Abstract
Peptide methionine sulfoxide reductase (MsrA), which repairs oxidized proteins, is present in most living organisms, and the cognate structural gene belongs to the so-called minimum gene set [Mushegian, A. R. & Koonin, E. V., (1996) Proc. Natl. Acad. Sci. USA 93, 10268–10273]. In this work, we report that MsrA is required for full virulence of the plant pathogen Erwinia chrysanthemi. The following differences were observed between the wild-type and a MsrA− mutant: (i) the MsrA− mutant was more sensitive to oxidative stress; (ii) the MsrA− mutant was less motile on solid surface; (iii) the MsrA− mutant exhibited reduced virulence on chicory leaves; and (iv) no systemic invasion was observed when the MsrA− mutant was inoculated into whole Saintpaulia ionantha plants. These results suggest that plants respond to virulent pathogens by producing active oxygen species, and that enzymes repairing oxidative damage allow virulent pathogens to survive the host environment, thereby supporting the theory that active oxygen species play a key role in plant defense.
Bacterial pathogenesis includes steps wherein the partners recognize each other and accordingly respond by activating specific sets of genes (1). Oxidative burst is a very well characterized response of mammal hosts toward bacterial infection (2). Oxidative burst happens once bacteria have been isolated after phagocytosis, thereby allowing the local concentration of active oxygen species (AOS) to reach antibacterial activity. With plant hosts, the situation remains unclear (3, 4). Biochemical investigations with plant cell cultures revealed two types of AOS responses (2–6). In incompatible interactions, i.e., those involving a plant and a pathogen that is not virulent on this plant, two phases of plant AOS release occur. In phase I, a transient and weak AOS release occurs and is considered non-specific because a wide range of biotic and abiotic stress will induce it. Phase II intervenes later (approximately 6 hr), lasts longer, and yields much higher AOS concentration (approximately 1–2 mM). In contrast, in compatible interactions, i.e., those yielding plant disease, only a phase I-like response is thought to take place.
AOS were recently proposed to orchestrate the so-called hypersensitive response (HR) of non-host, or resistant, plants infected by avirulent pathogens, i.e., in incompatible interactions (6, 7). HR culminates in local necrosis that surrounds and eventually blocks the invading avirulent pathogen. What about plants that are infected by virulent pathogens (i.e., in so-called compatible interactions) when no HR occurs and plant disease develops? Is the infected host unable to release AOS, as suggested by in vitro characterization of cell cultures? Are the virulent pathogens able to resist?
Erwinia chrysanthemi is often seen as a brute-force plant pathogen that macerates the plant cell wall of a wide array of dicotyledons, causing soft-rot disease without exhibiting much host specificity (8). Accordingly, it induces no HR, and virulence is thought to rely heavily, if not exclusively, on macerating enzymes. In this work, we report the surprising finding that peptide methionine sulfoxide reductase (MsrA), an enzyme repairing oxidative damage caused to protein (9–11), is important for virulence of E. chrysanthemi. This finding strongly supports the hypothesis that, in compatible interactions, virulent pathogens have developed protective systems against plant AOS.
MATERIALS AND METHODS
Strains, Phage, and Plasmids Used.
The wild-type E. chrysanthemi strain used was 3937. The original mutant strain was a derivative of 3937 and was previously listed as pin14 or RH7006 (12, 13). To avoid secondary mutations that could have been selected during storage of strain RH7006, the msrA∷MudIIPR3(CmR) mutation was transduced from RH7006 strain into a fresh wild-type 3937 background, yielding the MEH14 strain. Strain MEHK3 contains a msrA∷aphA3(KanR) mutation (see details below). Generalized transducing phage ΦEC2 was used to move mutations into different E. chrysanthemi 3937 strains. The Escherichia coli strains used were BL21 (Stratagene) for expression/purification protocols and TG1 for routine DNA manipulations. The cloning vectors used were pET22b+ (Novagen), pUC18 (14), and pLAFR3 (15) which, on insertion of the msrA gene, yielded the recombinant plasmids pTMS10, pUMS5, and pLAMS11, respectively. The pUMSK3 plasmid was derived from pUMS5 after inserting the aphA3 cassette (16) in the msrA gene (see constructions below).
Culture Conditions.
Luria–Bertani (LB) rich medium was used for routine bacterial growth. Ceria medium contains casamino acids (0.05%), KH2PO4 (8.5 g/liter), (NH4)2SO4 (1 g/liter), sodium citrate (0.5 g/liter), MgSO4 (0.1 g/liter), MnSO4 (10−6 M), FeSO4 (10−6 M), succinate (0.4%), and polygalacturonate (0.04%). pH value was set at 6.0 by using KOH. When required, tetracyclin (25 μg/ml), ampicillin (50 μg/ml), or kanamycin (25 μg/ml) was added. In the disk inhibition test with oxidative agents, FeSO4 was added to a final concentration of 50 μM.
