Abstract
In Saccharomyces cerevisiae, the Tor proteins mediate a wide spectrum of growth-related cellular processes in response to nutrients. The pleiotropic role of the Tor proteins is mediated, at least in part, by type 2A protein phosphatases (PP2A) and 2A-like protein phosphatases. Tor-mediated signaling activity promotes the interaction of phosphatase-interacting protein Tap42 with PP2A and 2A-like protein phosphatases. The distinct complexes formed between Tap42 and different phosphatases mediate various cellular events and modulate phosphorylation levels of many downstream factors in the Tor pathway in a Tor-dependent and rapamycin-sensitive manner. In this study, we demonstrate that the interaction between Tap42 and the catalytic subunits of PP2A (PP2Ac) is required for cell cycle-dependent distribution of actin. We show that mutations in PP2Ac and Tap42 that perturb the interaction cause random distribution of actin during the cell cycle and that overexpression of the Rho2 GTPase suppresses the actin defects associated with the mutants. Our findings suggest that the Tap42-PP2Ac complex regulates the actin cytoskeleton via a Rho GTPase-dependent mechanism. In addition, we provide evidence that PP2A activity plays a negative role in controlling the actin cytoskeleton and, possibly, in regulation of the G2/M transition of the cell cycle.
In Saccharomyces cerevisiae, two homologues of phosphatidylinositol 3-kinase, Tor1 and Tor2, mediate cell growth in response to nutrients (41). Tor-mediated signaling activity controls a wide spectrum of growth-related cellular processes, including translation initiation, ribosomal biogenesis, autophagy, protein degradation, and gene expression (1, 3, 8, 14, 35, 37). The macrolide antibiotic rapamycin, via its association with the immunophilin FKBP12, specifically binds to the Tor proteins and inhibits their function (17, 24). Inactivation of the Tor proteins either by genetic depletion or by rapamycin treatment ceases cell growth and arrests the cell cycle progression at early G1 phase (1, 27).
Tor1 and Tor2 are highly similar in their amino acid sequences. Mutations in either Tor1 or Tor2 that prevent it from binding to the rapamycin-immunophilin complex confer on yeast cells rapamycin resistance in a dominant manner (7, 32, 48), indicating that the two proteins are redundant in functions sensitive to rapamycin. However, Tor2 has a unique function that cannot be performed by Tor1. This unique function of Tor2 requires its kinase activity but is not sensitive to rapamycin inhibition (53). Genetic studies have suggested that the unique Tor2 function is required for organization of the actin cytoskeleton during the cell cycle (44). TOR2 mutants that display random distribution of actin during the cell cycle have been isolated (18). This type of defects is not observed in cells depleted for TOR1, nor can it be suppressed by TOR1 (18). Tor2 appears to control the actin cytoskeleton via a Rho GTPase-dependent mechanism. Overexpression of RHO1 and RHO2, two genes encoding Rho GTPases, is able to suppress the actin defects in tor2 mutants (42). In addition, it has been found that the activity of Rom2, a guanine nucleotide exchange factor of Rho GTPases, is significantly reduced in a tor2 mutant displaying actin cytoskeleton defects, indicating that Tor2 regulates Rho GTPases via Rom2 (42, 43). However, the mechanism by which Tor2 controls Rom2 remains elusive.
Type 2A protein phosphatases (PP2A) and 2A-like protein phosphatases have recently emerged as major downstream effectors of the Tor proteins (4, 10, 22, 36). In yeast, two closely related genes, PPH21 and PPH22, encode two functionally redundant catalytic subunits of PP2A (PP2Ac) (47), which exist primarily in cells as a heterotrimeric complex with two regulatory subunits, designated A and B. The product of the TPD3 gene serves as the A subunit, and two distinct proteins, encoded by CDC55 and RTS1, serve as alternative B subunits (16, 45, 52). Deletion of both PPH21 and PPH22 eliminates most of the cellular PP2A activity and drastically reduces cell growth (47). Deletion of a third gene, PPH3, in combination with a pph21 pph22 double deletion, is lethal (39). The PPH3 gene encodes a 2A-like phosphatase that differs from PP2A in its enzymatic properties and subunit composition (20). Yeast cells contain a second 2A-like phosphatase catalytic subunit, encoded by SIT4, which performs cellular functions distinct from those of Pph21 and Pph22 (49). Sit4 normally associates with a family of related proteins, termed Sap proteins, including Sap155, Sap185, Sap190, and possibly Sap4 (33). Sit4 complexed with any of the Sap proteins promotes progression through G1 via regulation of G1 cyclin production (12, 49).
The Tor proteins regulate PP2A and 2A-like phosphatases in yeast via an essential protein encoded by the TAP42 gene, which was isolated via its genetic interaction with SIT4 and PPH21 (10). When overexpressed, TAP42 suppresses the Ts− phenotype produced by sit4-102 and pph21-102, two mutant alleles that have been used extensively for studying the function of the Sit4 and Pph21 phosphatases (10, 12, 31, 49). Tap42 forms complexes with either PP2Ac, Pph21 and Pph22, or 2A-like phosphatase Sit4. These Tap42-containing complexes are structurally independent of the conventional holoenzyme of the phosphatases (10, 22). Tap42 is a phosphoprotein whose phosphorylation depends on the Tor proteins (22). The Tor-dependent phosphorylation of Tap42 appears to promote its interaction with phosphatases, since inactivation of the Tor proteins by rapamycin treatment or nutrient depletion prevents formation of the Tap42-phosphatase complexes (10).
The Tap42-phosphatase complexes appear to play a key role in the Tor pathway. Mutations in TAP42 have been shown to confer on yeast cells rapamycin resistance. In addition, Tap42 has been found to be associated with PP2Ac and Sit4 only in actively growing cells, not in cells entering stationary phase, indicating that formation of the Tap42-phosphatase complexes is essential for cell growth (10). Despite these findings, the precise role of these complexes in the Tor pathway and the effect of the Tap42 interaction on the activity of the phosphatases are not clear. Nevertheless, the observed straight correlation between the dissociation of phosphatases from Tap42 and their activation suggests that Tap42 may act as a phosphatase inhibitor (2, 5, 21).
Previous studies have revealed roles for the Tap42-Sit4 complex in Tor-mediated gene expression. Several transcription factors for genes involved in nutrient metabolism have been identified as the targets of this complex (2). However, the role of the Tap42-PP2Ac complex in the Tor pathway is largely unknown. To better understand this complex, we sought to identify PP2Ac mutants that are defective in its formation. We found that the mutant Pph21 phosphatase encoded by pph21-102, a well-characterized phosphatase mutant allele (31), was specifically defective in its interaction with Tap42. This finding, together with the fact that cells carrying this mutant gene were defective in actin cytoskeleton organization (31), raised the possibility that the Tap42-Pph21 complex was involved in controlling the polarized distribution of actin during the cell cycle. We confirmed this notion by demonstrating that mutations in Tap42 that perturb its interaction with Pph21 cause the same defects. Our results suggest that it is the Tap42-PP2Ac complex rather than the PP2A holoenzyme that is involved in the organization of the actin cytoskeleton. In addition, we provide evidence that PP2A activity negatively regulates the actin cytoskeleton via a Rho GTPase-dependent mechanism.
MATERIALS AND METHODS
Yeast strains and reagents.
The strains used in this study are listed in Table 1. Yeast cells were normally grown in YP medium containing 1% yeast extract and 2% peptone or synthetic complete medium lacking the appropriate amino acid(s) for selection. All media contain 2% glucose as a carbon source. Standard methods were used for yeast genetics and molecular cloning (13). The plasmids used in this study are listed in Table 2. Rapamycin (Sigma) was stored in 10% Tween 20-90% ethanol at a concentration of 1 mg/ml and was added to growth medium to a final concentration of 300 ng/ml. An anti-hemagglutinin (HA) monoclonal antibody (12CA5) and an anti-Tpd3 antibody were described previously (22). Anti-Tap42 and anti-Sit4 antibodies against recombinant six-histidine-tagged Tap42 and Sit4, respectively, were raised in rabbits.
