Wexler, and N. Arnheim. 1999. Analysis of germline mutation spectra at the Huntington's disease locus supports a mitotic mutation mechanism. Hum. Mol. Genet. 8:173-183. [DOI] [PubMed] [Google Scholar]
- 40.Lopez de Munain, A., A. M. Cobo, J. J. Poza, D. Navarrete, L. Martorell, F. Palau, J. I. Emparanza, and M. Baiget. 1995. Influence of the sex of the transmitting grandparent in congenital myotonic dystrophy. J. Med. Genet. 32:689-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luria, S. E., and M. Delbruck. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mankodi, A., M. P. Takahashi, H. Jiang, C. L. Beck, W. J. Bowers, R. T. Moxley, S. C. Cannon, and C. A. Thornton. 2002. Expanded CUG repeats trigger aberrant splicing of CIC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell 10:35-44. [DOI] [PubMed] [Google Scholar]
- 43.Martorell, L., K. Johnson, C. A. Boucher, and M. Baiget. 1997. Somatic instability of the myotonic dystrophy (CTG)n repeat during human fetal development. Hum. Mol. Genet. 6:877-880. [DOI] [PubMed] [Google Scholar]
- 44.Maurer, D. J., B. L. O'Callaghan, and D. M. Livingston. 1996. Orientation dependence of trinucleotide CAG repeat instability in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:6617-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meijer, H., E. de Graaff, D. M. Merckx, R. J. Jongbloed, C. E. de Die-Smulders, J. J. Engelen, J. P. Fryns, P. M. Curfs, and B. A. Oostra. 1994. A deletion of 1.6 kb proximal to the CGG repeat of the FMR1 gene causes the clinical phenotype of the fragile X syndrome. Hum. Mol. Genet. 3:615-620. [DOI] [PubMed] [Google Scholar]
- 46.Merrihew, R. V., R. G. Sargent, and J. H. Wilson. 1995. Efficient modification of the APRT gene by FLP/FRT site-specific targeting. Somat. Cell Mol. Genet. 21:299-307. [DOI] [PubMed] [Google Scholar]
- 47.Mila, M., S. Castellvi-Bel, A. Sanchez, C. Lazaro, M. Villa, and X. Estivill. 1996. Mosaicism for the fragile X syndrome full mutation and deletions within the CGG repeat of the FMR1 gene. J. Med. Genet. 33:338-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore, H., P. W. Greenwell, C.-P. Liu, N. Arnheim, and T. D. Petes. 1999. Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl. Acad. Sci. USA 96:1504-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.New, J. H., T. Sugiyama, E. Zaitseva, and S. C. Kowalczylowski. 1998. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391:407-410. [DOI] [PubMed] [Google Scholar]
- 50.Oussatcheva, E. A., V. I. Hashem, Y. Zou, R. R. Sinden, and V. N. Potaman. 2001. Involvement of the nucleotide excision repair protein UvrA in instability of CAG · CTG repeat sequences in Escherichia coli. J. Biol. Chem. 276:30878-30884. [DOI] [PubMed] [Google Scholar]
- 51.Panigrahi, G. B., J. D. Cleary, and C. E. Pearson. 2002. In vitro (CTG) · (CAG) expansions and deletions by human cell extracts. J. Biol. Chem. 277:13926-13934. [DOI] [PubMed] [Google Scholar]
- 52.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parniewski, P., A. Bacolla, A. Jaworski, and R. D. Wells. 1999. Nucleotide excision repair affects the stability of long transcribed CTG/CAG tracts in an orientation-dependent manner. Nucleic Acids Res. 27:616-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parsons, M. A., R. R. Sinden, and M. G. Izban. 1998. Transcriptional properties of RNA polymerase II within triplet repeat-containing DNA from the human myotonic dystrophy and Fragile X loci. J. Biol. Chem. 273:26998-27008. [DOI] [PubMed] [Google Scholar]
- 55.Pearson, C. E., and R. R. Sinden. 1996. Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry 35:5041-5043. [DOI] [PubMed] [Google Scholar]
- 56.Philips, A. V., L. T. Timchenko, and T. A. Cooper. 1998. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 280:737-740. [DOI] [PubMed] [Google Scholar]
- 57.Pluciennik, A., R. R. Iyer, P. Parniewski, and R. D. Wells. 2000. Tandem duplication. A novel type of triplet repeat instability. J. Biol. Chem. 275:28386-28397. [DOI] [PubMed] [Google Scholar]
- 58.Pluciennik, A., R. R. Iyer, M. Napierala, J. E. Larson, M. Filutowicz, and R. D. Wells. 2002. Long CTG CAG repeats from myotonic dystrophy are preferred sites for intermolecular recombination. J. Biol. Chem. 277:34074-34086. [DOI] [PubMed] [Google Scholar]
- 59.Richard, G.-F., G. M. Goellner, C. T. McMurray, and J. E. Haber. 2000. Recombination-induced CAG trinucleotide repeat expansions in yeast involve the MRE11-RAD50-XRS2 complex. EMBO J. 19:2381-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richard, G.-F., and F. Paques. 2000. Mini- and microsatellite expansions: the recombination connection. EMBO Rep. 1:122-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rolfsmeier, M. L., M. J. Dixon, and R. S. Lahue. 2000. Mismatch repair blocks expansions of interrupted trinucleotide repeats in yeast. Mol. Cell 6:1501-1507. [DOI] [PubMed] [Google Scholar]
