Abstract
Although it is now well documented that metazoans have evolved general transcription factor (GTF) variants to regulate their complex patterns of gene expression, there is so far no information regarding the existence of specific GTFs in plants. Here we report the characterization of a ubiquitously expressed gene that encodes a bona fide novel transcription factor IIB (TFIIB)-related protein in Arabidopsis thaliana. We have shown that this protein is the founding member of a plant-specific TFIIB-related protein family named pBrp (for plant-specific TFIIB-related protein). Surprisingly, in contrast to common GTFs that are localized in the nucleus, the bulk of pBrp proteins are bound to the cytoplasmic face of the plastid envelope, suggesting an organelle-specific function for this novel class of TFIIB-related protein. We show that pBrp proteins harbor conditional proteolytic signals that can target these proteins for rapid turnover by the proteasome-mediated protein degradation pathway. Interestingly, under conditions of proteasome inhibition, pBrp proteins accumulate in the nucleus. Together, our results suggest a possible involvement of these proteins in an intracellular signaling pathway between plastids and the nucleus. Our data provide the first evidence for an organelle-related evolution of the eukaryotic general transcription machinery.
Eukaryotic multisubunit DNA-dependent RNA polymerases (RNAP) are very sophisticated enzymes. Despite this complexity, these enzymes are not capable of selective promoter recognition and rely on a complex combination of auxiliary factors, collectively known as general transcription factors (GTFs), for transcription initiation to be achieved (57, 65, 83). Among those factors, two evolutionarily conserved GTFs are in the heart of promoter recognition in eukaryotes: the TATA-binding protein (TBP), which is common to the three RNAP systems (34), and a transcription factor B (TFIIB for RNAPII and Brf for RNAPIII) (15, 17, 31, 48). TBP, B-type factors, and RNAP are considered to form the minimal ancestor from which all three nuclear transcription systems arose (10). These factors do play a central role in transcription, since they are sufficient to direct specific transcription by RNAP on either supercoiled or modified DNA templates in vitro (39, 61).
Given the seminal role they play in transcription regulation in eukaryotes, TBP and B-type factors were initially thought to be ubiquitous. However, this notion has been recently challenged by the demonstration that metazoans evolved specific isoforms of GTFs to regulate their intricate patterns of gene expression. These homologs include several TBP-related factors (TRF1 in Drosophila and TLFs in all metazoans) (11, 18), as well as a novel human TFIIB/Brf-related protein (67, 75). Biochemical and in vitro analyses indicated that these proteins are probably involved in gene-selective and/or cell type-specific transcription, such as with tRNA genes for TRF1 (74), snRNA genes for TFIIB/Brf-related protein (67, 75), and genes involved in spermiogenesis for mouse TLF (50, 84). More recently, tissue-specific variants for the GTF TFIIA (60, 77) and for some of the TBP-associated factors (19, 35) were also identified in metazoans, where they are expected to play a role in developmental processes (26, 35). Thus, it might be speculated that the appearance of specific isoforms of GTFs during evolution provided to the eukaryotic cell a greater flexibility toward establishing energy-consuming cellular functions and potential for more sophisticated regulation of gene expression.
An important step in eukaryotic cell evolution has been the emergence of the plant cell lineage. Although they represent contrasting life forms, plants and animals were derived from a universal eukaryotic ancestor (30). The acquisition by the universal ancestor of a specific cytoplasmic organelle, the plastid, has been the crucial event that gave rise to the plant lineage. The plastid was derived from prokaryotes (cyanobacteria), whose fate became linked to the eukaryotic ancestor host (29, 30). Consistent with their prokaryotic origin, plastids have retained remnants of eubacterial DNA, known as plastid genomes (72). However, of the several thousand proteins required to build up plastids, fewer than 200 are still encoded by the plastid genomes. During plant cell evolution, the plastid genome size was strongly reduced, either by loss of organelle genes or by their translocation to the host nucleus (51). As a result of this massive gene transfer, plastid development is now strongly dependent on nuclear gene expression (8, 71) and needs tight coordination between nuclear and plastid gene expression. However, from the functional standpoint, it was considered that plastid acquisition occurred without profoundly affecting the host cell genetic systems, since the basic components of the eukaryotic-type translation and transcription apparatuses are well conserved between higher plants and animals (7, 14, 28, 45-47). This assumption has to be reconsidered now, since the recent completion of the Arabidopsis thaliana genome sequence revealed genes coding for putative largest and second-largest subunits of a novel eukaryotic-type RNA polymerase (3). This observation motivated us to search the Arabidopsis database for the presence of a putative GTF variant(s) that might have evolved to fulfill plant-specific functions.