Inverse PCR to Obtain Nucleotide Sequence.
Chromosomal DNA of the strain MEH14 was purified as described (17), subsequently restricted by the appropriate endonuclease, and ligated under dilute conditions (1 μg/ml) to favor intramolecular circularization. Restriction enzymes were AluI or HaeIII, which cut at the extremity of Mu DNA (18). PCR amplification was performed on this ligated DNA by using two diverging oligonucleotides, located either in the Mu S or in the Mu c extremity, as primers. Outward and inward oligonucleotides of extremity S were, respectively, S1 (5′-GCTAAAGTTTTCGCATTTATCGTG-3′) and S2 (5′-ACTACTGCTTTTTATTCATTACATGGG-3′). Outward and inward oligonucleotides of extremity c were, respectively, c1 (5′-CCGAATAATCCAATGTCCTCC-3′) and c2 (5′-GCTTGCAAGCCTGTAGTGCAA-3′). In one experiment, we used HpaI, which has no recognition site in the Mu DNA with oligonucleotides S1 and c1. Nucleotide sequence of the PCR product was obtained (Genome Express, Grenoble, France), and new rounds of inverse PCR allowed us to obtain ca. 2,276 nt of the pin14 locus. Nucleotide sequence of the msrA region of wild-type 3937 strain was obtained by direct PCR.
Construction of Plasmids.
A msrA-containing DNA fragment was obtained by PCR amplification by using E. chrysanthemi 3937 chromosomal DNA as template and oligonucleotides 5′ sense P14E, containing a EcoRI site (5′-CATCAGGAATTCACTTCCGAG-3′) and 3′ reverse complement P14B, containing a BamHI site (5′-TTTAACGGATCCTTTTCGCACAG-3′). P14E and P14B are located at approximately 400 nt and 280 nt, respectively, of the msrA ORF. PCR was performed for one step for 3 min at 94°C, followed by 30 cycles of 30 sec at 94°C, 30 sec at 55°C, and 60 sec at 72°C. The PCR product was restricted by using BamHI and EcoRI. The resulting restricted fragments were inserted into EcoRI–BamHI-restricted pUC18 or pLAFR3 vectors to yield pUMS5 and pLAMS11, respectively. The pUMSK3 was constructed by inserting the aphA3 cassette, obtained after Smal digestion of pUC18K (16) in the PmlI-digested pUMS5 plasmid. The PmlI site lies within the msrA gene at the position equivalent to the thirtieth amino acid. The pTMS10 plasmid was constructed as follows. The msrA ORF was amplified by PCR by using oligonucleotides 5′ sense P14N, containing NdeI site (5′-TATACATATGGTGATAGCGAATTTCGATAAG-3′) and a 3′ reverse complementing primer P14X with XhoI site (5′-GTGCTCGAGACCCTGCGGGGGCAGGCACA-3′). The PCR product was digested by NdeI and XhoI enzymes and ligated into the pET22b+ (Novagen) cut with NdeI and XhoI enzymes. msrA was expressed under the control of a T7 promoter in the E. coli BL21 strain, which contains a chromosomal copy of an isopropyl-d-thiogalactoside (IPTG)-inducible T7 RNA polymerase encoding gene. Moreover, the construction was done such that the encoded MsrA protein was fused with a (His)6 tag at its C terminus. All plasmid constructions were checked by DNA sequence analysis.
Construction of Strains.
MEH14 was obtained by ΦEC2-transducing msrA∷MudIIPR3 from the original RH7006 strain (12, 13) into strain 3937. The msrA∷MudIIPR3 mutation was checked by PCR analysis of CmR transductants by using either the pair of 5′ sense primers P143 (5′-CACCCGTGGTTGTAAAACACGTTTCGG-3′, located 137 nt upstream of the initiation codon of msrA) and reverse complementary 3′ S1 (see above), or the pair of 5′ sense primer c1 (see above) and 3′ reverse complementary P145 (5′-GCGGTTTCCTGCTCTGTGGTT-3′, located within msrA, 448 nt downstream of the initiation codon). MEHK3 strain was obtained by recombining the pUMSK3-carried msrA∷aphA3 mutation into the E. chrysanthemi 3937 chromosome. Plasmid pUMSK3 was introduced into strain 3937 by electroporation. One AmpR KanR clone was then inoculated into 4 ml LB liquid medium in the absence of antibiotics at 30°C for 24 hr. Twenty μl were used subsequently for inoculating a fresh 4-ml culture of LB. The operation was repeated six times. Samples of each subculture were spread onto LB plates containing kanamycin and replicated onto LB plates containing ampicillin. After six subcultures, KanR AmpS colonies were isolated. PCR analysis by using appropriate msrA primers (see above) allowed confirmation of the substitution of the wild-type msrA gene by the msrA∷aphA3 mutated allele.