TABLE 1.
Strains used in this studya
| Strain | Genotype | Source |
|---|---|---|
| Y661 | MATa (wild type [W303-1A]) | Laboratory stock |
| Y162 | MATa (HA)3PPH21 | This study |
| Y176 | MATatap42::HIS3 [tap42-11-pRS415 (CEN6 ARS4 LEU2)] | This study |
| Y531 | MATα pph22::HIS3 pph21::LEU2 | Laboratory stock |
| Y638 | MATα ura3::(HA)3pph22-102-URA3 pph22::HIS3 pph21::LEU2 | This study |
| Y648 | MATα ura3::(HA)3PPH22-URA3 pph22::HIS3, pph21::LEU2 | This study |
| Y689 | MATα ura3::(HA)3PPH21-URA3 pph22::HIS3 pph21::LEU2 | This study |
| Y690 | MATα ura3::(HA)3pph21-102-URA3 pph22::HIS3 pph21::LEU2 | This study |
| Y703 | MATa (HA)3PPH21 tap42::HIS3 [tap42-11-pRS415 (CEN6 ARS4 LEU2)] | This study |
All strains are derivatives of W303 (ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 GAL) (50).
TABLE 2.
Plasmids used in this studya
| Plasmid | Description | Source or reference |
|---|---|---|
| tap42-11-pRS415 (CEN6 ARS4 TRP1) | 10 | |
| pJY182 | TOR1S1972R-pRS314 (CEN6 ARS4 TRP1) | Laboratory stock |
| pJY296 | 3HA-PPH21-pRS406 (URA3) | Laboratory stock |
| pJY702 | 3HA-pph21-102-pRS406 (URA3) | This study |
| pJY714 | RHO2-pRS424 (2μm TRP1) | This study |
All plasmids are derivatives of pRS vectors (46).
Construction of the triple-HA epitope-tagged pph21-102 allele.
The plasmid-borne (HA)3pph21-102 gene [(HA)3pph21-102-pRS406; CEN URA3] was created by oligonucleotide-directed mutagenesis using a 5′ end triple-HA epitope-tagged PPH21, generated previously, as a template (26). The mutagenic primer was 5′-GTGGACGTGCTGCAGTTCAAGGAGAATG-3′. The resulting plasmid was digested with NcoI internally to URA3 and transformed into a pph21 pph22 double-deletion strain. This manipulation resulted in an integration of the (HA)3pph21-102 gene at the URA3 locus, thus creating the pph21 pph22 (HA)3pph21-102 strain (Y690). The pph21 pph22 double-deletion strain (Y531) was generated by selecting Leu+ His+ progeny from a cross between a pph21::LEU2 strain and a pph22::HIS3 strain created previously (22).
Construction of plasmid RHO2-pRS424 (pJY714).
A 1.4-kb DNA fragment containing the RHO2 open reading frame was amplified from yeast genomic DNA by PCR using high-fidelity Taq DNA polymerase (Roche). The PCR products were digested with SacI and XhoI, whose sites were introduced via the 5′ and 3′ primers, respectively, and cloned into pRS424 (46) at the corresponding sites. The 5′ primer for the PCR was 5′-ATACATGAGCTCCAACTGCTACACCATTTA −3′. The 3′ primer was 5′-ATACATCTCGAGCTATGGTTTGATGTGCCA-3′. The underlined regions are where the restriction sites lie.
Preparation of cell extracts.
Yeast cells were grown in appropriate medium to early log phase. Cells were collected, washed twice with ice-cold lysis buffer (50 mM Tris-Cl [pH 7.4], 50 mM NaF, 5 mM EDTA, 1 mM dithiothreitol [DTT], 5% glycerol), resuspended in the same buffer containing 1× protease inhibitor cocktails (Roche), and lysed by vortexing with glass beads. Cell lysates were diluted fourfold with wash buffer containing 50 mM Tris-Cl (pH 7.4), 50 mM NaF, 200 mM NaCl, 1 mM DTT, 1% Triton X-100, and 1× protease inhibitors. Insoluble cell debris was removed by centrifugation at 12,000 × g for 15 min, and the supernatants were then used for coimmunoprecipitation (see below). Protein concentration in the supernatants was determined by using the Bio-Rad protein assay.
Cross-linking anti-Tap42 antibody to protein A-Sepharose beads.
An aliquot of anti-Tap42 serum (1 ml) was diluted 10-fold with phosphate-buffered saline (PBS) and incubated with 1 ml of protein A-conjugated Sepharose beads (Zymed) at 4°C for 2 h with gentle shaking. The beads were washed three times with PBS buffer and twice with 0.2 M sodium borate (pH 9.0). After the final wash, the beads were resuspended in 10 ml of the sodium borate buffer. The cross-linking reaction was carried out by using dimethylpimelimidate (Pierce) as described before (15).
Coimmunoprecipitation.
To precipitate Tap42, an aliquot of cell extract containing 1 mg of protein was incubated with 20 μl of protein A beads conjugated with the anti-Tap42 antibody at 4°C for 3 h. After the incubation, beads were washed three times with wash buffer (50 mM Tris-Cl [pH 7.4], 200 mM NaCl, 1 mM DTT, 1% Triton X-100) and once with 20 mM Tris-Cl, pH 7.4, and boiled for 5 min in 70 μl of 2× sodium dodecyl sulfate (SDS) sample buffer. An aliquot (20 μl) of sample was fractionated with an SDS-10% polyacrylamide gel and transferred to a nitrocellulose membrane, which was then immunoblotted with anti-Tap42 (1:1,000 dilution), anti-Sit4 (1:1,000 dilution), or anti-HA epitope (12CA5 at 1:1,000 dilution) antibodies. To precipitate Tpd3, an aliquot of extract containing 1 mg of protein was incubated with 2 μl of anti-Tpd3 antibody at 4°C for 2 h. The Tpd3 protein-antibody complexes were then precipitated with protein A beads during a 90-min incubation at 4°C. Beads were washed three times with wash buffer and once with 20 mM Tris-Cl, pH 7.4, and boiled for 5 min in 70 μl of 2× SDS sample buffer. Western blot analysis was then performed as described above with anti-Tpd3 (1:2,000) and anti-HA epitope (12CA5; 1:1,000 dilution) antibodies.
Protein phosphatase assays.
Cell extracts for the phosphatase assays were made as described before (34). 32P-labeled calf thymus H1 histone (Sigma) was prepared by phosphorylation with the catalytic subunit of cyclic AMP-dependent protein kinase (PKA) (Sigma) in reaction buffer containing 50 mM Tris-Cl (pH 7.4), 10 mM MgCl2, 10 mM 2-mercaptoethanol, 100 μM ATP, and [γ-32P]ATP (10 mCi/mmol). After incubation at 30°C for 4 h, the reaction was terminated by the addition of PKA inhibitor (100 nM; Sigma). Unincorporated labeled nucleotides were removed from the reaction mixture with a Micro Bio-spin column (Bio-Rad) by following the manufacturer's instructions. The H1 histone phosphatase activity in the cell extracts was determined as described before (9). Bacterially expressed type 1 phosphatase inhibitor 2 (I-2; New England Laboratory) was used at a concentration of 10 nM.
Fluorescence microscopy.
