Abstract
The human U1 snRNP-specific U1A protein autoregulates its own production by binding to and inhibiting the polyadenylation of its own pre-mRNA. Previous work demonstrated that a short sequence of U1A protein is essential for autoregulation and contains three distinct activities, which are (i) cooperative binding of two U1A proteins to a 50-nucleotide region of U1A pre-mRNA called polyadenylation-inhibitory element RNA, (ii) formation of a novel homodimerization surface, and (iii) inhibition of polyadenylation by inhibition of poly(A) polymerase (PAP). In this study, we purified and analyzed 11 substitution mutant proteins, each having one or two residues in this region mutated. In 5 of the 11 mutant proteins, we found that particular amino acids associate with one activity but not another, indicating that they can be uncoupled. Surprisingly, in three mutant proteins, these activities were improved upon, suggesting that U1A autoregulation is selected for suboptimal inhibitory efficiency. The effects of these mutations on autoregulatory activity in vivo were also determined. Only U1A and U170K are known to regulate nuclear polyadenylation by PAP inhibition; thus, these results will aid in determining how widespread this type of regulation is. Our molecular dissection of the consequences of conformational changes within an RNP complex presents a powerful example to those studying more complicated pre-mRNA-regulatory systems.
The U1 small nuclear ribonucleoprotein (snRNP) is the most abundant member of the spliceosomal snRNPs in vertebrate cells. Human U1 snRNP is required for splicing of pre-mRNA and is composed of the 164-nucleotide (nt) U1 small nuclear RNA (snRNA) and 10 polypeptides, 3 of which are specific to U1 snRNP (34). One of these U1 snRNP-specific proteins, U1A, contains two conserved RNA recognition motifs (RRMs) characteristic of the largest family of RNA binding proteins (reviewed in references 3, 25, and 31). Independent of the other U1 snRNP proteins, the N-terminal 101 residues of U1A (U1A1-101), containing one of these RRMs, is sufficient to bind to stem-loop 2 (SL2) of U1 snRNA (22, 27) and the U1A-SL2 complex has been the subject of intense biochemical and structural studies. Indeed, of the more than 1,000 RRMs known, the N-terminal RRM of U1A is the best understood at the biochemical and structural levels. RRMs are about 80 amino acids in length and consist of a β1α1β2β3α2β4 structure in which the four β strands form a sheet buttressed by two α helices (13, 23; see Fig. 1A). Usually, the RRM is sufficient for RNA binding activity; however, in the case of U1A, additional flanking sequences in the form of a third α helix, helix C (residues 92 to 98), are necessary (1, 7, 11, 14, 15, 19). In stark contrast to the N-terminal RRM, the C-terminal RRM of U1A has low affinity for RNA and no cellular RNA targets have been identified (21).
FIG. 1.
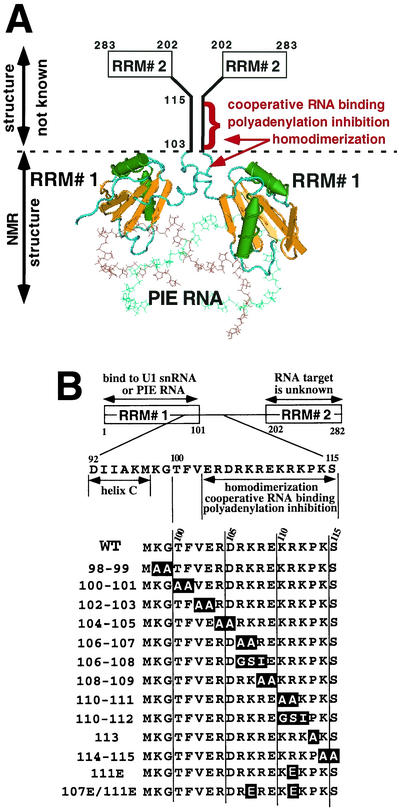
Structural features of the (U1A)2-PIE RNA complex and the U1A mutations that are the subject of this work. (A) Shown is the ribbon structure of two molecules of U1A (residues 1 to 102) bound to PIE RNA during autoregulation. Note that the atomic structure of residues 103 to 283, which includes a C-terminal RRM (RRM# 2), has not been determined. The three activities map to residues 103 to 115, and the homodimerization domain extends N terminal to about residue 95. (B) Shown are the domain structure of U1A and the sequences of the mutant U1A proteins that are the subject of this work. The mutated residues are boxed. On the left are the names of the mutant U1A proteins, which correspond to the amino acid positions of the mutations. WT, wild type.
U1 snRNP is involved in early steps of spliceosome formation and binds to the 5′ splice site of the pre-mRNA (reviewed in reference 18). The function of U1 snRNP-bound U1A in splicing is unknown, and it is possible that U1A is not even essential for the splicing reaction because in vitro splicing in HeLa cell nuclear extracts can still proceed in the absence of U1A (33) and the Saccharomyces cerevisiae U1A homolog is not an essential gene (20). The U1A protein also functions in 5′ and 3′ splice site communication, although the molecular nature of this remains unclear (9, 28). Aside from its role in U1 snRNP function, snRNP-free U1A autoregulates its own expression level by a negative feedback mechanism in which the polyadenylation of its own pre-mRNA is inhibited (2). The 3′ untranslated region (UTR) of the human U1A pre-mRNA contains a 50-nt sequence, designated the polyadenylation-inhibitory element (PIE) RNA, whose sequence and structure are conserved in vertebrates. PIE RNA consists of two asymmetric 7-nt loops flanked by short base-paired sequences that each bind one molecule of U1A protein (see Fig. 1A). Although one of the loops, when studied in isolation, has a 27-fold lower affinity for U1A than the other loop, it was demonstrated that two molecules of U1A bind with high affinity (Kd, ∼0.1 nM) to PIE RNA, which is indicative of cooperative RNA binding (2, 29). The resulting (U1A)2-PIE RNA complex inhibits addition of the poly(A) tail to the U1A pre-mRNA by specifically inhibiting the enzyme poly(A) polymerase (PAP) (8). Inhibition of polyadenylation requires both the C-terminal 20 residues of PAP and residues 103 to 115 of U1A (9). By using a similar inhibitory mechanism, U1A can also affect expression of the immunoglobulin heavy-chain gene by regulating poly(A) tail addition, in this case, however, through multiple novel U1A binding sites that deviate significantly from the consensus (26).
Determination of the structures of both free U1A1-101 and RNA-bound U1A2-102 by X-ray crystallography (23, 24) and nuclear magnetic resonance (NMR) analysis (1, 7, 14, 15, 19, 32) showed that helix C undergoes a large 135° conformational change upon binding to an RNA containing SL2 of U1 snRNA or as a monomer to PIE RNA (see Fig. 1A). In the (U1A)1-RNA complex, this conformational change prevents dissociation of U1A, consistent with the observation that mutation or deletion of helix C destabilizes the complex (1, 7, 11). PIE RNA also contributes to complex formation because fluorescence resonance energy transfer analysis by tagging of PIE RNA identified an inherent bend that brings the two U1A proteins close together (5, 6). The conformational change in helix C also leads to the formation of a homodimerization surface, as seen in both the NMR structure of the (U1A)2-PIE RNA complex (32) and fluorescence resonance energy transfer analysis by tagging of the U1A protein (4).