Purification of MsrA Protein.
A culture of E. coli BL21/pTMS10 was grown in LB added with ampicillin. At OD600 of 1, IPTG was added to a final concentration of 1 mM, and growth was continued for an additional 2 hr. Cells were harvested by centrifugation and resuspended in phosphate buffer (20 mM, pH 7.2), centrifuged again and resuspended in phosphate buffer (20 mM, pH 7.2) added with β-mercaptoethanol (10.7 mM), NaCl (0.5 M), and MgCl2 (5 mM). Cells were lysed by being passed through a French press, the resulting lysate was centrifuged, and the supernatant applied to a nickel-containing HiTrapTM chelating resin column (Pharmacia). The column was washed once with 5 ml of buffer A (NaCl 0.5M/phosphate buffer 20 mM, pH 7.2), then with 15 ml of buffer A added with 0.1 M imidazole. The MsrA-(His)6 protein was eluted by a linear gradient of 0.1–0.5 M imidazol in buffer A. Fractions were analyzed by SDS/PAGE with Coomassie blue staining. MsrA-(His)6 protein was identified by immunoblot using anti-(His)6 tag (Invitrogen) by using the protocol previously described (19).
Enzymatic Activity Assays.
Kinetic analysis of MsrA activity was performed at 25°C by monitoring H-transfer from NADPH to Met(O) or N-Ac-Met(O), by recording variation in OD340. The reaction mixture consisted of 50 mM Tris⋅HCl, pH 7.4/0.2 mM NADPH/78.43 μg/ml thioredoxin/3.04 ng/ml thioredoxin reductase (kindly provided by C. Williams, University of Michigan, Ann Arbor, MI), and 3.3 mM Met(O) or N-Ac-Met(O) as substrates (20).
Motility Test.
An inoculum containing 105 cells from an overnight culture, grown at 30°C, was infiltrated into a fresh low agar (0.4%) containing Ceria medium plates. After 24-hr incubation at 30°C, the extent of motility was evaluated by measuring the area of colonization around the inoculation site. A typical experiment includes testing simultaneously three samples of the same culture, each on a different plate, and averaging the area values obtained.
Disk Inhibition Assay.
Cells were grown up to OD600 value of 1.0 in Ceria medium. A 0.25-ml fraction was mixed with 2.5 ml soft agar and poured onto Ceria medium-containing plates. Filter disks were impregnated with 400 mM H2O2 and put on top of the bacterial lawns. The control experiment was done with a filter soaked in water. Plates were incubated overnight at 30°C.
Virulence Tests.
Virulence test on chicory leaves was done as follows. Cultures were grown overnight in Ceria medium at 30°C. OD600 was measured and 105 cells were infiltrated into the chicory leaves, which were subsequently incubated at 30°C into large glass Petri dishes containing moistened filter paper. The area of maceration was measured after 24 hr at 30°C. In a typical experiment, 20 leaves were inoculated per strain tested. Care was taken to test wild-type and mutated strains on leaves from the same chicory head. Virulence tests on potted Saintpaulia ionantha were carried out as previously described (21).
RESULTS
Sequence Analysis of the pin14 Locus.
Candidate virulence factor genes were previously isolated after MudIIPR3 transposon mutagenesis of strain 3937 (12, 13). A collection of so-called pin loci (for plant inducible) was obtained, the expression of which required plant extract to be added to culture medium, and the inactivation of which impaired the virulence of E. chrysanthemi. The present study focuses on the so-called pin14 locus. The pin14 locus was transduced from strain RH7006 back into the wild-type strain 3937, yielding strain MEH14 (see Materials and Methods for details on strain history). Analysis of the altered nucleotide sequence and of its counterpart in the wild-type strain showed a gene consisting of a 642 nt ORF that would encode a protein of 213 amino acids with predicted molecular mass of 23.7 kDa. A potential ribosome-binding site, reading AGGAG, was found 6 nt upstream from a GTG potential initiation codon. No potential transcription terminator was found downstream of the UAA stop codon. Comparison of the predicted amino acid sequence to protein and DNA data banks identified a series of homologs existing in a wide spectrum of organisms belonging to mammals (20), plants, yeast (22), archea, or eubacteria (23). The size of the homologs was approximately 200 residues except in a few cases, where this sequence appeared as a discrete domain within a larger polypeptide (24). These sequences are collectively referred to as peptide methionine sulfoxide reductase (9–11). Because, as shown below, we demonstrated that the pin14 locus encodes a protein exhibiting peptide methionine sulfoxide reductase activity, it is designated msrA after its E. coli homolog (25–27).