Yeast cells were grown to early log phase at 23°C and shifted to 37°C for 4 h. The cells were fixed directly in growth medium with 3.7% formaldehyde at room temperature for 30 min. The cells were then collected and resuspended in PBS containing 3.7% formaldehyde. After incubation at room temperature for 1 h, the cells were washed three times with PBS and stained with Alexa-conjugated phalloidin (Molecular Probes) as described before (38). Stained cells were visualized with a Zeiss Axiovert microscope by either differential interference contrast (DIC) or fluorescence microscopy.
RESULTS
The Pph21 phosphatase in the pph21-102 cells is defective in its interaction with Tap42.
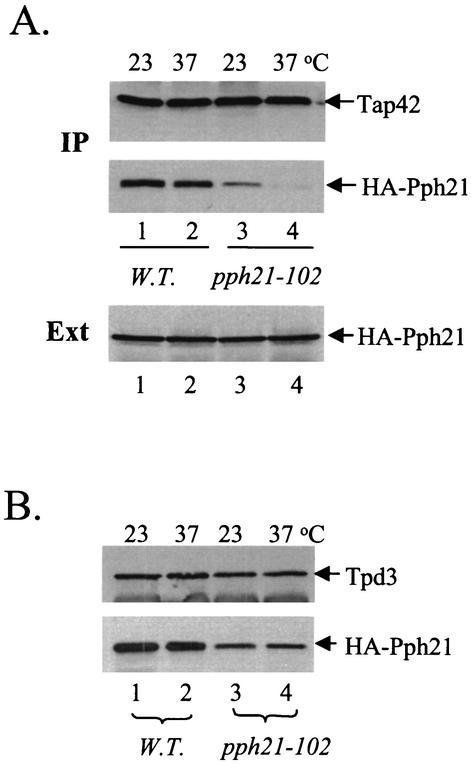
TAP42 was originally isolated as a high-copy-number suppressor of the sit4-102 Ts− allele, in which the mutant gene encodes a Sit4 protein containing a glutamine-to-lysine substitution at position 38 (E38K) (49). We have recently found that this mutant Sit4 protein (Sit4E38K) is specifically defective in its interaction with Tap42 (H. Wang et al., submitted for publication). The equivalent amino acid substitution (E102K) is present in the mutant Pph21 and Pph22 phosphatases in the pph21-102 and pph22-102 cells, respectively, and renders the cells temperature sensitive (31). Importantly, overexpression of TAP42 also suppresses the Ts− phenotype of the pph21-102 and pph22-102 cells (10). It is thus conceivable that the mutant Pph21 and Pph22 phosphatases (Pph21E102K and Pph22E102K), like Sit4E38K, are defective for their interaction with Tap42. To test this notion, we introduced a triple-HA-epitope-tagged wild-type or pph21-102 mutant gene into a pph21 pph22 double-deletion strain. The resulting strain containing the HA-tagged wild-type PPH21 gene (HA-PPH21 pph22) behaved like the isogenic wild-type strain at both 23 and 37°C. However, the strain containing the HA-tagged pph21-102 gene (pph21-102 pph22) was temperature sensitive in that it ceased growth at 37°C and grew with a slightly reduced rate at 23°C compared to the HA-PPH21 pph22 strain. Cells of both strains were grown to early log phase at 23°C and then shifted to 37°C for 2 h. The interaction of Pph21 with Tap42 was determined by coimmunoprecipitation. As shown in Fig. 1A, we found that Pph21 was efficiently copurified with Tap42 from extracts made from wild-type cells grown at either 23 or 37°C. In contrast, significantly less Pph21was copurified with Tap42 from extracts made from the pph21-102 cells grown at 23°C (Fig. 1A, middle, compare lanes 1 and 3). Following a 2-h incubation at 37°C, the amount of Pph21 in the Tap42 precipitate was barely detectable (compare lanes 3 and 4). Despite this, we found that the levels of expression of Pph21 in the mutant cells were comparable to those in the wild-type cells (Fig. 1A, bottom), suggesting that the significant reduction in the interaction of Pph21 with Tap42 in the mutant cells was not due to a lower level of expression of this protein. Thus, this result demonstrates that Pph21E102K, encoded by the pph21-102 allele, is defective for interaction with Tap42. Using the same experimental setting, we found that the Pph22E102K phosphatase was also impaired in its interaction with Tap42 (data not shown).
FIG. 1.
The Pph21 phosphatase in the pph21-102 allele is defective in its interaction with Tap42. (A) Wild-type (W.T.; Y689; lanes 1 and 2) and mutant cells (Y690; lanes 3 and 4) were grown to early log phase at 23°C and then shifted to 37°C for 2 h. Cell extracts were made and precipitated with anti-Tap42 antibody cross-linked to protein A beads. The presence of Tap42 (top) and HA-Pph21 (middle) in the precipitates was detected by Western blot analysis with anti-Tap42 and anti-HA antibodies, respectively. The amount of HA-Pph21 in the total cell extracts (Ext) was also determined (bottom). IP, immunoprecipitation. (B) Cell extracts from the same cells were incubated with anti-Tpd3 antibody. The precipitates were then analyzed by Western blotting to detect the presence of Tpd3 and HA-Pph21 with anti-Tpd3 and anti-HA antibodies, respectively.
In yeast, Pph21 and Pph22 exist primarily as a heterotrimeric complex with two other subunits, an A subunit encoded by TPD3 and a B subunit encoded by either CDC55 or RTS1 (16, 45, 52). Since the Pph21E102K phosphatase was defective in its association with Tap42, we asked whether the same mutant protein was impaired in its interaction with the A subunit, the product of the TPD3 gene. We thus examined the interaction of Pph21 with Tpd3 in the pph21-102 cells by coimmunoprecipitation. As shown in Fig. 1B, we found that the amount of Pph21E102K that copurified with Tpd3 was less than that of the wild-type Pph21 protein (compare lanes 1 and 3), suggesting that Pph21E102K is defective in its association with Tpd3. However, the Pph21-Tpd3 interaction in the mutant cells didn't appear to be affected by the temperature shift, suggesting that the Ts− phenotype of the cells is unlikely to be a consequence of a failure in formation of the PP2A holoenzyme.
Tap42 is required for the cell cycle-dependent polarized distribution of actin.