- 62.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 63.Sargent, R. G., R. V. Merrihew, R. Nairn, G. Adair, M. Meuth, and J. H. Wilson. 1996. The influence of a (GT)29 microsatellite sequence on homologous recombination in the hamster adenine phosphoribosyltransferase gene. Nucleic Acids Res. 24:746-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sargent, R. G., M. Brenneman, and J. H. Wilson. 1997. Repair of site-specific double-strand breaks on a mammalian chromosome by homologous and illegitimate recombination. Mol. Cell. Biol. 17:267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sargent, R. G., R. L. Rolig, A. E. Kilburn, G. M. Adair, J. H. Wilson, and R. S. Nairn. 1997. Recombination-dependent deletion formation in mammalian cells deficient in the nucleotide excision repair gene ERCC1. Proc. Natl. Acad. Sci. USA 94:13122-13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sargent, R. G., J. L. Meservy, B. D. Perkins, A. E. Kilburn, Z. Intody, G. M. Adair, R. S. Nairn, and J. H. Wilson. 2000. Role of the nucleotide excision repair gene ERCC1 in formation of recombination-dependent rearrangements in mammalian cells. Nucleic Acids Res. 28:3771-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sarkar, P. S., H.-C. Chang, F. B. Boudi, and S. Reddy. 1998. CTG repeats show bimodal amplification in E. coli. Cell 95:531-540. [DOI] [PubMed] [Google Scholar]
- 68.Schmidt, K. H., C. M. Abbott, and D. R. Leach. 2000. Two opposing effects of mismatch repair on CTG repeat instability in Escherichia coli. Mol. Microbiol. 35:463-471. [DOI] [PubMed] [Google Scholar]
- 69.Schmucker, B., W. G. Ballhausen, and R. A. Pfeiffer. 1996. Mosaicism of a microdeletion of 486 bp involving the CGG repeat of the FMR1 gene due to misalignment of GTT tandem repeats at chi-like elements flanking both breakpoints and a full mutation. Hum. Genet. 98:409-414. [DOI] [PubMed] [Google Scholar]
- 70.Schumacher, S., I. Pinet, and M. Bichara. 2001. Modulation of transcription reveals a new mechanism of triplet repeat instability in Escherichia coli. J. Mol. Biol. 307:39-49. [DOI] [PubMed] [Google Scholar]
- 71.Schweitzer, J. K., and D. M. Livingston. 1997. Destabilization of CAG trinucleotide repeat tracts by mismatch repair mutations in yeast. Hum. Mol. Genet. 6:349-355. [DOI] [PubMed] [Google Scholar]
- 72.Shinohara, A., and T. Ogawa. 1998. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391:404-407. [DOI] [PubMed] [Google Scholar]
- 73.Sinden, R. R. 1999. Biological implications of the DNA structures associated with disease-causing triplet repeats. Am. J. Hum. Genet. 64:346-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith, G. K., J. Jie, G. E. Fox, and X. Gao. 1995. DNA CTG triplet repeats involved in dynamic mutations of neurologically related gene sequences form stable duplexes. Nucleic Acids Res. 23:4303-4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Timchenko, N. A., Z. J. Cai, A. L. Welm, S. Reddy, T. Ashizawa, and L. T. Timchenko. 2001. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J. Biol. Chem. 276:7820-7826. [DOI] [PubMed] [Google Scholar]
- 76.Tiscornia, G., and M. S. Mahadevan. 2000. Myotonic dystrophy: the role of the CUG triplet repeats in splicing of a novel DMPK exon and altered cytoplasmic DMPK mRNA isoform ratios. Mol. Cell 5:959-967. [DOI] [PubMed] [Google Scholar]
- 77.Usdin, K., and K. J. Woodford. 1995. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 23:4202-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Usdin, K., and E. Grabczyk. 2000. DNA repeat expansions and human disease. Cell. Mol. Life Sci. 57:914-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van den Broek, W. J., M. R. Nelen, D. G. Wansink, M. M. Coerwinkel, H. te Riele, P. J. Groenen, and B. Weiringa. 2002. Somatic expansion behavior of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 11:191-198. [DOI] [PubMed] [Google Scholar]
- 80.Waring, J. D., and R. G. Korneluk. 1998. Genetic studies of the myotonic dystrophy CTG repeat, p. 131-146. In R. D. Wells and S. T. Warren (ed.), Genetic instabilities and hereditary neurological diseases. Academic Press, Inc., San Diego, Calif.
- 81.Wells, R. D., and S. T. Warren (ed.). 1998. Genetic instabilities and hereditary neurological diseases. Academic Press, Inc., San Diego, Calif.
- 82.Wohrle, D., I. Kennerknecht, M. Wolf, H. Enders, S. Schwemmle, and P. Steinbach. 1995. Heterogeneity of DM kinase repeat expansion in different foetal tissues and further expansion during cell proliferation in vitro: evidence for a causal involvement of methyl-directed DNA mismatch repair in triplet repeat stability. Hum. Mol. Genet. 4:1147-1153. [DOI] [PubMed] [Google Scholar]
- 83.Wong, L. J., T. Ashizawa, D. G. Monckton, C. T. Caskey, and C. S. Richards. 1995. Somatic heterogeneity of the CTG repeat in myotonic dystrophy is age and size dependent. Am. J. Hum. Genet. 56:114-122. [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang, L., E. P. Leeflang, J. Yu, and N. Arnheim. 1994. Studying human mutations by sperm typing: instability of CAG trinucleotide repeats in the human androgen receptor gene. Nat. Genet. 7:531-535. [DOI] [PubMed] [Google Scholar]