The E. chrysanthemi msrA ORF is surrounded by two long intergenic regions of 380 nt and 295 nt upstream and downstream, respectively. Upstream, the beginning of an ORF in the opposite orientation is found while an ORF running in the same direction as msrA is found downstream. This genetic organization is similar to that found in the E. coli msrA-centered region with the exception that a REP sequence is found in the intergenic region downstream of E. coli msrA but not in the E. chrysanthemi counterpart.
Peptide Methionine Sulfoxide Reductase Activity Associated with MsrA.
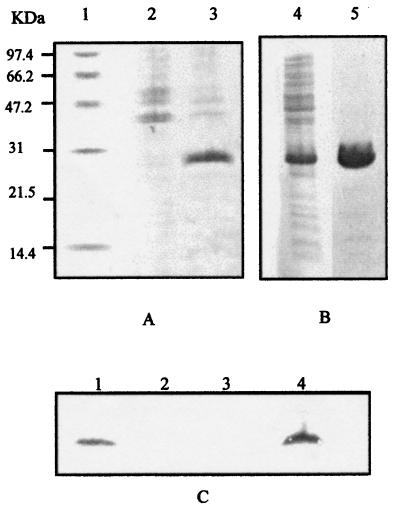
Cells of E. coli BL21/pTMS10 were grown in the presence of the inducer IPTG to overproduce MsrA-(His)6 protein. Fig. 1A shows that a strain containing pTMS10 synthesized a protein of approximately 27 kDa. This protein was recognized by antibodies directed against the (His)6 tag (not shown). The MsrA-(His)6 was purified to homogeneity by using a nickel affinity column (Fig. 1B). It was submitted to partial Edman degradation, and the NH2 terminal amino acid sequence (VIANFDK) obtained was the same as the nucleotide sequence-derived amino acid sequence. Pure MsrA-(His)6 was subsequently assayed for activity by using Met(O) and N-Ac-Met(O) as substrates and thioredoxin as a reductant. Specific activities obtained were of the same magnitude as those reported with the E. coli enzyme, i.e., 38 nmol/mg per min and 32 nmol/mg per min for Met(O) and N-Ac-Met(O) substrates, respectively (22, 26). N-Ac-Met(O) is a model for peptide link; this showed that MsrA can reduce either free oxidized-methionine or protein-contained oxidized methionine.
Figure 1.
Electrophoretic characterization of MsrA protein. (A) SDS/PAGE analysis of E. coli cultures grown in LB medium with added IPTG (1 mM). Proteins were visualized by Coomassie staining. Lane 1: size markers; lane 2: E. coli BL21/pET22b+; lane 3: E. coli BL21/pTMS10. (B) SDS/PAGE analysis of the purification of MsrA-(His)6. Lane 1: BL21/pTMS10 extract; lane 2, MsrA-(His)6 protein eluted from the Ni-NTA resin after treatment with imidazol. (C) Immunoblot analysis of E. chrysanthemi strains 3937 (lane 1), MEH14 (lane 2), MEH14/pLAFR3 (lane 3), MEH14/pLAMS11 (lane 4) by using anti-MsrA antibodies.
Peptide Methionine Sulfoxide Reductase Is Important for E. chrysanthemi Virulence.
Virulence tests were carried out by using chicory leaves as plant material. Initial characterization was conducted by using MEH14 wherein MudIIPR3 could cause a polar effect on downstream gene expression. Therefore, the msrA gene was cloned in a pLAFR3 low copy number vector, giving rise to pLAMS11. Immunoblot analysis showed that MEH14 strain carrying the pLAMS11 plasmid contained roughly wild-type levels of MsrA protein (Fig. 1C). MEH14/pLAMS11 was much more virulent than MEH14/pLAFR3 because, (i) the complemented strain caused maceration of twice as many leaves as the mutant, and (ii) lesion size was on average larger with the complemented strain than with the mutant; comparison of lesion size produced by MEH14/pLAFR3 and MEH14/pLAMS11 gave highly significant differences through the χ2 test, i.e., χ2 = 19.05, P < 0.01. (Table 1). This result confirmed the importance of msrA for virulence.
Table 1.
Virulence tests on chicory leaves
| Strain | Number of leaves with lesion, % | Size of lesions, cm2 |
|---|---|---|
| MEH14/pLAFR3 | 39 | 3.48 ± 1.71 |
| MEH14/pLAMS11 | 87.5 | 7.26 ± 4.72 |
Each chicory leaf was inoculated with 105 bacterial cells. ±, standard deviation.