The functional implications of this structural work became evident with the assigning of three biochemically defined activities to residues 103 to 115 of U1A that are essential for autoregulation: (i) cooperative binding of two U1A proteins to PIE RNA, (ii) formation of a novel homodimerization surface, and (iii) inhibition of polyadenylation (16). It was assumed that these activities are tightly linked for the following reasons. Cooperative RNA binding could be readily explained by the formation of a homodimerization surface that accounted for ∼30% (1,190 Å) of the protein-protein and protein-RNA contacts in the complex (32). Likewise, polyadenylation inhibition could be readily explained by the same homodimerization surface and can be accurately mimicked by a dimeric peptide that was conformationally constrained in a similar head-to-head parallel orientation (16). Experimental evidence of this tight linkage came from the finding that all three activities were strongly reduced in two U1A mutant proteins that each contain three residue substitutions in the 102-to-115 region.
Here we undertook a systematic fine-scale mutational and biochemical analysis of U1A residues 102 to 115 and flanking residues. Five of the 11 mutant U1A proteins analyzed demonstrate that particular amino acids associate with one function but not another, indicating that these functions can be uncoupled. Even more surprisingly, three mutant proteins show increased activity, suggesting that the autoregulatory system is under selective pressure not to be too strong. The effects of these mutations on autoregulatory activity in vivo were also determined. Only U1A and U170K are known to regulate nuclear polyadenylation by PAP inhibition; thus, these results will aid in determining how widespread this type of regulation is. Our molecular dissection of the consequences of conformational changes within an RNP complex presents a powerful example to those studying more complicated pre-mRNA-regulatory systems.
MATERIALS AND METHODS
Plasmids.
All of the plasmids containing the wild-type U1A (U1Awt) and mutant U1A proteins were encoded by cDNAs lacking the U1A binding sites in the 3′ UTR. The U1A expression constructs used to make stable cell lines were derived from the pIRESPuro3 plasmid (Clontech), in which a Tet-OFF promoter replaced the constitutively active cytomegalovirus promoter. pIRESPuro3 is a bicistronic expression vector that produces two polypeptides from one transcript; one is the U1A protein, and the other is the puromycin resistance protein.
Proteins and RNA substrates.
The U1Awt and mutant U1A proteins were expressed in and purified from BL21 cells as previously described, first by nickel chromatography with nitrilotriacetic acid-agarose (Qiagen), followed by Mono S chromatography on an AKTA system (Pharmacia) (10). RNA substrates used in the in vitro assays were made by in vitro transcription as previously described, and if necessary, the RNAs were gel purified prior to use (16).
Cell culture, stable cell lines, and extract preparation.
Cell lines stably expressing U1Awt and mutant versions of U1A protein were derived from HeLa Tet cells (Clontech), which stably express the reverse tetracycline repressor (rTA), by using G418 as the selection reagent. The growth medium contained Dulbecco modified Eagle medium, 10% fetal calf serum, penicillin, streptomycin, 0.2 μg of G418 per ml to maintain the expression of rTA, and various amounts of puromycin and doxycycline (DOX). In the absence of DOX, the Tet-OFF promoter is active, whereas in the presence of DOX, rTA represses the activity of the Tet-OFF promoter. Stable cells were selected in complete growth medium containing 0.8 μg of puromycin per ml, whereas stable expression was maintained by growing them in medium containing 0.8 to 4 μg of puromycin per ml, depending on the level of expression desired. Note that puromycin was omitted when DOX was present in the medium. The level of expression of the tagged U1A proteins was confirmed by Western blotting.
Nuclear and cytoplasmic fractionation was performed as follows. One 10-cm-diameter plate of HeLa cells was harvested, pelleted, and washed once in 1× phosphate-buffered saline. The pellet was resuspend in buffer A (20 mM Tris [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100), rotated for 5 min, and centrifuged at 5,000 × g for 3 min. The supernatant (cytoplasmic extract) was removed, and the pellet was resuspended in buffer C (420 mM KCl, 10% glycerol, 20 mM Tris [pH 7.5], 0.1 mM EDTA) and incubated with agitation for 10 min. After centrifugation for 5 min at 5,000 × g, the supernatant (nuclear extract) was collected. Protein concentration was determined by using Bio-Rad reagent.
Western blotting and in vitro assays.
Experiments to determine polyadenylation inhibition, homodimerization, and RNA binding (electrophoretic mobility shift assays [EMSAs]) were performed as previously described (16). Western blots were probed with an anti-U1A antibody as previously described (26). Western blots were reprobed with an anti-U2AF65 antibody to confirm equal loading and transfer of samples.
RESULTS
Rationale for strategy of approach.
We undertook a mutagenic analysis of U1A residues 98 to 115 with the following goals: (i) to better understand the functional basis of three biochemically defined activities (Fig. 1A) and how they affect each other and U1A autoregulation and (ii) to determine the sequence constraints on autoregulation that would allow a more straightforward assessment of the polyadenylation-regulatory potential of other proteins with similar sequences. Note that limited, rather than exhaustive, mutagenesis was performed because the goal was not to determine the function of each individual amino acid. In most cases, we changed two amino acids at a time (Fig. 1B) because we expected that cooperativity and homodimerization would depend on an extensive series of protein-protein interactions that were unlikely to be significantly affected by single amino acid changes. We also made and analyzed mutations in residues 98 to 101 because they would likely contribute to at least some of these activities. We did not mutagenize residues 92 to 97 of helix C, however, because they are needed for the binding of U1A as a monomer to RNA (1, 11, 19), which would overly complicate the analysis. Additionally, we made and analyzed several single-residue changes at positions 107, 111, and 113 for the following reasons. Proline 113 is near the extreme end of the conserved region and would be expected to disrupt the predicted α-helical structure of residues 102 to 112. Mutation of lysine 107, arginine 111, or both would disrupt two groups of three basic residues that had been predicted to be critical for polyadenylation inhibition. During our database searches, we found it necessary and useful to include at least one group of three uninterrupted basic residues in a consensus polyadenylation-inhibitory sequence because this greatly shortened the list of proteins with significant matches (10). Because there was no experimental basis for the inclusion of these three basic residues, mutation of positions 107 and 111 would be the first direct test of this inclusion.
Recombinant mutant U1A proteins all bind similarly to SL2 RNA.