Except for the presence of duplicated TBP genes (12, 28), which encode functionally indistinguishable TBP proteins (32, 78), there has been so far no information regarding the existence in plants of GTF isoforms. In order to address this point, we have searched the Arabidopsis genome sequence for TBP and/or B-type GTF variants. We did not find TBP variants other than the two previously described paralog proteins, but we found a novel gene related to TFIIB/Brf in the Arabidopsis genome. Its full-length cDNA was cloned and sequenced, and we demonstrated that the corresponding protein represents the founding member of a bona fide plant-specific TFIIB-related protein family (hereafter referred to as plant-specific TFIIB-related protein [pBrp]). The bulk of pBrp proteins localize to the cytosolic face of plastid envelope, suggesting that the existence of this novel TFIIB-related factor is intriguingly related to the presence of this organelle in plant cells. The molecular characteristics of pBrp as well as its localization are consistent with the hypothesis that this novel TFIIB-related protein represents a transcription factor involved in intercompartmental signal-induced communication between plastids and the nucleus. Thus, land plant evolution seems to be characterized not only by a massive transfer of cyanobacterial plastid-origin genes to the nucleus. The nuclear transcription machinery itself evolved a specific TFIIB-related factor in order to fulfill the requirements for plastid acquisition, indicating an organelle-dependent evolution of the eukaryotic transcription machinery.
MATERIALS AND METHODS
Methods in bioinformatics and molecular biology.
Gene searching was done on the Arabidopsis genome sequence database or the nonredundant databases by using the TBLASTN program (2) with the amino acid sequences of human TFIIB (31) or A. thaliana pBrp as query sequences. For cloning of the A. thaliana PBRP full-length cDNA, PCR was performed on an Arabidopsis cDNA library by using the primers 5′-GCGGTTTGTTTACTCTTGTAG-3′ and 5′-GCCCAAGATACACACAAAGAC-3′. The spinach and maize pBrp orthologs were isolated by screening of λZap libraries (Clontech, Palo Alto, Calif.), using a labeled DNA fragment corresponding to the core domain region as the probe. The DNA probe was labeled with [α32P]dATP by using a nick translation kit (New England Biolabs). Prehybridization was performed in a solution of 6× SSPE (0.1% sodium dodecyl sulfate [SDS], 0.02% polyvinylpyrrolidone, 0.02% Ficoll, 50 μg of salmon sperm DNA per ml), and hybridization was performed in a solution of 3× SSPE at 55°C for 18 h. The filters were washed three times with 2× SSPE-0.1% SDS for 10 min at 55°C and three times with 0.1% SSPE-0.1% SDS for 10 min at 55°C. Several positives were obtained and were ligated into the plasmid pBluescript II KS(−) (Stratagene), cloned by transformation into strain XL1-Blue (Stratagene), and sequenced. Phylogenetic studies were performed with the core domain sequence comparison as described previously (25). Secondary-structure predictions were performed as described previously (20, 27). Total RNA was extracted from different frozen tissues as described previously (54). For Northern analysis, RNAs were separated on formaldehyde gels and blotted on nitrocellulose membranes (Schleicher & Schuell) according to the supplier's protocol. High-stringency hybridization was performed by following the manufacturer's protocol.
Protein isolation and gel retardation assays.