Peptide Methionine Sulfoxide Reductase Is Important for Systemic Invasion of S. ionantha Whole Plants.
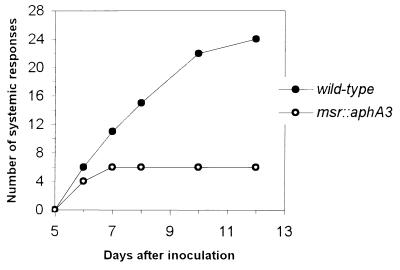
To know more precisely the role of MsrA in E. chrysanthemi virulence, comparison of wild-type and MsrA− strains was carried out by using whole potted plants. The aphA3 cassette, which is designed to avoid polar effects on downstream gene expression (16), was inserted into the cloned msrA gene and the resulting msrA∷aphA3 allele recombined into the 3937 chromosome, yielding MEHK3 strain. The behavior of the MsrA− mutant was compared with that of the parental strain 3937 after inoculation of S. ionantha potted plants (Fig. 2). In all plants, inoculation of both strains resulted in a patch of maceration at the inoculation site after 1 day. The rotting symptom continued to spread throughout the leaf and its petiole and, after 6 days, maceration expanding to a second petiole was detected in 6 of the 24 plants infected with the wild-type and in 4 of the 24 plants infected with the mutant. After 10 days, the infection with the wild-type strain resulted in a collapse of the whole plant, whereas with the mutant there was no symptom progression, and the initial lesion dried out. This result showed the importance of a functional MsrA in the spread of bacterial cells throughout the infected plant.
Figure 2.
Evolution of symptoms induced by E. chrysanthemi 3937 (wt) and its mutant derivative MEHK3 (msrA∷aphA3) strains on S. ionantha plants. For each strain, 24 plants (one leaf per plant) were inoculated. Response was considered as systemic when at least two leaves with their petioles were macerated.
Peptide Methionine Sulfoxide Reductase Affects E. chrysanthemi Resistance to Oxidative Stress.
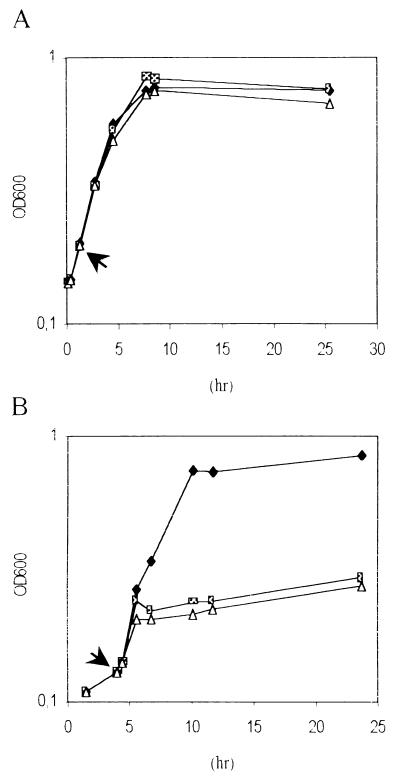
An experiment using liquid cultures revealed hypersensitivity of the MEH14 strain to oxidative agents because it stopped growing in the presence of paraquat 2 μM, whereas the wild-type strain resisted up to 5 μM (Fig. 3). Note that the numbering of colony-forming units paralleled the results obtained by reading OD600 values (not shown). Interestingly, by using a disk inhibition test, the difference in susceptibility to oxidants (paraquat or H2O2) was clearly visible only if FeSO4 (50 μM) was added to the medium (not shown). Fe2+ ions are well known to catalyze AOS formation by means of the Fenton reaction (4). Sensitivity to H2O2 of 3937/pLAFR3, MEH14/pLAFR3 and MEH14/pLAMS11 was tested by the disk assay. Comparison of the areas of growth inhibition demonstrated that the hypersensitivity of MEH14/pLAFR3 was caused by the lack of a functional msrA gene, thereby establishing the importance of MsrA in protecting the cells from oxidative agents (Table 2).
Figure 3.
Effect of paraquat on the growth of (A) E. chrysanthemi 3937 (wt) and (B) MEH14 (msrA∷MudIIPR3) strains. Strains were grown in Ceria medium. OD600 were recorded. Addition of paraquat is indicated by an arrow. ♦: no paraquat; ▩: 2 μM paraquat; and ▵: 5 μM paraquat. Three repetitions gave similar results. Values for one typical experiment are shown.
Table 2.