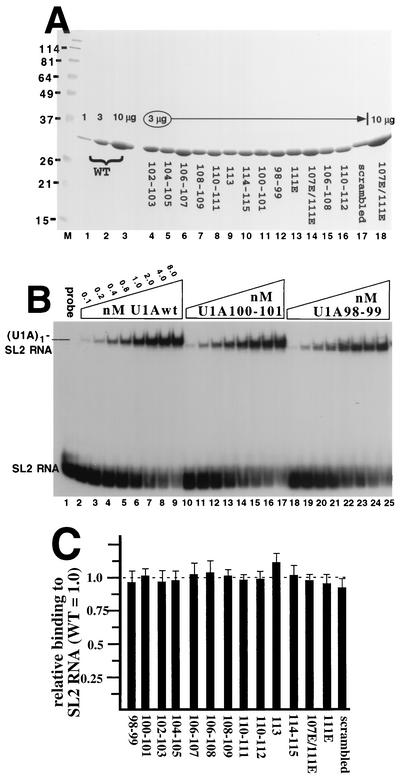
To aid in purification, all of the U1A proteins in Fig. 1B contained a C-terminal histidine tag that we had previously shown does not affect U1A activity in any of the assays used in this work (9, 16). The U1A proteins were expressed in BL21 bacterial cells and purified to >95% homogeneity by Ni-nitrilotriacetic acid chromatography, followed by Mono S chromatography. The quality and uniformity of the preparations were verified with a Coomassie-stained protein gel (Fig. 2A). To compare cooperative RNA binding of the mutant U1A proteins with that of U1Awt, it was essential that all of the U1A proteins bind with similar affinity to RNAs with a single binding site. EMSA of each U1A protein bound to radiolabeled SL2 RNA was done, and an example of some of the results is shown in Fig. 2B. All of the EMSAs were quantitated by phosphorimager analysis, and the values were used to generate the graph in Fig. 2C, where it is evident that each mutant U1A protein bound to SL2 RNA with the same relative affinity as U1Awt.
FIG. 2.
Relative SL2 RNA binding activities of the mutant U1A proteins compared to that of U1Awt. (A) Coomassie-stained gel of purified, recombinant U1A proteins. Lanes 1 to 3 contain 1, 3, and 10 μg, respectively, of U1Awt protein. Lanes 4 to 17 contain 3 μg of the various mutant U1A proteins, as indicated. Lane 17 contains the previously characterized scrambled mutant U1A protein (16). For comparison, lane 18 contains 10 μg of the U1A107E/111E mutant protein. The values on the left are in kilodaltons and represent the positions of the molecular weight markers (lane M). WT, wild type. (B) Autoradiogram of an EMSA of selected U1A proteins bound to 32P-radiolabeled SL2 RNA, which is the site of U1A binding on U1 snRNA. Each lane contains 1.0 nM SL2 RNA, except lane 1, which lacks added U1A protein. Lanes 2 to 25 contain increasing amounts of U1Awt or mutant U1A protein, and the nanomolar concentration and type of U1A are indicated. On the left are indicated the positions and identities of the complexes. (C) Graphic summary of the EMSAs in which the y axis shows the SL2 RNA binding activities of the mutant U1A proteins relative to that of U1Awt, which was set to 1.0. Also shown are the standard deviations, which were less than 15% for each mutant protein. Each mutant protein was tested in at least three independent EMSAs.
Analysis of cooperative binding to PIE RNA.
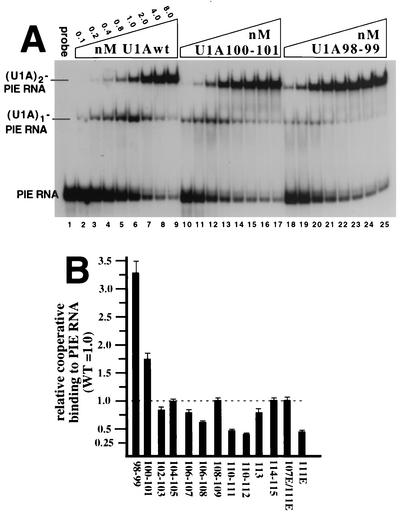
It has previously been demonstrated that the binding constants (Kd's) of the two individual binding sites on PIE RNA for U1A differ by about 27-fold. This was done by measuring the Kd of each site in the absence of the other site, which was inactivated by mutation (29). However, in the case of the wild-type PIE RNA, where both binding sites are present, the second molecule of U1A binds with nearly the same affinity as the first molecule, indicating cooperative RNA binding and suggesting a direct interaction between the two RNA-bound U1A proteins. We had previously shown that residues 102 to 115 are important for cooperativity because scrambled mutant U1A (in which the order of these residues is scrambled) resulted in complete loss of cooperative RNA binding (16) with no significant effect on binding as a monomer to SL2 RNA. To examine this region of U1A in more detail, we analyzed cooperative RNA binding by performing an EMSA of each mutant U1A protein bound to radiolabeled PIE RNA (Fig. 3A and B). Four of the mutant U1A proteins had no reduction, and seven mutant U1A proteins had a moderate reduction in cooperativity compared to U1Awt. These results were consistent with the expectation that cooperativity would be based on an extensive set of interactions across the entire homodimerization region. The fact that none of the mutant proteins was as severely down in cooperativity as scrambled mutant U1A (which binds at a 4% level relative to U1Awt) indicated that pairs of residues in this region make partial but not essential contributions to cooperativity.
FIG. 3.
Relative cooperative RNA binding activities of mutant U1A proteins compared to that of U1Awt. (A) Shown is an autoradiogram of an EMSA of selected U1A proteins bound to 32P-radiolabeled PIE RNA. Each lane contains 1 nM PIE RNA, except lane 1, which contains no added U1A protein. Lanes 2 to 25 contain increasing amounts of mutant U1A protein or U1Awt, and the nanomolar concentration and type of U1A are indicated. On the left are indicated the positions and identities of the complexes. (B) Graphic summary of the EMSAs in which the y axis shows the PIE RNA binding activities of the mutant proteins in forming the (U1A)2-PIE RNA complex relative to that of U1Awt, which was set to 1.0. Also shown are the standard deviations, which were less than 15% for each mutant protein. Each mutant protein was tested in at least three independent EMSAs. WT, wild type.