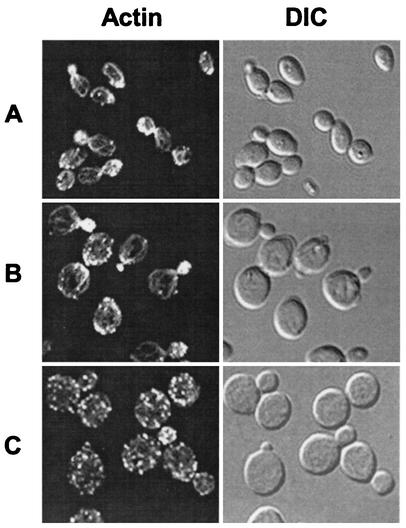
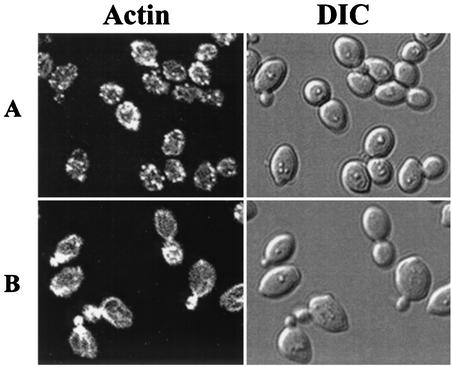
Previous studies have demonstrated that the pph21-102 cells are defective in the polarized distribution of actin during the cell cycle (31). Our finding that the mutant cells are defective for formation of the Tap42-Pph21 complex raised the possibility that the Tap42-Pph21 complex was required for organization of the actin cytoskeleton during the cell cycle. To test this notion, we examined the assembly of the actin cytoskeleton in tap42 Ts− allele tap42-11, which was isolated previously (10). Exponentially growing tap42-11 cells were shifted from 23 to 37°C for 4 h. Cells before and after the shift were fixed, stained with fluorescent phalloidin, and imaged. As shown in Fig. 2, when grown at 23°C, the tap42-11 cells had an increased size and round cell morphology in comparison with wild-type cells (Fig. 2A and B, DIC) (10). Despite this, these mutant cells displayed a normal distribution of actin in all phases of the cell cycle (Fig. 2B). In 73% of the unbudded cells actin patches were found to form the actin cap at the site of the future bud. In 91% of the cells containing small and midsize buds actin patches accumulated in the daughter cells and, in most cases, actin cables that ran in parallel to the longitudinal axis of the cell were clearly visible. In 57% of the large-budded cells actin patches were concentrated at the mother-bud neck (Table 3). Following the incubation at 37°C, however, actin cables vanished and the normal asymmetric distribution of the actin patches during the cell cycle was replaced with a randomized distribution pattern (Fig. 2C). The percentages of cells displaying a polarized distribution of actin in the stages of unbudded, small- and midsize-budded, and large-budded cells fell to 13, 22, and 2%, respectively (Table 3). These observations suggest that Tap42 is required for the cell cycle-dependent polarized distribution of actin. It is noteworthy that after a 4-h incubation at the nonpermissive temperature, a significant portion (∼50%) of the tap42-11 cells arrested as unbudded cells, indicating a late G1 arrest, while many others (31%) arrested as large-budded cells, a characteristic of a G2/M arrest.
FIG. 2.
Tap42 is required for the cell cycle-dependent polarized distribution of actin. Exponentially growing yeast cells were shifted from 23 to 37°C for 4 h. Before (A and B) and after (C) the shift the cells were fixed, stained with fluorescent phalloidin, and imaged in fluorescence or DIC mode. (A) Wild-type cells (Y661) at 23°C; (B) tap42-11 mutant cells (Y176) at 23°C; (C) tap42-11 mutant cells (Y176) at 37°C.
TABLE 3.
Quantitative analysis of actin defects in the tap42 and pph21 mutant cellsa
Exponentially growing yeast cells were shifted from 23 to 37°C for 4 h. Before and after the shift the cells were fixed and stained for actin. The distribution of actin in the cells was examined with a confocal microscope, and the percentages of cells displaying polarized actin distribution at three different stages, unbudded, small to midsize budded, and large budded, of the cell cycle were quantified. At each stage, ≥200 cells for each strain were counted for actin distribution. The small- and midsize-budded cells are defined as those with a cross section bud diameter two-thirds that of the mother cell or less. The large-budded cells are those with a bud diameter more than two-thirds that of the mother cell. A polarized actin distribution is defined by the presence of an actin cap in the unbudded cells, by concentrated actin patches presented in the bud of the small- and midsize-budded cells, and by concentrated actin patches present at the mother-bud neck of the large-budded cells.
+, present; −, absent.
The association between Tap42 and PP2Ac is defective in the tap42-11 cells.
The finding that both the tap42-11 and pph21-102 cells are defective in organization of the actin cytoskeleton suggests that the Tap42-PP2Ac complex is involved in the process. Since the Pph21E102K phosphatase in the pph21-102 cells is defective for its interaction with Tap42, we asked whether the Tap42 protein in the tap42-11 cells was also impaired in its interaction with Pph21. Accordingly, we replaced the PPH21 gene in both the wild-type and tap42-11 mutant strains with a triple-HA-epitope-tagged PPH21 by two-step gene replacement (40). The interaction of Tap42 with the HA epitope-tagged Pph21 in the resulting strains was then determined by coimmunoprecipitation. As shown in Fig. 3, we found that Pph21 was effectively coprecipitated with Tap42 in extracts made from wild-type cells grown at either 23 or 37°C (compare lanes 1 and 2). However, significantly less Pph21 was coprecipitated with Tap42 in extracts made from the mutant cells grown at 23°C (compare lanes 3 and 1). Following a 2-h incubation at the nonpermissive temperature, the amount of Pph21 associated with Tap42 was reduced to a level barely detectable. The reduction in the amount of Pph21 associated with Tap42 in the mutant cells was not due to loss of expression of Pph21 or Tap42, as we found that the expression levels of both Pph21 and Tap42 in the mutant cells were essentially the same as those in the wild-type ones (Fig. 3B). Hence, the mutant Tap42 protein in the tap42-11 cells is defective for interaction with Pph21. Using the same experimental setting, we found that the mutant Tap42 protein was also impaired in its ability to interact with Pph22 (data not shown). Since inactivation of Tap42 is expected to affect neither the catalytic activity of the Pph21 and Pph22 phosphatases nor their interaction with Tpd3, our results suggest that it is a defect in the Tap42-PP2Ac complex rather than one in the PP2A holoenzyme that causes the abnormal distribution of actin in the tap42-11 cells.
FIG. 3.
The Tap42 protein in the tap42-11 mutant cells is defective for interaction with phosphatases. Exponentially growing wild-type (W.T.; Y162) and tap42 mutant (Y703) cells were shifted from 23 to 37°C for 2 h. Cell extracts were made from cells before (lanes 1 and 3) and after (lanes 2 and 4) the shift and incubated with anti-Tap42 antibody cross-linked to protein A beads. The presence of Tap42 (top), HA-Pph21(middle), and Sit4 (bottom) in the precipitates (A) and in the cell extracts (Ext; B) was detected by Western blot analysis with anti-Tap42, anti-HA, and anti-Sit4 antibodies, respectively. IP, immunoprecipitation.
Unlike the pph21-102 cells, which arrest at G2/M upon incubation at the nonpermissive temperature, most of the tap42-11 cells arrest at G1 phase following a 4-h incubation at 37°C (10). We reasoned that the G1 arrest might be due to a defect in the interaction of the mutant Tap42 protein with Sit4, whose function is required for the G1/S transition of the cell cycle (49). We thus further examined the interaction between Tap42 and Sit4 in the tap42-11 cells. The Tap42 precipitates from the above experiment were further analyzed for the presence of Sit4 by Western blotting. As shown in Fig. 3A (bottom), we found that Sit4 was effectively copurified with Tap42 from extracts made from wild-type cells grown at either 23 or 37°C (compare lanes 1 and 2). However, less Sit4 was copurified with Tap42 from extracts made from the mutant cells grown at 23°C (compare lanes 3 and 1). Following incubation of the mutant cells at 37°C for 2 h, the amount of Sit4 copurified with Tap42 was reduced to an insignificant level (lane 4). Again, the absence of the Sit4-Tap42 interaction in the mutant cells grown at 37°C was not due to loss of Sit4 protein expression, since we found that the expression levels of Sit4 were essentially the same before and after the temperature shift (Fig. 3B, bottom). Thus, the mutant Tap42 protein in the tap42-11 cells is defective for interaction with Sit4. This observation suggests that the G1 arrest of the tap42-11 cells is likely to be a consequence of the inability of Tap42 to interact with Sit4 at the nonpermissive temperature. The finding that the sit4-102 pph21-102 double mutant behaves like the tap42-11 mutant is in agreement with this conclusion (10).
The pph21 pph22 double-deletion cells display a normal actin cytoskeleton.