Effect of H2O2 treatment on E. chrysanthemi growth
| Strain | Area of inhibition, cm2 |
|---|---|
| 3937/pLAFR3 | 4.57 ± 0.17 |
| MEH14/pLAFR3 | 10.89 ± 1.47 |
| MEH14/pLAMS11 | 4.84 ± 0.18 |
Disks were impregnated with H2O2 (400 mM) and laid on top of bacterial lawns. The amount of growth inhibition was calculated as the area of growth inhibition minus the disk area. Experiments were made by using Ceria medium with added tetracycline and FeSO4 (50 μM). Means and standard deviation (indicated by ±) of triplicate determinations are from two independently grown cultures.
Peptide Methionine Sulfoxide Reductase Affects E. chrysanthemi Spreading on Solid Surfaces.
Bacterial spreading on low-agar medium (0.4%) was found to depend on a functional MsrA. Colony spreading on low-agar Ceria medium plates revealed that MEH14/pLAFR3 spread less efficiently around the site of inoculation than 3937/pLAFR3 (Table 3). The involvement of msrA in this behavior was demonstrated by the ability of the pLAMS11 plasmid to complement motility defects (Table 3).
Table 3.
MsrA is important for E. chrysanthemi spreading on solid surfaces
| Strain | Area of growth/cm2 |
|---|---|
| 3937/pLAFR3 | 1.85 ± 0.2 |
| MEH14/pLAFR3 | 0.64 ± 0.1 |
| MEH14/pLAMS11 | 3.6 ± 0.6 |
Strains were inoculated on 0.4% agar Ceria medium plates and incubated for 24 h at 30°C. Area of growth zones away from the initial inoculation point was measured. Means and standard deviation (indicated by ±) of triplicate determinations are from three independently grown cultures.
DISCUSSION
Virulence of the plant pathogen E. chrysanthemi has been studied mainly in relation to its ability to produce extracellular macerating enzymes (8). To date, iron-scavenging systems are the only other known virulence-associated function (28, 29). To identify new factors, a genetic strategy is to search among genes whose expression is activated selectively when the bacterium multiplies either within the infected host or in synthetic media mimicking the host internal milieu (30). F.V.G. and Beaulieu applied this strategy and looked for genes that would be activated when the bacterium multiplies in the presence of plant extracts (12, 13). They isolated a collection of so-called pin genes, of which 10 were selected as bona fide new virulence factors, i.e., they did not affect systems involved in synthesizing, regulating, or secreting degradative enzymes, or in iron assimilation. In the present study, one of these loci, pin14, was identified as being msrA, encoding peptide methionine sulfoxide reductase.
In so-called incompatible interactions between plant and avirulent pathogens, AOS are thought to be instrumental in inducing the HR (6). In contrast, in compatible interactions plant-released AOS are thought to be of little physiological relevance, and, in particular, are considered not to reach levels high enough to be antimicrobial (4). Yet, transgenic potatoes, in which elevated levels of H2O2 were artificially generated by glucose oxidase, exhibited good resistance to soft-rot disease caused by Erwinia carotovora (31), showing that an artificially induced oxidative burst could be used to prevent bacterial infection in compatible interactions. In the present study, we report that full virulence of E. chrysanthemi requires peptide methionine sulfoxide reductase, an enzyme that repairs oxidized proteins. Thus, in contrast to the current view, this finding suggests that, in susceptible interaction, plant AOS release occurs at a level such that antioxidative repair systems are necessary for virulent pathogens to survive and/or to maintain their destructive power.
The msrA mutant was unable to cause systemic invasion of whole plants. One possible reason for this is that populations of msrA mutants do not survive well in plant tissues, and the absence of systemic infection could be caused by a decline in population. However, no obvious difference in bacterial fitness was observed when cultures of wild-type and msrA mutant strains were compared under various laboratory conditions (minimal or rich media, starvation, etc.) or in coculture with tobacco cells (data not shown). A second possibility is that isolated cells of msrA mutants do not survive well in plant tissues. This hypothesis can be related to the finding that in E. coli, widely different levels of resistance to H2O2 are attained depending on whether one deals with individual cells or with high cell-density populations (32). Thus, a mutated cell leaving the site of inoculation, wherein high cell density afforded protection, would be killed before it could colonize other parts of the plant. This hypothesis could explain the absence of systemic invasion observed with infected potted Saintpaulia. A third possibility is that oxidation damages virulence factors which, in the absence of MsrA repairing function, become limiting for bacterial development throughout the plant. The physiological substrate(s) of MsrA in the cell is presently unknown. However, at least one putative specific target was identified in this study, i.e., motility. Flagellar-based motility was identified as being important for virulence of the closely related E. carotovora (33). However, in the present work as well as in the initial characterization of the pin14 mutant (13), E. chrysanthemi msrA cells appeared motile under light microscopy, suggesting that flagellar-dependent motion is not affected by msrA mutation. In contrast, plate assays revealed a reduced efficiency of the msrA mutant to colonize plates away from the inoculation site. A peculiar type of motility might be under msrA control, and/or some other process is at work in the bacterial spreading on solid surfaces (e.g., chemotaxis, adhesion, etc.). We tested twitching motility, as reported for the human pathogen Pseudomonas aeruginosa (34), but we were unable to detect such a mode of motion with the wild-type E. chrysanthemi 3937 strain (not shown). Therefore, our ongoing studies aim at characterizing the MsrA-dependent mechanism underlying spreading of E. chrysanthemi on solid surfaces. In particular, it will be extremely interesting to know if alteration of the MsrA-dependent spreading provides a basis for understanding the inability of the mutant to invade systemically S. ionantha plants.