With this in mind, we were surprised to find that two mutant U1A proteins, U1A98-99 and U1A100-101, exhibited a significant increase in cooperativity (EMSA in Fig. 3A and graph in Fig. 3B). The anomalous behavior of U1A98-99 and U1A100-101 was not due to a specific set of EMSA conditions because similar results were obtained when we varied the MgCl2, NaCl, competitors, cross-linking, and percentage of acrylamide (data not shown). An alternative explanation is that changes in the homodimerization activity of free mutant U1A protein, rather than RNA-bound U1A, were responsible for this anomalous behavior. By using a battery of assays (immunoprecipitation, coselection, and gel filtration), we had previously shown that U1Awt has no detectable homodimerization activity in the absence of RNA (16). By using these same assays with these two up mutant proteins in the absence of RNA, we observed no detectable homodimerization activity (data not shown), thus ruling out this alternative explanation. This increase in cooperativity is also visible in the gel shift pattern of the EMSA. For example, there were four points in the titration curve of U1Awt where the (U1A)1-PIE RNA complex was more abundant than the (U1A)2-PIE RNA complex (Fig. 3A, lanes 2 to 5). In contrast, there were no points in the titration curve of the U1A98-99 mutant protein, where the (U1A)1-PIE RNA complex was more abundant than the (U1A)2-PIE RNA complex. Thus, each incremental increase in the U1A98-99 mutant protein favored the notion that the added protein preferentially binds the (U1A)1-PIE RNA complex, shifting it to the (U1A)2-PIE RNA complex instead of binding free PIE RNA. The U1A100-101 mutant protein also showed a significant increase in cooperativity, although less than the U1A98-99 mutant protein. Stated in a different way, our results indicate that residues 98 to 101 act to dampen cooperativity, suggesting that one of their functions is to reduce the negative feedback inhibition in the U1A autoregulatory system. Finally, we note the puzzling observation that the U1A111E mutant protein is down 50% in cooperativity but the double-mutant protein U1A107E/111E is like the wild type. Possible explanations for this result are presented in the Discussion.
Analysis of polyadenylation inhibition.
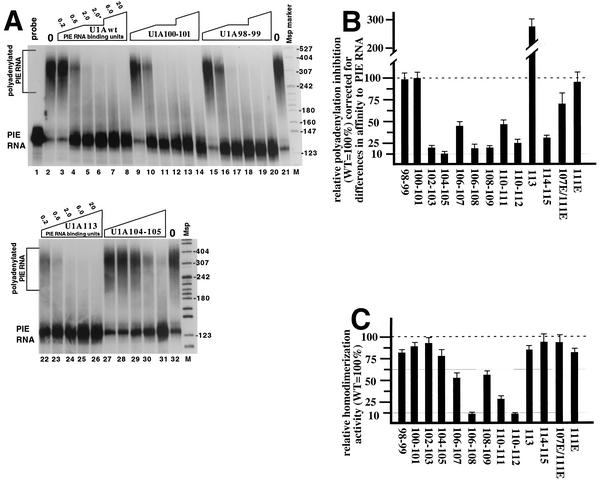
We next tested the mutant U1A proteins for the ability to inhibit polyadenylation in HeLa cell nuclear extracts. It has previously been shown that only the (U1A)2-PIE RNA complex, but not (U1A)1-PIE RNA, is active in polyadenylation inhibition (8, 29). Since each mutant U1A protein bound with a different affinity as two molecules to PIE RNA, we expressed the amounts of added U1A as PIE RNA binding units to reflect these differences. For each mutant U1A protein or U1Awt, one PIE RNA binding unit was chosen to be the amount needed to shift 50% of PIE RNA to the (U1A)2-PIE RNA complex. Thus, polyadenylation inhibition could be plotted as a function of the amount of (U1A)2-PIE RNA complex in the reaction, giving us a direct measure of the inhibitory activity of that complex independent of its binding affinity for PIE RNA. Figure 4A is an example of polyadenylation inhibition of the U1A98-99 and 100-101 mutant proteins (lanes 9 to 20), demonstrating that these mutant proteins had an inhibitory activity similar to that of U1Awt even though they were up mutant in cooperativity. Figure 4A, lanes 27 to 31, is an example of polyadenylation inhibition of the U1A104-105 mutant protein, which was severely down in inhibitory activity even though it bound PIE RNA with a cooperativity similar to that of U1Awt. The U1A113 mutant protein (lanes 22 to 26) was of particular interest because it was the only mutant protein that actually inhibited polyadenylation better than U1Awt. Mutation of proline 113 to alanine would remove the disruption of the predicted α helix of this region; thus, we speculate that the function of this conserved proline is to reduce the negative feedback inhibition in the U1A autoregulatory system.
FIG. 4.
Relative polyadenylation-inhibitory and homodimerization activities of mutant proteins compared to those of U1Awt. (A) Polyadenylation-inhibitory activity of selected mutant U1A proteins relative to that of U1Awt. The autoradiogram is of a denaturing 8% acrylamide gel used to separate the products of a polyadenylation assay by using HeLa nuclear extracts, 32P-radiolabeled PIE RNA, and increasing amounts of exogenously added U1A. Lane 1 is the RNA probe in the absence of nuclear extract. Lanes 2, 21, and 32 are the complete polyadenylation assay in the absence of exogenously added U1A protein. Lanes 3 to 20 and 22 to 31 contain increasing amounts of various types of U1A protein, as indicated. The amount of U1A is in PIE RNA binding units, where 1.0 represents the amount of U1A needed to obtain 50% of the (U1A)2-PIE RNA complex, as determined by EMSA from Fig. 3. As denoted by the asterisk, lanes 6, 12, and 18 used the same amount of U1A as lanes 5, 11, and 17, respectively, except that a different dilution of U1A was done. The two lanes marked M contain 32P-labeled MspI markers from MspI-digested pBR322, and the sizes of the markers are indicated in nucleotides on the right. (B) Graphic summary of the polyadenylation inhibition assays. The y axis shows the polyadenylation-inhibitory activities of the mutant U1A proteins relative to that of U1Awt, which was set to 100%. Note that the values were corrected to reflect differences in affinity to PIE RNA as described in the text and as shown in Fig. 4A. Note that standard deviations were less than 15% for all of the mutant proteins, which were tested in at least three independent experiments. (C) Graphic summary of the homodimerization assay results. The y axis shows the homodimerization activities of the mutant U1A proteins relative to that of U1Awt, which was set to 100%. The assay is described in the text. Note that the standard deviations were less than 15% for all of the mutant proteins, which were tested in at least three independent experiments. WT, wild type.
The graph in Fig. 4B summarizes the polyadenylation-inhibitory activities of the mutant U1A proteins relative to that of U1Awt after correction for differences in affinity to PIE RNA. Mutation of residues in the 102-to-112 region all had moderate or severe effects on polyadenylation inhibition that contrasted with the smaller effects of these residues on cooperative PIE RNA binding (Fig. 3B). Mutations in residues 98 to 101 did not affect polyadenylation-inhibitory activity. These results indicate that the polyadenylation inhibition region spans a smaller number of residues (102 to 115) than the homodimerization or cooperativity region (see Discussion). As discussed earlier, the changes to residues 107 and 111 were designed to test the importance of three basic contiguous residues for inhibition of polyadenylation. Mutation of arginine 111 had no effect on polyadenylation inhibition, while the double mutation at 107 and 111 had a modest but significant reduction in polyadenylation inhibition, suggesting that at least one stretch of three basic contiguous residues is important for polyadenylation inhibition, although not strongly so.
Analysis of homodimerization activity.