Studies on the Tap42-phosphatase complexes have revealed a correlation between dissociation of a phosphatase from Tap42 and its activation (2, 5, 21). Since the Pph21 phosphatase disassociates from Tap42 when the pph21-102 cells are incubated at the nonpermissive temperature (Fig. 1), it is possible that the actin defects associated with the mutant cells are caused by an unregulated Pph21 phosphatase rather than by the inactivation of Pph21. To test this notion, we examined the actin cytoskeleton in cells depleted of both PPH21 and PPH22 (pph21 pph22). We reasoned that if the actin defects in the pph21-102 cells were caused by inactivation of the phosphatase, we would expect to observe the same defects in cells depleted of both Pph21 and Pph22. However, this was not the case. The pph21 pph22 double-deletion cells grew slowly at 23°C and ceased to grow at 37°C; thus, they were temperature sensitive. Nevertheless, we failed to detect any significant defects in organization of the actin cytoskeleton at all phases of the cell cycle under both conditions (Fig. 4 and Table 3). It is worth noting that the pph21-102 allele used in this study has a pph22 background (pph21-102 pph22); therefore, the PP2A activity in the pph21-102 pph22 cells can hardly be lower than that in the pph21 phh22 double-deletion strain, indicating that the actin defects in the pph21-102 cells are not caused by the absence of PP2A activity.
FIG. 4.
The pph21 pph22 double-deletion cells display a normal distribution of actin. Exponentially growing pph21 pph22 cells (Y531) were shifted from 23 to 37°C for 4 h. The cells before (23°C) and after (37°C) the shift were fixed, stained with fluorescent phalloidin, and imaged in fluorescence or DIC mode.
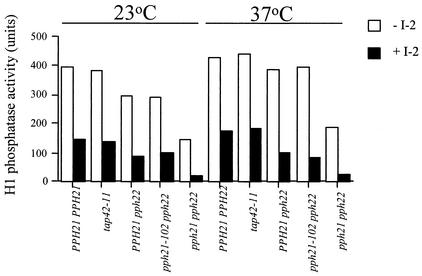
To further confirm that the actin defects associated with the pph21-102 and tap42-11 mutant cells were not caused by inactivation of PP2A, we examined PP2A activity in the mutant cells. As shown in Fig. 5, we found that, at both the permissive and nonpermissive temperatures, PP2A activity in the tap42-11 cells was similar to that in the wild-type cells (PPH21 PPH22) grown under the same conditions. Similarly, PP2A activity in the pph21-102 cells was comparable to that in their isogenic wild-type cells (PPH21 pph22) at both 23 and 37°C. On the other hand, the activity of PP2A was found to be largely absent in pph21 pph22 double-deletion cells at either temperature. These results demonstrated that both the tap42-11 and pph21-102 cells retained a normal PP2A activity when grown at the nonpermissive temperature, suggesting that the actin defects in these mutant cells are not caused by loss of PP2A activity. It is thus likely that an unregulated PP2A activity is responsible for the mutant phenotypes associated with the pph21-102 and tap42-11 cells.
FIG. 5.
The pph21-102 and tap42-11 cells display a normal PP2A activity at both the permissive and nonpermissive temperatures. Yeast cells of strains Y162 (PPH21 PPH22), Y176 (tap42-11), Y648 (PPH21 pph22), Y638 (pph21-102 pph22), and Y531 (pph21 pph22) were grown to early log phase at 23°C and then shifted to 37°C for 2 h. Cells before and after the shift were lysed, and phosphatase activity in the lysates was determined by an in vitro phosphatase assay using phosphorylated H1 histone as the substrate. The H1 histone phosphatase activity was measured in the presence (solid bars) or absence (open bars) of 10 nM type 1 phosphatase inhibitor 2 (I-2). PP2A activity was defined as the activity that was insensitive to I-2. The activities shown are the averages of three independent experiments. Units shown are arbitrary.
Overexpression of RHO2 suppresses the actin defects in the tap42-11 cells.
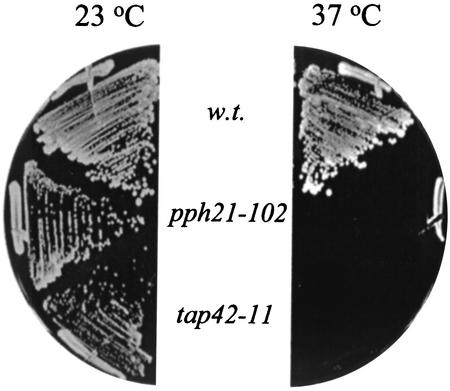
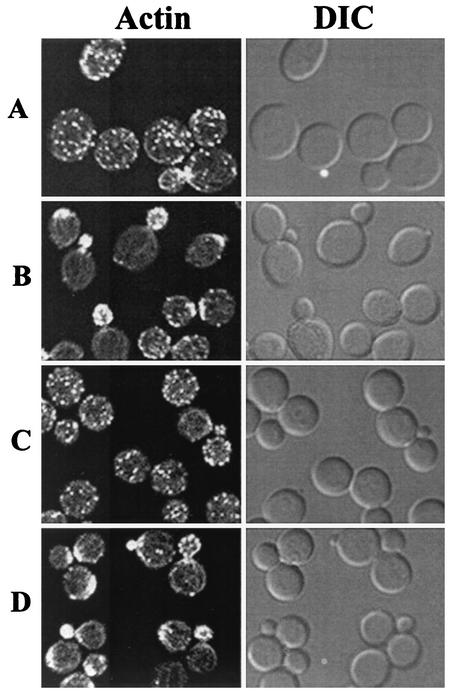
In yeast, Rho GTPases are key regulators in signaling pathways that link extracellular signals to the cell cycle-dependent polarized distribution of actin (43). Overexpression of RHO1 or RHO2 is found to suppress the actin defects associated with the tor2 mutants (42). Since the Tap42-Pph21 complex acts downstream of the Tor proteins, it is thus possible that the Tap42 complex controls organization of the actin cytoskeleton via the same mechanism. To test this notion, we asked whether overexpression of RHO2 was able to suppress the actin defects in the tap42-11 cells grown at the nonpermissive temperature. The mutant cells were transformed with a high-copy-number plasmid containing a RHO2 gene, and transformants were monitored for growth at the nonpermissive temperatures (30, 34, and 37°C) and for the cell cycle-dependent organization of the actin cytoskeleton. As shown in Fig. 6, we found that overexpression of RHO2 failed to suppress the growth defect of the tap42-11 cells at the nonpermissive temperatures (shown only for 37°C). This result was not unexpected because the mutant Tap42 protein was also defective for its interaction with Sit4, which has an essential function in the G1/S transition (Fig. 3) (49). However, when the distribution pattern of actin was examined, we found that overexpression of RHO2 restored the normal distribution of actin in the mutant cells grown at 37°C (Fig. 7, compare panels A and B). The percentages of cells displaying polarized actin distribution at the stages of unbudded, small- and midsize-budded, and large-budded cells were 57, 72, and 41%, respectively, in cells expressing RHO2 (versus 13, 22, and 2%, respectively, in cells without RHO2 overexpression) (Table 3). This result suggests that, like the Tor2 protein, the Tap42-Pph21 complex controls the actin cytoskeleton via a Rho GTPase-dependent mechanism.
FIG. 6.
Overexpression of RHO2 cannot suppress the growth defect of the tap42-11 and pph21-102 mutant cells. Wild-type (w.t.; Y661) and pph21-102 (Y690) and tap42-11 (Y176) mutant cells overexpressing RHO2 (pRS424-RHO2) were streaked on plates containing synthetic complete medium lacking Trp and incubated at either 23 (left) or 37°C (right) for 3 days.
FIG. 7.