To produce tagged recombinant proteins in Escherichia coli, the open reading frames of A. thaliana pBrp and A. thaliana TFIIB2 were amplified by PCR with primers designed from the corresponding cDNA sequences. The Arabidopsis TFIIB2 cDNA was PCR amplified with the primers 5′-CGCGGATCCATGTCGGATGCGTATTGTACGG-3′ and 5′-GCTCTAGAAGGACTTGACAGGTTTTTCAGAT-3′. All primers used for PCR amplification contained a BamHI (5′ primer) and a SalI (3′ primer) restriction site, respectively, so the amplified fragments could be cloned in frame with a His6 tag into the BamHI and SalI sites of the pQE30 plasmid (Qiagen). All recombinant proteins were expressed in E. coli strain BL21 (Novagen). Bacteria were transformed with the corresponding vectors and grown in Luria-Bertani medium at 37°C to an optical density at 600 nm of ∼0.6, the production of recombinant proteins was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and the bacteria were grown for an additional 3 h at 37°C. Bacteria were harvested by centrifugation and lysed by mild sonication at 4°C in 20 mM HEPES (pH 7.9)-500 mM KCl-0.1% (vol/vol) Nonidet P-40-10% (vol/vol) glycerol-5 mM imidazole-0.1 mM phenylmethylsulfonyl fluoride (PMSF). The lysates were cleared by centrifugation and loaded onto a 0.5-ml column of nitrilotriacetic acid-agarose containing immobilized Ni2+ ions (Qiagen). The column was washed with 10 column volumes of lysis buffer supplemented with 20 mM imidazole and eluted with a 5-ml linear gradient of imidazole (20 to 300 mM) in lysis buffer. Fractions containing the recombinant proteins were pooled and dialyzed against 20 mM HEPES (pH 7.9)-100 mM KCl-10% (vol/vol) glycerol-1 mM dithiothreitol-0.1 mM EDTA-0.1 mM PMSF. Electrophoretic mobility shift DNA-binding experiments were performed in saturating amounts of bovine serum albumin as described previously (49). The protein components (25 nM A. thaliana TBP2, 40 nM human TFIIB, 50 nM A. thaliana TFIIB2, and 40 nM A. thaliana pBrp), as indicated in the figure legends, were incubated with 1 nM labeled DNA probe containing the adenovirus major late promoter (positions −40 to +20 relative to the transcription initiation site) for 30 min at 30°C. The complexes formed were then separated by electrophoresis through a 4.5% polyacrylamide gel containing Tris-borate-EDTA buffer supplemented with 4% glycerol. Electrophoresis was performed at 100 V until the bromophenol blue dye reached the bottom of the gel. The gels were dried and exposed overnight to films (Kodak).
Antibodies and Western blotting.
A rabbit antiserum was raised against the purified A. thaliana pBrp protein according to the protocol of the manufacturer (Eurogentec). A 1:3,000 dilution of polyclonal anti-pBrp antibodies was used to perform Western blot analysis of the indicated protein extracts. Dilutions of 1:25,000, 1:50,000, 1:2,000, and 1:3,000 of anti-OE24 (38), anti-IE37 (38), anti-L4 (76), and anti-SIG3 (62) polyclonal antibodies, respectively, were used to perform Western blot analysis of the protein extracts. A 1:500 dilution of 8WG16 anti-RPB1 monoclonal antibody was used. The goat anti-rabbit and anti-mouse (Bio-Rad) secondary antibodies were used to detect the proteins with a chemiluminescence detection system (ECL; Amersham Pharmacia Biotech Inc).
Transient and stable expression in Arabidopsis and confocal microscopy analysis.