We also analyzed these mutant proteins for homodimerization activity by using a previously described coselection assay in which recombinant, unlabeled U1A containing a histidine tag was used to select 35S-labeled U1A lacking a histidine tag made by in vitro translation off a U1A mRNA lacking the PIE RNA sequence (16). For this assay, both the His-tagged U1A protein and the 35S-labeled U1A protein were either all wild type or all mutant. In agreement with this previous work, the U1A106-108 and U1A110-112 mutant proteins were strongly defective (more than fivefold) in homodimerization, indicating that the assay was working (autoradiograms not shown). As summarized in Fig. 4C, the homodimerization results indicate that all of the single- and two-amino-acid mutant proteins were similar to U1Awt or modestly down (2-fold), except for the U1A110-111 mutant protein (down 3.3-fold). The lack of any strong effects on homodimerization contrast with the effects on cooperativity and polyadenylation inhibition, including the fact that none of the mutant proteins showed an increase in homodimerization. This suggests that residues 98 to 115 contribute in a similar way to stabilization of the homodimerization surface.
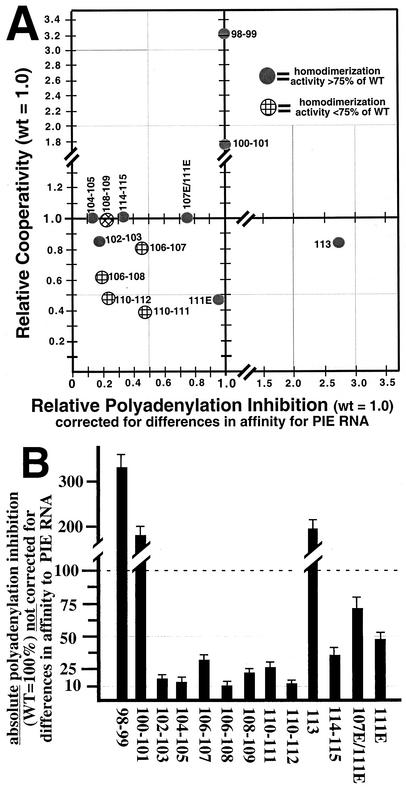
The graph in Fig. 5A summarizes the data by plotting relative cooperativity versus relative polyadenylation inhibition, where the values for relative polyadenylation inhibition have been adjusted for differences in binding to PIE RNA (as measured in the experiment whose results are shown in Fig. 3B). This type of graph is useful for facile assessment of the differences between mutant proteins that affect only cooperativity or only polyadenylation inhibition and mutant proteins that affect both activities. Thus, mutant U1A proteins falling on the y axis (U1A98-99, U1A100-101, and U1A111E) affect only cooperativity while mutant proteins falling on the x axis affect only polyadenylation inhibition. With only one exception (the U1A108-109 mutant protein), these x and y axis mutant proteins had no significant change in homodimerization activity and thus were truly affecting only one activity. We also note that four of the five mutant proteins affecting both activities, i.e., mutant proteins in the lower left quadrant, also were significantly down in homodimerization. Although the type of graph shown in Fig. 5A is useful for dissecting changes in polyadenylation inhibition activity separately from changes in cooperativity, the ability of U1A to inhibit polyadenylation in vivo is a combination of these two activities. Thus, Fig. 5B is a graph of the absolute polyadenylation-inhibitory activity of the mutant proteins (U1Awt is set to 100%) and the values are not corrected for differences in affinity of binding to PIE RNA. Figure 5B can therefore be used to predict the inhibitory activity of each mutant protein in vivo.
FIG. 5.
Summary of mutant U1A proteins. (A) The x axis shows the polyadenylation-inhibitory activities of the mutant proteins relative to that of U1Awt, which was set to 1.0, and the values were adjusted for differences in affinity to PIE RNA (taken from Fig. 3B). The y axis shows the cooperative RNA binding activities of the mutant proteins relative to that of U1Awt, which was set to 1.0. Solid circles represent mutant U1A proteins having homodimerization activity similar (>75%) to that of U1Awt, whereas hatched circles represent mutant U1A proteins with lower (<75%) homodimerization activity than U1Awt. (B) The y axis shows the absolute polyadenylation-inhibitory activities of the mutant proteins, where that of U1Awt was set to 100%. Note that unlike in panel A, the values here were not corrected for differences in affinity of binding to PIE RNA. WT and wt, wild type.
Inhibitory activity in vivo.
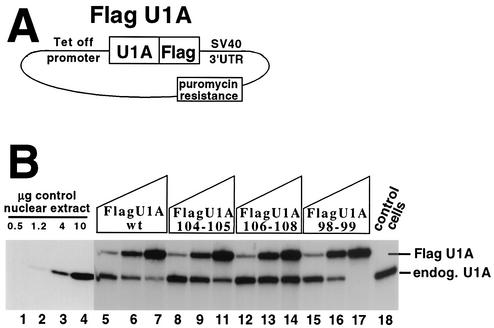
We had previously shown that overexpression of U1Awt in vivo by using a vector lacking the PIE RNA U1A binding sites leads to downregulation of endogenous U1A levels (2). Note that the U1A protein stably expressed from this type of transgene is not subject to autoregulation because it lacks PIE RNA in its 3′ UTR. We adapted this approach to determine the relative ability in vivo of the mutant U1A proteins to autoregulate endogenous U1A expression. As diagrammed in Fig. 6A, we produced HeLa cells stably expressing U1Awt and mutant U1A proteins under the control of a tetracycline-controlled promoter (the Tet-OFF system from Invitrogen) by using puromycin as the selection agent. The tetracycline promoter system has the advantage that varying the amount of DOX, an inhibitor of the tetracycline promoter, in the medium allows control of the levels of the FLAG-tagged U1A proteins.
FIG. 6.
In vivo inhibitory activity of selected mutant U1A proteins on the expression of endogenous U1A. (A) Schematic of the plasmid harboring FLAG epitope-tagged U1A under the control of a tetracycline (Tet-OFF) promoter used to make puromycin-resistant stable cell lines. Note that the vector does not contain the PIE RNA U1A binding sites and so would not be a target for U1A autoregulation. SV40, simian virus 40. (B) Western blot with an anti-U1A antibody used to measure the inhibitory effect of the tagged U1A proteins on the expression of endogenous (endog.) U1A protein. Lanes 5 to 17 contain 10 μg of nuclear extract from cells stably expressing different amounts of tagged U1A protein, as indicated above the lanes. Lanes 5 to 17 are four sets of three lanes each where each set is from a single stable cell line clone expressing the tagged U1A protein as indicated, where the amount of tagged U1A was controlled by addition of increasing amounts of DOX, an inhibitor of the Tet-OFF promoter. Similar results were obtained by analysis of other stable cell line clones (data not shown). Lanes 1 to 4 and 18 contain material from control cells stably expressing the plasmid shown in panel A that lacks the FLAG-U1A coding region. Lanes 1 to 4 contain different amounts of nuclear extract to gauge the sensitivity of the enhanced-chemiluminescence-based Western blot. Anti-U1A antibody 856 was used in the Western blot as previously described (26). Probing of the same blot with an anti-U2AF65 antibody confirmed equal loading and transfer of the nuclear proteins (data not shown).