Overexpression of RHO2 restores the normal distribution of actin in the tap42-11 and pph21-102 mutant cells. Yeast cells expressing RHO2 or vector alone were grown at 23°C to exponential phase and shifted to 37°C. After a 4-h incubation at 37°C, the cells were fixed, stained with fluorescent phalloidin, and imaged in fluorescence and DIC modes. (A) tap42-11 cells expressing vector alone; (B) tap42-11 cells expressing RHO2; (C) pph21-102 cells expressing vector alone; (D) pph21-102 cells expressing RHO2.
Similarly, we found that overexpression of RHO2 suppressed the actin defects in the pph21-102 cells grown at 37°C (Table 3; Fig. 7, compare panels C and D). However, as for the tap42-11 cells, overexpression of RHO2 was unable to suppress the growth defect of the pph21-102 cells at 37°C (Fig. 6). This result, again, suggests that the Tap42-Pph21 complex controls the distribution of actin via a Rho GTPase-dependent mechanism. In addition, it also indicates that the Ts− phenotype of the pph21-102 cells is not caused by defects in the actin cytoskeleton.
The organization of the actin cytoskeleton is sensitive to rapamycin.
Previous studies have demonstrated that the interaction between Tap42 and phosphatases is regulated by the Tor proteins and is sensitive to rapamycin (10). The finding that the Tap42-PP2Ac complex is required for organization of the actin cytoskeleton suggests that this particular cellular event ought to be rapamycin sensitive. To test this notion, we examined the distribution of actin in wild-type cells treated with rapamycin. We found that the drug-treated cells displayed a random distribution of actin in all phases of the cell cycle. The actin cables normally observed in untreated cells were completely absent (Fig. 8A). Nevertheless, rapamycin failed to perturb the normal distribution of actin in cells expressing TOR1S1972R, a rapamycin-resistant TOR1 gene (Fig. 8B). These observations indicate that organization of the actin cytoskeleton in yeast is sensitive to rapamycin and that a functional TOR1 alone is able to sustain a normal actin cytoskeleton.
FIG. 8.
The cell cycle-dependent organization of the actin cytoskeleton is sensitive to rapamycin treatment. Wild-type yeast cells expressing a control vector (pRS314) (A) or pJY182 (TOR1S1972R-pRS314) (B) were grown to early log phase at 23°C and treated with 300 nM rapamycin for 90 min. Cells were then fixed, stained with fluorescent phalloidin, and imaged in fluorescence and DIC modes.
DISCUSSION
The Tap42-PP2Ac complex is required for organization of the actin cytoskeleton.
Previous studies on the pph21-102 allele revealed that the mutant cells displayed an abnormal distribution of actin throughout the cell cycle (31). This observation led to the suggestion that PP2A was required for organization of the actin cytoskeleton in yeast, for pph21-102 was believed to be a loss-of-function allele. A similar conclusion was drawn from the study of the pph22 temperature-sensitive alleles (11). However, the finding that the pph21-102 allele is defective for the interaction between the mutant Pph21 phosphatase and Tap42 (Fig. 1) raises the possibility that the Tap42-PP2Ac complex plays a role in controlling the actin cytoskeleton. This notion is confirmed by our demonstration that the cell cycle-dependent polarized distribution of actin is perturbed in the tap42-11 cells, in which the mutant Tap42 protein is defective for interaction with PP2Ac (Fig. 2). Since the Tap42-PP2Ac complex is independent of the PP2A holoenzyme, mutations in Tap42 are expected to affect neither the formation of the PP2A holoenzyme nor the phosphatase activity of the holoenzyme. Therefore, our finding further suggests that it is the defects in the Tap42-PP2Ac complex rather than the defects in the PP2A holoenzyme that perturb the actin cytoskeleton. We thus conclude that the Tap42-PP2Ac complex is involved in controlling the organization of the actin cytoskeleton during cell cycle progression.
Recent studies of the Tap42-phosphatase complexes have revealed a straight correlation between the dissociation of the phosphatases from Tap42 and their activation (2, 5, 21). This observation has led to the suggestion that Tap42 downregulates the activity of the phosphatases with which it associates. In this regard, the dissociation of PP2Ac from Tap42 that occurs when the pph21-102 and tap42-11 cells are grown at the nonpermissive temperature is expected to result in an activation of the phosphatase. It is thus likely that the actin defects associated with the mutant cells are caused by an upregulated PP2A activity rather than by a loss of PP2A activity. This conclusion is in accordance with the finding that both tap42-11 and pph21-102 cells display a normal PP2A activity at the nonpermissive temperature (Fig. 5) and is further supported by the observation that inactivation of both Pph21 and Pph22 does not cause any defects in the organization of the actin cytoskeleton (Fig. 4). Therefore, our results suggest a negative role for PP2A activity in the organization of the actin cytoskeleton during the cell cycle, in contrast to the positive role suggested by the previous studies (31).
Tap42-PP2Ac controls the actin cytoskeleton via a Rho GTPase-dependent mechanism.
Formation of the Tap42-PP2Ac complex occurs in actively growing cells, in which the Tor proteins are active, but not in cells treated with rapamycin (10), suggesting that Tap42 associates with PP2Ac in a Tor-dependent and rapamycin-sensitive manner. Therefore, the finding that the organization of the actin cytoskeleton requires the Tap42-PP2Ac complex indicates that this process is downstream of the Tor proteins and ought to be rapamycin sensitive. Indeed, we found that cells treated with rapamycin exhibited a perturbed actin cytoskeleton (Fig. 8). However, in this case, we cannot rule out the possibility that the effect of rapamycin is due to inhibition of the Tor-dependent protein synthesis (1). Since Tap42 has been shown to be required for protein synthesis (10), is it possible that the defects in the actin cytoskeleton associated with tap42-11 and pph21-102 are caused indirectly by a reduction in protein synthesis? Several lines of evidence argue against this possibility. First, a study on the pph21-102 allele has demonstrated that the mutant cells maintain a level of protein synthesis similar to that of wild-type cells a few hours after being shifted to the nonpermissive temperature and continue to increase in size despite an arrest at G2/M (31). Second, unlike rapamycin-treated cells, which arrest at early G1 phase as small unbudded cells as a consequence of a reduction in protein synthesis (1), the tap42-11 cells arrest either as large unbudded or large-budded cells after a 4-h incubation at the nonpermissive temperature, during which the size of the cells keeps increasing (Fig. 2, DIC, compare panels B and C) (10). The large unbudded morphology is a characteristic of cells undergoing a late G1 arrest and is likely to be a consequence of the defect in formation of the Tap42-Sit4 complex in the mutant cells rather than a consequence of reduction in protein synthesis (Fig. 2B). Finally, the actin defects in both the tap42-11 and pph21-102 cells are suppressed by overexpression of RHO2 (Fig. 7), suggesting that the Tap42-Pph21 complex controls the actin cytoskeleton via a Rho GTPase-dependent mechanism.
Intriguingly, it is thought that regulation of the actin cytoskeleton is Tor2 specific and that Tor1 does not have a role; defects in the actin cytoskeleton have been found to be associated only with mutations in TOR2 and are not suppressed by the presence of TOR1 (18). On the other hand, our finding that the cell cycle-dependent distribution of actin requires the Tap42-PP2Ac complex, which in turn requires the shared function of the Tor proteins (10), suggests that this process is regulated redundantly by Tor1 and Tor2. Therefore, it is possible that regulation of the actin cytoskeleton is not the unique Tor2 function. In accordance with this view, it has been found that the mutant Tor2 proteins that are defective in actin cytoskeleton organization are unable to provide the shared function, implying that they are also defective in the shared function (18). In addition, given the fact that the Tor proteins regulate phosphorylation levels of many cellular proteins via the Tap42-phosphatase complexes (2, 5, 21), it is possible that Tor signaling activity controls the phosphorylation of a factor(s) involved in modulating Rho GTPases. In this view, the finding that Rom2 exhibits a Tor2-dependent activity toward Rho GTPases suggests that this protein may be the target of the Tap42-PP2Ac complex (42).