To insert the A. thaliana PBRP promoter in front of the uidA (β-glucuronidase [GUS]) reporter gene, a 1,053-bp PCR-amplified PstI-XbaI promoter fragment (positions −1053 to −35 relative to the translation initiation codon) was ligated into the PstI-XbaI sites of the promoterless pBI101 plasmid (Clontech). To make the A. thaliana PBRP:EYFP (enhanced yellow fluorescent protein) gene fusion constructs, an A. thaliana PBRP full-length cDNA (devoid of the stop termination codon) was amplified with primers containing a BamHI (5′ primer) and a SalI (3′ primer) restriction site. The PCR product was ligated in EcoRV-digested pBluescript II KS(−) (Stratagene) and was controlled for point mutations by sequencing. To produce the final gene construct, a BamHI-XbaI partial digest containing A. thaliana PBRP cDNA was ligated in frame with the XbaI-SacI EYFP gene (Clontech) into the BamHI-SacI sites of the binary vector derived from plasmid pBI121 (Clontech). To clone the Nt-PBRP:EYFP gene fusion construct, an N-terminal A. thaliana PBRP cDNA fragment (corresponding to A. thaliana pBrp amino acids 1 to 150) was amplified with primers containing a BamHI (5′ primer) and a SalI (3′ primer) restriction site and was ligated in EcoRV-digested pBluescript II KS(−) (Stratagene) and controlled by sequencing. The gene fusion construct was then ligated into the binary vector by using a similar strategy. All binary vectors were transferred into Agrobacterium tumefaciens cells by standard procedures, and stable transformation was performed as previously described (9). GUS staining was performed on stable transformants as previously described (37). For the transient transformation, A. thaliana wild-type or fus6 mutant (Nottingham Arabidopsis Stock Centre, Nottingham, United Kingdom) (16) seeds were grown in liquid culture in MS medium for 2 days. Just after emergence and greening of the cotyledons, wild-type or mutant seedlings were exposed to the transformed A. tumefaciens for 30 min. Seedlings were then dried, and the coculture was left for 2 days in the light under standard growth conditions. Transiently transformed seedlings were analyzed by confocal laser scanning microscopy, using a Leica TCS-SP2 operating system. Green fluorescent protein (GFP) and chlorophyll fluorescences were excited and collected sequentially (400 Hz, line by line) by using the 488-nm line of an argon laser for GFP excitation and the 633-nm line of a helium-neon laser for chlorophyll excitation. Fluorescence emission was collected from 500 to 535 nm for GFP and from 643 to 720 nm for chlorophyll.
Plant cell fractionation.
Intact spinach chloroplasts were prepared by homogenization of 1 kg of leaves from young plants in 3 liters of GR medium (0.33 M sorbitol, 2 mM EDTA, 1 mM MnCl2, 1 mM MgCl2, 1 mM tetrasodium pyrophosphate, 0.1% defatted bovine serum albumin, 50 mM HEPES-NaOH [pH 6.8]) as described previously (5). The leaves were homogenized in a Waring blender. All of the steps were carried out in a cold room. The filtered homogenate was centrifuged for 10 min at 6,300 × g (on a JA-10 rotor [Beckman]), and the crude chloroplast pellet was purified further by isopycnic centrifugation onto a Percoll gradient (22). The intact chloroplasts were then used for immunochemical assays or subfractionation and protease treatment. Intact Arabidopsis chloroplasts were purified as described previously (43). Nongreen plastids from cauliflower buds (Brassica oleracea) were purified onto Percoll gradients as described previously (1). Intact nuclei from either Arabidopsis and spinach leaves or Arabidopsis suspension culture cells were purified as described previously (80). The stroma, envelopes, and thylakoids were purified from lysed chloroplasts by centrifugation through a sucrose step gradient as described previously (21). The hypotonic medium as well as the different sucrose layers contained the protease inhibitors 1 mM PMSF, 5 mM ɛ-amino caproic acid, 1 mM benzamidine HCl, and 1% general protease inhibitor cocktail (Sigma). Protease treatment of isolated intact spinach chloroplasts was performed as described previously (38). Thermolysin from Bacillus thermoproteolyticus (Calbiochem) was used to probe envelope polypeptides accessible from the cytosolic face of the outer envelope membrane. Intact chloroplasts (1 mg of chlorophyll per ml) were incubated for 30 min at 4°C with increasing amounts of the protease (from 0, 10, 50, 200, and 400 μg/ml). The digestion was terminated by the addition of EGTA, and the treated chloroplasts were purified again through a Percoll gradient containing EGTA. The treated, intact chloroplasts were recovered and were analyzed by Western blotting for the presence of pBrp, OE24, and L4 proteins. For agglutination assays, 15 μl of chloroplast suspension (0.1 mg of chlorophyll per ml) was incubated on a glass slide with 50 μl of medium and either preimmune serum or anti-pBrp antibodies diluted to 1/100 (38).