Stable cell lines expressing the various U1A proteins were produced, and clones expressing higher levels were subjected to further analysis. In the absence of DOX, expression of tagged U1A was maximal and similar to that of the other tagged U1A proteins (lanes 7, 11, 14, and 17). However, the tagged U1A proteins differed in the ability to repress endogenous U1A levels, as quantified by phosphorimager analysis of the Western blot. To demonstrate that these differences were due to the levels of tagged U1A and not an artifact of the stable-cell selection process, we titrated in various amounts of DOX to reduce the levels of tagged U1A. In every case, the endogenous U1A levels were restored to near normal levels (Fig. 6, lanes 5, 8, 12, and 15). On the basis of Fig. 5A and B, mutant protein U1A98-99 is predicted to have enhanced inhibitory activity in vivo because of its increased binding affinity for PIE RNA (not because of any changes in its intrinsic polyadenylation-inhibitory activity). Consistent with this prediction, mutant protein U1A98-99 strongly reduced the levels of endogenous U1A compared to those of FLAG-U1Awt. Inhibition was still observed when tetracycline promoter activity was reduced by addition of DOX. In contrast, high levels of stably expressed U1A104-105 had only a modest inhibitory effect on endogenous U1A while the U1A106-108 mutant protein had essentially no inhibitory effect. Thus, the biochemical activities of these mutant U1A proteins correlate with their abilities to inhibit U1A expression in vivo.
DISCUSSION
We have presented a mutagenic and biochemical analysis of a domain in the U1A protein previously shown to contain three distinct biochemical activities necessary for autoregulation by inhibition of polyadenylation. The goals were to better understand how these three activities are interconnected with each other and to determine their boundaries in the protein. We also expected to learn whether a particular activity is dispensable for regulation of polyadenylation, giving us a clearer criterion by which to judge whether other proteins containing domains similar in sequence also regulate polyadenylation. Besides improving our mechanistic understanding of this domain and of U1A autoregulation, several rather surprising conclusions emerged, as discussed in detail below.
Boundaries of the three activities.
These results begin to map down to the single-residue level which sets of amino acids are responsible for which type of interaction or activity. The polyadenylation-inhibitory region is the best mapped of the three activities because it can be accurately mimicked with a dimeric peptide containing residues 102 to 115 (16). Thus, the polyadenylation inhibition region spans a smaller number of residues (102 to 115) than the homodimerization (94 to 112) or cooperativity (residues 94 to 113) region. We include residues 94 to 97 in the homodimerization region because they form a homodimer surface in the NMR structure of the (U1A1-102)2-PIE RNA trimeric complex (32). The other end of the homodimerization domain is likely to be proline 113, which would break the predicted coil-coil structure. Although our data indicate that the cooperativity region spans residues 98 to 113, the cooperativity region may also extend N terminal of residue 98. As discussed earlier, mutation of residues N terminal of 98 affects U1A binding as a monomer to RNA (1, 11, 19), making interpretation of their effect on cooperativity problematic.
Although it appears that the determinants controlling these three activities overlap considerably, the data do not support this because none of the two- or one-residue mutations had strong effects on homodimerization (with the exception of mutant protein U1A110-111), while many of these same mutant proteins had strong up or down effects on polyadenylation inhibition and cooperativity. The fact that there is almost no correlation suggests that alternative explanations need to be considered, such as the possibilities that (i) other regions of U1A or PIE RNA make larger contributions to homodimerization and cooperativity and (ii) these interactions have other, unrelated, functions yet to be discovered.
Uncoupling of polyadenylation inhibition and cooperativity.
We expected difficulty in uncoupling cooperative RNA binding, polyadenylation inhibition, and homodimerization because the former two should be direct consequences of homodimerization that arises from an overlapping ∼20-residue-long surface area. It is also known that the register or alignment of the homodimerization surface is critical to its inhibitory activity because a dimeric peptide containing two copies of these same residues, in which each copy is N terminally linked to the other copy, accurately mimics the polyadenylation inhibition seen with the entire protein (16). No inhibition is observed in a matching dimeric peptide when the two copies are C terminally linked. Thus, subtle mutations in this region would be expected to drastically alter its inhibitory activity. Additionally, each of the three previously characterized mutations in this region was strongly reduced in all three activities. Despite these arguments, uncoupling was relatively easy to obtain because nearly half (5 of 11) of the mutant proteins exhibited significant changes in one activity with little or no change in the other activity (Fig. 5A). We therefore propose that specificity determinants arising from the tertiary structure of the homodimerization surface contribute more than the α-helical secondary structure to these activities. Resolution of this awaits structural data on a longer fragment of RNA-bound U1A both with and without the inhibited PAP domain.
The idea that the register or alignment of the homodimer surface is important for function offers an explanation for the puzzling observation that the U1A111E mutant protein is down 50% in cooperativity whereas the double-mutant protein U1A107E/111E is not down. Unlike the U1A111E mutant protein, the double-mutant protein has a perfect 10-residue stretch of alternating positive and negative amino acids that may electrostatically pair with each other via slippage of the alignment of the two RNA-bound U1A proteins. This is also consistent with the homodimerization data (Fig. 4C), in which the U1A111E mutant protein is down in activity compared to the wild-type or double-mutant protein.
Homodimerization activity was nearly unaffected by all of the single- and two-residue mutations. This contrasts with the significant loss seen with the previously published three-residue substitutions (U1A 106-108 and U1A110-112). The fact that some of the two-residue mutations overlap the three-residue mutations suggests that a threshold number of contacts had been disrupted in the three-residue mutant proteins. Notably, the two-residue mutations that did show the strongest reduction clustered in the 106-to-112 region (Fig. 4C) in the heart of the presumed homodimerization surface. The simplest explanation for this clustering is that the center, rather than the edges, of the homodimerization surface plays a stronger role in stabilizing the complex.
U1A autoregulation and homeostasis.
In a surprising number of cases (3 of 13), substitution mutations actually resulted in increased activity in vitro, and testing in vivo of one of these three up mutant proteins confirmed this result. Our interpretation is that the U1A autoregulatory complex is suboptimal in terms of the ability to inhibit polyadenylation and that certain residues act to keep the complex from overregulating U1A levels. This is consistent with our recent work demonstrating that the polyadenylation-regulatory domain (PRD) of U1A is weak when directly compared with the known PRDs of U170K or the putative PRDs of other proteins (17). The data presented here show the ease and frequency with which such up mutations are found and strongly suggest that the autoregulatory system is under selective pressure not to be too strong. In hindsight, this result is perhaps not so surprising because the conservation of U1A autoregulation implies a requirement for homeostasis of U1A levels where either over- or underregulation would be deleterious to the cell.
Mutations that increase cooperative RNA binding.