Rho1 and Rho2 GTPases control the actin cytoskeleton via the Pkc1-mitogen-activated protein kinase (MAPK) pathway in yeast (43). Since overexpression of RHO2 suppresses the actin defects associated with the pph21, tap42, and tor2 mutants, it is expected that the Pkc1-MAPK pathway is downregulated in these mutants (Fig. 7) (6, 19). However, it has been recently found that inactivation of the Tor proteins by rapamycin induces hyperactivation of the Pkc1-MAPK pathway and consequently actin depolarization (51). Furthermore, the rapamycin-induced actin depolarization requires Sit4 and Tap42, since inactivation of Sit4 or mutations in TAP42 nullify the effect of the drug (51). It thus appears that the Tor proteins negatively regulate the Pkc1-MAPK pathway via the Tap42-Sit4 complex, a notion that is inconsistent with our findings. One way to explain this paradox is to suggest that the Tor proteins are able to control the Pkc1-MAPK pathway via two different mechanisms. In support of this, it has been shown that rapamycin has its effect at the cell surface, presumably by altering cell wall integrity (25, 51), whereas the downregulation of the Pkc1-MAPK1 pathway in the tor2 mutants was found to be independent of the cell surface alterations (6). Since rapamycin-induced hyperactivation of the Pkc1-MAPK pathway can be suppressed under osmotically stable conditions, it is likely that the hyperactivation of the Pkc1-MAPK pathway in the cells treated with rapamycin is caused by the cell wall remodeling during the transition into the quiescent phase (25, 51). In accordance with this notion, it has been found that the hyperactivation of the Pkc1-MAPK pathway induced by rapamycin is transient (25). Constitutive activation of the Pkc1-MAPK pathway, as happens in the sit4 deletion cells and in the tap42-11 cells grown at the permissive temperature, not only fails to perturb the actin cytoskeleton but also prevents rapamycin from doing so (51). It is thus appears that rapamycin-induced actin depolarization is caused by a transient activation of the Pkc1-MAPK pathway as a consequence of changes in cell wall integrity.
The role of the Tap42-PP2Ac complex in the G2/M transition.
In addition to the defects in the organization of the actin cytoskeleton, the pph21-102 cells are also defective in the G2/M transition, which is characterized by a G2/M block at the nonpermissive temperature. Since the pph21-102 cells are impaired in the function of the Tap42-PP2Ac complex rather than in that of the PP2A holoenzyme (Fig. 1), the G2/M block associated with the mutant cells suggests a role for the Tap42-PP2Ac complex in controlling the G2/M transition. This notion is supported by the fact that overexpression of TAP42 is able to suppress the Ts− phenotype, and by inference the G2/M block, caused by the pph21-102 allele (10, 31). Although the terminal phenotype of the tap42-11 cells grown at the nonpermissive temperature is a predominant G1 arrest (∼50% of cells arrests as large unbudded cells in our strain background), a significant portion (∼31%) of the cells arrest with a large bud, a characteristic of a G2/M arrest (H. Wang, unpublished observation). The G1 arrest associated with the mutant cells is likely to be a consequence of the defects in the Tap42-Sit4 complex, which is required for the G1/S transition (Fig. 3) (H. Wang, submitted for publication). In accordance with this, we have recently found that when the tap42-11 cells treated with hydroxyurea, which were arrested at S phase, were released into hydroxyurea-free medium at the nonpermissive temperature, the majority (∼67%) of the cells were arrested as large-budded cells. This observation suggests that, upon release from S phase, the mutant cells were unable to progress into G1 phase at the nonpermissive temperature (data not shown). Although it is possible that the G2/M block in the tap42-11 and pph21-102 cells is caused by activation of the morphogenesis checkpoint in response to the defects in the actin cytoskeleton (30), our finding that overexpression of RHO2 suppresses the actin defects but not the Ts− phenotype of the pph21-102 cells argues against it. Taking these results together, we suggest that the Tap42-PP2Ac complex is involved in controlling the G2/M transition of yeast cells. Since the Tap42-PP2Ac complex acts downstream of the Tor proteins, our finding further suggests a role for the Tor signaling pathway in controlling entry into mitosis in the yeast cell cycle.
It has been suggested that in yeast PP2A plays a positive role in entry into mitosis, a conclusion that is contradictory to the findings from other systems, in which PP2A has been shown to play a negative role in the same process (23, 28, 29, 31). Since the conclusion is largely based on the studies of the pph21-102 allele, our finding that this mutant is specifically defective in the formation of the Tap42-PP2Ac complex rather than the PP2A holoenzyme offers a different interpretation for the role of PP2A activity in the G2/M transition. For the same reasons that led us to suggest that PP2A activity negatively regulates the actin cytoskeleton, the involvement of the Tap42-PP2Ac complex in the G2/M transition prompts us to propose a negative role for PP2A activity in this process. Further study of the role of the Tap42-PP2Ac complex in the G2/M transition will allow us to test this notion. In addition, it is worth noting that, in Xenopus laevis, the PP2A activity that negatively regulates the G2/M transition has been found to be elicited by a novel form of PP2A, termed INH, rather than the PP2A holoenzyme (28).
The role of Tap42 in PP2A regulation.
Mutational analyses of the genes for PP2Ac have established the role of PP2Ac in three important cellular processes, namely, organization of the actin cytoskeleton, regulation of the G2/M transition, and control of cell wall integrity (11, 31). Since cell wall integrity is regulated by the Pkc1-MAPK pathway, in which Rho1 and Rho2 GTPases partake (43), the connection between the Tap42-PP2Ac complex and the Rho GTPases indicates a role for the complex in this process. As such, Tap42 is involved in all the processes in which PP2Ac has an established role. Since Tap42 has been found to be associated with only ∼5% of PP2Ac under normal conditions (10), the fact that the previous studies of PP2Ac reveal only the function of the Tap42-PP2Ac complex, not that of the PP2A holoenzyme, underscores the importance of Tap42 in PP2Ac regulation.
Acknowledgments
We are indebted to Guilleromo Romero for providing us his microscope facilities and assisting us in their usage. We thank Jack Yalowich, Ferruccio Galbiati, and Lisa Schneper for critical review of this manuscript.
This work was supported by the CMRF fund from University of Pittsburgh School of Medicine and by the Startup fund to Y.J.
REFERENCES
- 1.Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite, and M. N. Hall. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7:25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689-692. [DOI] [PubMed] [Google Scholar]
- 3.Berset, C., H. Trachsel, and M. Altmann. 1998. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95:4264-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertram, P. G., J. H. Choi, J. Carvalho, W. Ai, C. Zeng, T. F. Chan, and X. F. Zheng. 2000. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 275:35727-35733. [DOI] [PubMed] [Google Scholar]
- 5.Bertram, P. G., C. Zeng, J. Thorson, A. S. Shaw, and X. F. Zheng. 1998. The 14-3-3 proteins positively regulate rapamycin-sensitive signaling. Curr. Biol. 8:1259-1267. [DOI] [PubMed] [Google Scholar]
- 6.Bickle, M., P. A. Delley, A. Schmidt, and M. N. Hall. 1998. Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 17:2235-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cafferkey, R., P. R. Young, M. M. McLaughlin, D. J. Bergsma, Y. Koltin, G. M. Sathe, L. Faucette, W. K. Eng, R. K. Johnson, and G. P. Livi. 1993. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol. Cell. Biol. 13:6012-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13:3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen, P., S. Alemany, B. A. Hemmings, T. J. Resink, P. Stralfors, and H. Y. Tung. 1988. Protein phosphatase-1 and protein phosphatase-2A from rabbit skeletal muscle. Methods Enzymol. 159:390-408. [DOI] [PubMed] [Google Scholar]
- 10.Di Como, C. J., and K. T. Arndt. 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10:1904-1916. [DOI] [PubMed] [Google Scholar]
- 11.Evans, D. R., and M. J. Stark. 1997. Mutations in the Saccharomyces cerevisiae type 2A protein phosphatase catalytic subunit reveal roles in cell wall integrity, actin cytoskeleton organization and mitosis. Genetics 145:227-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-Sarabia, M. J., A. Sutton, T. Zhong, and K. T. Arndt. 1992. SIT4 protein phosphatase is required for the normal accumulation of SWI4, CLN1, CLN2, and HCS26 RNAs during late G1. Genes Dev. 6:2417-2428. [DOI] [PubMed] [Google Scholar]
- 13.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Academic Press, Inc., New York, N.Y.