In recent years, molecular analysis of virulence determinants in plant and mammalian pathogens revealed that both types of pathogens share a number of highly related mechanisms or strategies (35). These mechanisms include protein secretion systems (e.g., type II, type III), regulatory circuits (e.g., two-component, quorum sensing), and iron-capture systems. The present work reveals that MsrA is a new component of the basic microbial pathogenic program. Indeed, MsrA was found to be important for virulence in three mammalian pathogens, e.g., Streptococcus pneumoniae, Neisseria gonorrheae, and uropathogenic E. coli (36). Interestingly, in the case of mammalian pathogens, MsrA was proposed to be required for the functioning of adhesins. Such a proposal might be related to the observed reduced motility of the msrA mutant because adhesion and motility depend on related cell-bound extracellular appendices. Is MsrA specialized in repairing surface layers? A direct physical interaction seems unlikely because MsrA is cytoplasmic and adhesins or pilins, for instance, locate outside of the cells. A possibility is that MsrA has dual functions as a general repair system and as a regulator of extracellular appendices production. Alternatively, production of these appendices might be under the control of a regulator that is exquisitely sensitive to oxidative stress and preferentially repaired by MsrA.
Methionine sulfoxide reductase has been known for a long time to be present in all living organisms. The presence of msrA in the minimal gene set compatible with modern cellular life was therefore not unexpected (38). However, despite a broad knowledge of its biochemistry, the cellular role of methionine sulfoxide reductase remained poorly understood. A previous study revealed a role in protecting E. coli from oxidative stress under laboratory conditions (26). It is interesting to note that phenotypic screening based on conditions found in natural habitats, i.e., either plant host in the present study or animal in other studies (36), revealed a physiological role of methionine sulfoxide reductase in microbial pathogenesis. Similar strategies are likely to be required to identify the role of genes with no predicted function identified by whole genome sequencing approaches (37).
Acknowledgments
We are grateful to Dr. C. Williams (University of Michigan, Ann Arbor, MI) for a generous gift of thioredoxin reductase. Thanks to M. Bounias for his help in statistics, to Prof. Saghi (University of Fès, Morocco) for his help in allowing M.E.H. to come to France, to V. Mejean for suggestions, and to the members of the Erwinia group for valuable discussions. This work was made possible by grants from the Centre National de la Recherche Scientifique (ACC-SV6) and from the University of Aix-Marseille II, France. F.V.G. is a Charge de Recherche at the Institut National Recherche Agronomique. M.E.H. is on leave from the University of Fès, Morocco, and was supported by grants from the Communauté des Communes de Marseille, from a Centre National de la Recherche Scientifique–Centre National Recherche (Morocco) joined program, and from the Fondation pour la Recherche Médicale.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: AOS, active oxygen species; HR, hypersensitive response; msrA, methionine sulfoxide reductase; LB medium, Luria–Bertani medium; IPTG, isopropyl-d-thiogalactoside.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ012716).