Mutation of residues 98 to 101 significantly increases the cooperative RNA binding of U1A, a result all the more startling because the wild-type protein already exhibits a seemingly strong 27-fold cooperative effect, a rather impressive increase in cooperativity compared with other RNA binding proteins in which this parameter has been carefully measured (12, 30, 35). What is the rationale for this increase in cooperativity? One obvious explanation is that this surface contains unfavorable interactions that are removed by mutation. This is not supported by the NMR structure of the (U1A2-102)2-PIE RNA trimolecular complex, which showed that Met97 and Gly99 constitute the heart of a hydrophobic patch that should hold these two proteins together (32). We caution that the structural details of helix C and the hydrophobic patch may change because of end effects owing to their proximity to the end of the truncated U1A protein used for NMR. An alternative explanation is that interaction of residues 97 to 100 imposes interactions of residues C terminal to residue 100 that are less favorable for cooperative RNA binding. Stated in a different way, the register or alignment of the homodimer surface critically affects cooperative RNA binding. Additional biochemical and mutagenic approaches, along with the NMR structure of longer U1A molecules, are needed to clarify this issue. We note here that one limitation is that gel shift assays only measure the overall binding constant, and further work is needed to determine whether the association or dissociation rates are affected.
Mutations that increase polyadenylation inhibition.
Mutation of proline 113 to alanine significantly increased the ability of U1A to inhibit polyadenylation, with almost no effect on cooperativity or homodimerization. The proline and its position at the end of the conserved region are found in all vertebrate U1A proteins. The proline-to-alanine change would permit continuation of the predicted α-helical structure, suggesting that the proline maintains interactions of nearby residues unfavorable to polyadenylation inhibition. This idea is not consistent with observations that mutations in the adjacent upstream (108 to 112) and downstream (114 to 115) residues reduce the ability of U1A to inhibit polyadenylation. Alternatively, the binding pocket of PAP for this homodimerization surface may not be able to accommodate an extended α helix, necessitating the presence of proline 113. This would also explain the curious inability of dimeric peptides joined through the C terminus (equivalent to the proline 113 position) to inhibit polyadenylation whereas the same dimeric peptides joined at the N terminus were potent inhibitors of polyadenylation (16).
Although the in vivo inhibitory activity of the mutant proteins correlated with their biochemical activities, care must be taken not to overinterpret the in vivo data. The effect of the tagged mutant U1A proteins on the final levels of endogenous U1A can arise from several different activities, none of which are mutually exclusive. For example, the ability of a mutant U1A protein to compete with or replace the endogenous U1A protein in the autoregulatory complex is an essential part of the in vivo assay. Indeed, it was possible (although we have not observed this) that the endogenous U1A levels would actually increase because the mutant protein could enter the endogenous autoregulatory complex, thereby displacing either one or both of the endogenous U1A proteins in a given autoregulatory complex, but not be able to inhibit polyadenylation. In this scenario, the U1A104-105 mutant protein had the best chance of exhibiting this behavior but did not do so. In an alternative scenario, if the mutant U1A protein had normal polyadenylation-inhibitory activity but was defective in cooperative binding, then its ability to compete with endogenous U1A by entering the autoregulatory complex would be limited, resulting in little change in the levels of endogenous U1A. These limitations on the in vivo assay will be difficult to circumvent because one would have to eliminate the endogenous U1A protein. Our preliminary efforts to do this by RNA interference have indicated that reduced U1A levels are toxic to mammalian cells.
Lessons for pre-mRNA processing.
At one level, the U1A autoregulatory system is relatively simple because only U1A, PAP, and PIE RNA are necessary for reconstitution. Yet its mechanistic basis includes features found in more complicated regulatory systems. A 100° bend in PIE RNA imposes architectural constraints on the RNP complex and, coupled with a large 135° conformational change in helix C upon binding of U1A, results in the recruitment of a second U1A, leading to additional interactions and regulatory capabilities. This is all the more striking because only 18 amino acids and a small RNA sequence direct this whole process. Thus, principles of RNA recognition and reorganization used in this complex will no doubt be found in more complicated regulatory systems involving pre-mRNA processing.
Implications for the generalization of this type of regulation.
It has recently been shown that U1A can regulate the expression of the immunoglobulin heavy-chain (μ) gene by binding nonconsensus sequences on the μ pre-mRNA (26). In comparison to the U1A pre-mRNA, the μ pre-mRNA undergoes a complex series of processing steps involving competition between alternative polyadenylation sites and neighboring splice sites and the processing machineries that bind to them. At a superficial level, the secondary structure of the U1A binding sites on the μ pre-mRNA do not apparently match that seen with SL2 or PIE RNA. We are currently analyzing the secondary and tertiary structures of μ pre-mRNA to learn how U1A recognizes and regulates this heterologous pre-mRNA, which will enable us to judge whether U1A regulates other genes in a similar manner.
Acknowledgments
We thank Cathy Phillips and other members of the Gunderson lab for useful critique and discussion. We also thank members of the Rutgers University RNA club from the Peltz, Kiledjian, and Kinzy labs for useful discussion. We thank Iain Mattaj for the kind gift of U1A antibody 856.
This work was supported by National Institutes of Health grant 1R01-GM57286 and American Heart Association grant 9850071T to S.I.G.