- 14.Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96:14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harlow, E. D., and D. Lane. 1988. Antibodies: a laboratory manual, p. 519-552. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Healy, A. M., S. Zolnierowicz, A. E. Stapleton, M. Goebl, A. A. DePaoli-Roach, and J. R. Pringle. 1991. CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol. Cell.Biol. 11:5767-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heitman, J., N. R. Movva, and M. N. Hall. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253:905-909. [DOI] [PubMed] [Google Scholar]
- 18.Helliwell, S. B., I. Howald, N. Barbet, and M. N. Hall. 1998. TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics 148:99-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helliwell, S. B., A. Schmidt, Y. Ohya, and M. N. Hall. 1998. The Rho1 effector Pkc1, but not Bni1, mediates signalling from Tor2 to the actin cytoskeleton. Curr. Biol. 8:1211-1214. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann, R., S. Jung, M. Ehrmann, and H. W. Hofer. 1994. The Saccharomyces cerevisiae gene PPH3 encodes a protein phosphatase with properties different from PPX, PP1 and PP2A. Yeast 10:567-578. [DOI] [PubMed] [Google Scholar]
- 21.Jacinto, E., B. Guo, K. T. Arndt, T. Schmelzle, and M. N. Hall. 2001. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell 8:1017-1026. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, Y., and J. R. Broach. 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 18:2782-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinoshita, N., H. Ohkura, and M. Yanagida. 1990. Distinct, essential roles of type 1 and 2A protein phosphatases in the control of the fission yeast cell division cycle. Cell 63:405-415. [DOI] [PubMed] [Google Scholar]
- 24.Koltin, Y., L. Faucette, D. J. Bergsma, M. A. Levy, R. Cafferkey, P. L. Koser, R. K. Johnson, and G. P. Livi. 1991. Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol. Cell. Biol. 11:1718-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krause, S. A., and J. V. Gray. 2002. The protein kinase C pathway is required for viability in quiescence in Saccharomyces cerevisiae. Curr. Biol. 12:588-593. [DOI] [PubMed] [Google Scholar]
- 26.Kunkel, T. A., K. Bebenek, and J. McClary. 1991. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 204:125-139. [DOI] [PubMed] [Google Scholar]
- 27.Kunz, J., R. Henriquez, U. Schneider, M. Deuter-Reinhard, N. R. Movva, and M. N. Hall. 1993. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73:585-596. [DOI] [PubMed] [Google Scholar]
- 28.Lee, T. H., M. J. Solomon, M. C. Mumby, and M. W. Kirschner. 1991. INH, a negative regulator of MPF, is a form of protein phosphatase 2A. Cell 64:415-423. [DOI] [PubMed] [Google Scholar]
- 29.Lee, T. H., C. Turck, and M. W. Kirschner. 1994. Inhibition of cdc2 activation by INH/PP2A. Mol. Biol. Cell 5:323-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lew, D. J. 2000. Cell-cycle checkpoints that ensure coordination between nuclear and cytoplasmic events in Saccharomyces cerevisiae. Curr. Opin. Genet. Dev. 10:47-53. [DOI] [PubMed] [Google Scholar]
- 31.Lin, F. C., and K. T. Arndt. 1995. The role of Saccharomyces cerevisiae type 2A phosphatase in the actin cytoskeleton and in entry into mitosis. EMBO J. 14:2745-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorenz, M. C., and J. Heitman. 1995. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J. Biol. Chem. 270:27531-27537. [DOI] [PubMed] [Google Scholar]
- 33.Luke, M. M., F. Della Seta, C. J. Di Como, H. Sugimoto, R. Kobayashi, and K. T. Arndt. 1996. The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol. Cell. Biol. 16:2744-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nickels, J. T., and J. R. Broach. 1996. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 10:382-394. [DOI] [PubMed] [Google Scholar]
- 35.Noda, T., and Y. Ohsumi. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273:3963-3966. [DOI] [PubMed] [Google Scholar]
- 36.Peterson, R. T., B. N. Desai, J. S. Hardwick, and S. L. Schreiber. 1999. Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycin associated protein. Proc. Natl. Acad. Sci. USA 96:4438-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powers, T., and P. Walter. 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10:987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pringle, J. R., A. E. M. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565-602. [DOI] [PubMed] [Google Scholar]
- 39.Ronne, H., M. Carlberg, G. Z. Hu, and J. O. Nehlin. 1991. Protein phosphatase 2A in Saccharomyces cerevisiae: effects on cell growth and bud morphogenesis. Mol. Cell. Biol 11:4876-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothstein, R. 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194:281-301. [DOI] [PubMed] [Google Scholar]
- 41.Schmelzle, T., and M. N. Hall. 2000. TOR, a central controller of cell growth. Cell 103:253-262. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt, A., M. Bickle, T. Beck, and M. N. Hall. 1997. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 88:531-542. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt, A., and M. N. Hall. 1998. Signaling to the actin cytoskeleton. Annu. Rev. Cell Dev. Biol. 14:305-338. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt, A., J. Kunz, and M. N. Hall. 1996. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc. Natl. Acad. Sci. USA 93:13780-13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shu, Y., H. Yang, E. Hallberg, and R. Hallberg. 1997. Molecular genetic analysis of Rts1p, a B′ regulatory subunit of Saccharomyces cerevisiae protein phosphatase 2A. Mol. Cell. Biol. 17:3242-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sneddon, A. A., P. T. Cohen, and M. J. Stark. 1990. Saccharomyces cerevisiae protein phosphatase 2A performs an essential cellular function and is encoded by two genes. EMBO J. 9:4339-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stan, R., M. M. McLaughlin, R. Cafferkey, R. K. Johnson, M. Rosenberg, and G. P. Livi. 1994. Interaction between FKBP12-rapamycin and TOR involves a conserved serine residue. J. Biol. Chem. 269:32027-32030. [PubMed] [Google Scholar]
- 49.Sutton, A., D. Immanuel, and K. T. Arndt. 1991. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol. Cell. Biol. 11:2133-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 51.Torres, J., C. J. Di Como, E. Herrero, and M. A. De La Torre-Ruiz. 2002. Regulation of the cell integrity pathway by rapamycin-sensitive TOR function in budding yeast. J. Biol. Chem. 277:43495-43504. [DOI] [PubMed] [Google Scholar]
- 52.van Zyl, W., W. Huang, A. A. Sneddon, M. Stark, S. Camier, M. Werner, C. Marck, A. Sentenac, and J. R. Broach. 1992. Inactivation of the protein phosphatase 2A regulatory subunit A results in morphological and transcriptional defects in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:4946-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng, X. F., D. Florentino, J. Chen, G. R. Crabtree, and S. L. Schreiber. 1995. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell 82:121-130. [DOI] [PubMed] [Google Scholar]