References
- 1.Finlay B B, Falkow S. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scandalios J G. Oxidative Stress and the Molecular Biology of Antioxidant Defenses. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [Google Scholar]
- 3.Lamb C, Dixon R A. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251–257. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 4.Baker C J, Orlandi E W. Annu Rev Phytopathol. 1995;33:299–321. doi: 10.1146/annurev.py.33.090195.001503. [DOI] [PubMed] [Google Scholar]
- 5.Legendre L, Rueter S, Heinstein P F, Low P S. Plant Physiol. 1993;102:233–240. doi: 10.1104/pp.102.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine A, Tenhaken R, Dixon R, Lamb C. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 7.Lindgren P B. Annu Rev Phytopathol. 1997;35:129–152. doi: 10.1146/annurev.phyto.35.1.129. [DOI] [PubMed] [Google Scholar]
- 8.Barras F, Van Gijsegem F, Chatterjee A K. Annu Rev Phytopathol. 1994;32:201–234. [Google Scholar]
- 9.Brot N, Weissbach L, Werth J, Weissbach H. Proc Natl Acad Sci USA. 1981;78:2155–2158. doi: 10.1073/pnas.78.4.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abrams W R, Weinbaum G, Weissbach L, Weissbach H, Brot N. Proc Natl Acad Sci USA. 1981;78:7483–7486. doi: 10.1073/pnas.78.12.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brot N, Weissbach H. Arch Biochem Biophys. 1983;223:271–281. doi: 10.1016/0003-9861(83)90592-1. [DOI] [PubMed] [Google Scholar]
- 12.Beaulieu C, Van Gijsegem F. J Bacteriol. 1990;172:1569–1575. doi: 10.1128/jb.172.3.1569-1575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beaulieu C, Van Gijsegem F. Mol Plant–Microb Interact. 1992;5:340–346. [Google Scholar]
- 14.Yanisch-Perron C, Vieira J, Messing Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 15.Staskawicz B, Dahlbeck D, Keen N T, Napoli C. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ménard R, Sansonetti P J, Parsot C. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley Interscience; 1987. [Google Scholar]
- 18.Symonds N, Toussaint A, Van De Putte P, Howe M H. Phage Mu. Plainview, NY: Cold Spring Harbor Lab. Press; 1987. [Google Scholar]
- 19.Py B, Chippaux M, Barras F. Mol Microbiol. 1993;7:785–793. doi: 10.1111/j.1365-2958.1993.tb01169.x. [DOI] [PubMed] [Google Scholar]
- 20.Moskovitz J, Weissbach H, Brot N. Proc Natl Acad Sci USA. 1996;93:2095–2098. doi: 10.1073/pnas.93.5.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reverchon S, Expert D, Robert-Baudouy J, Nasser W. J Bacteriol. 1997;179:3500–3508. doi: 10.1128/jb.179.11.3500-3508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moskovitz J, Berlett B S, Poston J M, Stadtman E R. Proc Natl Acad Sc USA. 1997;94:9585–9889. doi: 10.1073/pnas.94.18.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maniloff J. Proc Natl Acad Sci USA. 1997;93:10004–10006. doi: 10.1073/pnas.93.19.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corpet F, Gouzy J, Kahn D. Nucleic Acids Res. 1998;26:323–326. doi: 10.1093/nar/26.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahman M A, Nelson H, Weissbach H, Brot N. J Biol Chem. 1992;267:15549–15551. [PubMed] [Google Scholar]
- 26.Moskovitz J, Rahman M A, Strassman J, Yancey S O, Kushner S R, Brot N, Weissbach H. J Bacteriol. 1995;177:502–507. doi: 10.1128/jb.177.3.502-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahman M A, Brot N, Weissbach H. Cell Mol Biol. 1992;38:529–542. [PubMed] [Google Scholar]
- 28.Expert D, Enard C, Masclaux C. Trends Microbiol. 1996;4:232–236. doi: 10.1016/0966-842X(96)10038-X. [DOI] [PubMed] [Google Scholar]
- 29.Masclaux C, Expert D. Plant J. 1995;7:121–128. [Google Scholar]
- 30.Heithoff D M, Conner C P, Mahan M J. Trends Microbiol. 1997;5:509–513. doi: 10.1016/S0966-842X(97)01153-0. [DOI] [PubMed] [Google Scholar]
- 31.Wu G, Shortt B J, Lawrence E B, Levine E B, Fitzsimmons K C, Shah D M. Plant Cell. 1995;7:1357–1368. doi: 10.1105/tpc.7.9.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma M, Eaton J W. Proc Natl Acad Sci USA. 1992;89:7924–7928. doi: 10.1073/pnas.89.17.7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulholland V, Hinton J C, Sidebotham J, Toth I K, Hyman L J, Perombelon M C, Reeves P J, Salmond G P. Mol Microbiol. 1993;9:343–356. doi: 10.1111/j.1365-2958.1993.tb01695.x. [DOI] [PubMed] [Google Scholar]
- 34.Darzins A. Mol Microbiol. 1994;11:137–153. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 35.Rahme L G, Stevens E J, Wolfort S F, Shao J, Tompkins R G, Ausubel F M. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 36.Wizemann T M, Moskovitz J, Pearce B J, Cundell D, Arvidson C G, So M, Weissbach H, Brot N, Masure R. Proc Natl Acad Sci USA. 1996;93:7985–7990. doi: 10.1073/pnas.93.15.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mushegian A R, Koonin E V. Proc Natl Acad Sci USA. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]