REFERENCES
- 1.Avis, J. M., F. H.-T. Allain, P. Howe, G. Varani, K. Nagai, and D. Neuhaus. 1996. Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding. J. Mol. Biol. 257:398-411. [DOI] [PubMed]
- 2.Boelens, W. C., E. J. R. Jansen, W. J. van Venrooij, R. Stripecke, I. W. Mattaj, and S. I. Gunderson. 1993. The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell 72:881-892. [DOI] [PubMed] [Google Scholar]
- 3.Burd, C. G., and G. Dreyfuss. 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265:615-621. [DOI] [PubMed] [Google Scholar]
- 4.Clerte, C., and K. B. Hall. 2000. Spatial orientation and dynamics of the U1A proteins in the U1A-UTR complex. Biochemistry 39:7320-7329. [DOI] [PubMed] [Google Scholar]
- 5.Grainger, R. J., A. I. H. Murchie, D. G. Norman, and D. M. J. Lilley. 1997. Severe axial bending of RNA induced by the U1A binding element present in the 3′ untranslated region of the U1A mRNA. J. Mol. Biol. 273:84-92. [DOI] [PubMed] [Google Scholar]
- 6.Grainger, R. J., D. G. Norman, and D. M. J. Lilley. 1999. Binding of U1A protein to the 3′ untranslated region of its pre-mRNA. J. Mol. Biol. 288:585-594. [DOI] [PubMed] [Google Scholar]
- 7.Gubser, C. C., and G. Varani. 1996. Structure of the polyadenylation regulatory element of the human U1A pre-mRNA 3′-untranslated region and interaction with the U1A protein. Biochemistry 35:2253-2267. [DOI] [PubMed] [Google Scholar]
- 8.Gunderson, S. I., K. Beyer, G. Martin, W. Keller, W. C. Boelens, and I. W. Mattaj. 1994. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell 76:531-541. [DOI] [PubMed] [Google Scholar]
- 9.Gunderson, S. I., S. Vagner, M. Polycarpou-Schwarz, and I. W. Mattaj. 1997. Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev. 11:761-773. [DOI] [PubMed] [Google Scholar]
- 10.Gunderson, S. I., Polycarpou-Schwarz, M., and I. W. Mattaj. 1998. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell 1:255-264. [DOI] [PubMed] [Google Scholar]
- 11.Hall, K. B. 1994. Interaction of RNA hairpins with the human U1A N-terminal RNA binding domain. Biochemistry 33:10076-10088. [DOI] [PubMed] [Google Scholar]
- 12.Heaphy, S., C. Dingwall, I. Ernberg, M. J. Gait, S. M. Green, J. Karn, A. D. Lowe, M. Singh, and M. A. Skinner. 1990. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell 60:685-693. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman, D. W., C. C. Query, B. L. Golden, S. W. White, and J. D. Keene. 1991. RNA-binding domain of the A protein component of the U1 small nuclear ribonucleoprotein analyzed by NMR spectroscopy is structurally similar to ribosomal proteins. Proc. Natl. Acad. Sci. USA 88:2495-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howe, P. W., F. H. Allain, G. Varani, and D. Neuhaus. 1998. Determination of the NMR structure of the complex between U1A protein and its RNA polyadenylation inhibition element. J. Biomol. NMR 11:59-84. [DOI] [PubMed] [Google Scholar]
- 15.Jovine, L., C. Oubridge, J. M. Avis, and K. Nagai. 1996. Two structurally different RNA molecules are bound by the spliceosomal protein U1A using the same recognition strategy. Structure 4:621-631. [DOI] [PubMed] [Google Scholar]
- 16.Klein-Gunnewiek, J. M. T., R. I. Hussein, Y. van Aarssen, D. Palacios, R. de Jong, W. J. van Venrooij, and S. I. Gunderson. 2000. Fourteen residues of the U1 snRNP-specific U1A protein are required for homodimerization, cooperative RNA binding, and inhibition of polyadenylation. Mol. Cell. Biol. 20:2209-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko, B., and S. I. Gunderson. 2002. Identification of new poly(A) polymerase-inhibitory proteins capable of regulating pre-mRNA polyadenylation. J. Mol. Biol. 318:1189-1206. [DOI] [PubMed] [Google Scholar]
- 18.Kramer, A. 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65:367-409. [DOI] [PubMed] [Google Scholar]
- 19.Kranz, J. K., and K. B. Hall. 1998. RNA binding mediates the local cooperativity between the beta-sheet and the C-terminal tail of the human U1A RBD1 protein. J. Mol. Biol. 275:465-481. [DOI] [PubMed] [Google Scholar]
- 20.Liao, X. C., J. Tang, and M. Rosbash. 1993. An enhancer screen identifies a gene that encodes the yeast U1 snRNP A protein: implications for snRNP protein function in pre-mRNA splicing. Genes Dev. 7:419-428. [DOI] [PubMed] [Google Scholar]
- 21.Lu, J., and K. B. Hall. 1995. An RBD that does not bind RNA: NMR secondary structure determination and biochemical properties of the C-terminal RNA binding domain from the human U1A protein. J. Mol. Biol. 247:739-752. [DOI] [PubMed] [Google Scholar]
- 22.Lutz-Freyermuth, C., C. C. Query, and J. D. Keene. 1990. Quantitative determination that one of two potential RNA-binding domains of the A protein component of the U1 small nuclear ribonucleoprotein complex binds with high affinity to stem-loop II of U1 RNA. Proc. Natl. Acad. Sci. USA 87:6393-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagai, K., C. Oubridge, T. H. Jessen, J. Li, and P. R. Evans. 1990. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature 348:515-520. [DOI] [PubMed] [Google Scholar]
- 24.Oubridge, C., N. Ito, P. R. Evans, C. H. Teo, and K. Nagai. 1994. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372:432-438. [DOI] [PubMed] [Google Scholar]
- 25.Perez-Canadillas, J.-M., and G. Varani. 2001. Recent advances in RNA-protein recognition. Curr. Opin. Struct. Biol. 11:53-58. [DOI] [PubMed] [Google Scholar]
- 26.Phillips, C., S. Jung, and S. I. Gunderson. 2001. Regulation of nuclear poly(A) addition controls the expression of immunoglobulin M secretory mRNA. EMBO J. 20:6443-6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scherly, D., W. Boelens, W. J. van Venrooij, N. A. Dathan, J. Hamm, and I. W. Mattaj. 1989. Identification of the RNA binding segment of human U1A protein and definition of its binding site on U1 snRNA. EMBO J. 8:4163-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarn, W. Y., and J. A. Steitz. 1995. Modulation of 5′ splice site choice in pre-messenger RNA by two distinct steps. Proc. Natl. Acad. Sci. USA 92:2504-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Gelder, C. W. G., S. I. Gunderson, E. J. R. Jansen, W. C. Boelens, M. Polycarpou-Schwarz, I. W. Mattaj, and W. J. van Venrooij. 1993. A complex secondary structure in U1A pre-mRNA that binds two molecules of U1A protein is required for regulation of polyadenylation. EMBO J. 12:5191-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Ryk, D. I., and S. Venkatesan. 1999. Real-time kinetics of HIV-1 Rev-Rev response element interactions. J. Biol. Chem. 274:17452-17463. [DOI] [PubMed] [Google Scholar]
- 31.Varani, G., and K. Nagai. 1998. RNA recognition by RNP proteins during RNA processing. Annu. Rev. Biophys. Biomol. Struct. 27:407-445. [DOI] [PubMed] [Google Scholar]
- 32.Varani, L., S. I. Gunderson, I. W. Mattaj, L. E. Kay, D. Neuhaus, and G. Varani. 2000. The NMR structure of the 38 kDa U1A protein-PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein. Nat. Struct. Biol. 7:329-335. [DOI] [PubMed] [Google Scholar]
- 33.Will, C. L., S. Rumpler, J. Klein-Gunnewiek, W. J. van Venrooij, and R. Luhrmann. 1996. In vitro reconstitution of mammalian U1 snRNPs active in splicing: the U1-C protein enhances the formation of early (E) spliceosomal complexes. Nucleic Acids Res. 24:4614-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Will, C. L., and R. Luhrmann. 1997. Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol. 9:320-328. [DOI] [PubMed] [Google Scholar]
- 35.Witherell, G. W., Wu, H.-N., and O. C. Uhlenbeck. 1990. Cooperative binding of R17 coat protein to RNA. Biochemistry 29:11051-11057. [DOI] [PubMed] [Google Scholar]