Cell-free degradation assay and inhibitor treatment.
Leaf cell extract was prepared as described previously (59). For these experiments, 1-week-old light-grown Arabidopsis seedlings were ground in liquid nitrogen and resuspended in degradation buffer (25 mM Tris [pH 7.5], 10 mM MgCl2, 5 mM dithiothreitol, 10 mM NaCl, and 10 mM ATP). Cell debris was removed by rapid centrifugation at 10,000 × g, and equal amounts of extract (supernatant) were transferred to individual tubes and incubated at room temperature for the indicated time. For inhibitor studies, cell extract was first incubated in the presence of the respective inhibitors for 1 h before the addition of the recombinant proteins. The recombinant proteins were detected by Western blotting with an anti-GFP monoclonal antibody (Invitrogen). The cell extract was then incubated for the time indicated, and the reaction was terminated by adding an equal volume of 2× gel loading buffer. For inhibitor studies on whole Arabidopsis plants, transfected seedlings were vacuum infiltrated for 5 min with either 100 μM MG132 or the corresponding inhibitor solvent (2% dimethyl sulfoxide [DMSO]) and were left in culture for 3 h before analysis by confocal laser scanning microscopy. For inhibitor studies on Arabidopsis suspension cells, cell cultures were subcultured for 3 h in a medium containing either MG132 (100 μM) or solvent DMSO before the cells were recovered by centrifugation and the nuclei were purified as described previously (80). The samples were then analyzed by Western blotting with the anti-pBrp, anti-RPB1, anti-IE37, and anti-LSU antibodies.
Nucleotide sequence accession number.
The accession number of the A. thaliana PBRP cDNA is AJ295068.
RESULTS
Characterization of a bona fide novel TFIIB-related protein in A. thaliana.
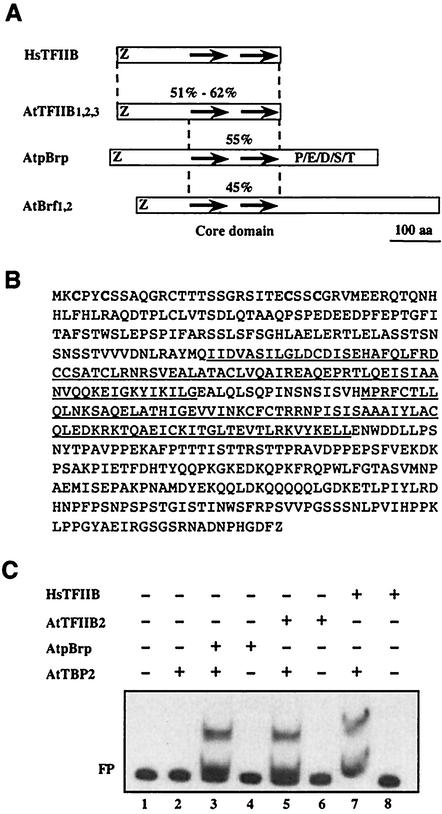
We searched the A. thaliana database for the presence of both TBP- and B-type GTF variants. We could not identify any TBP-related factor in the Arabidopsis genome other than the two paralogs previously identified (28), thus indicating that plants do not harbor any TBP-related protein. In contrast, using human TFIIB as the query sequence, we identified several genes coding for putative TFIIB-related proteins in the Arabidopsis genome (Fig. 1A). In our search, we selected only genes encoding putative proteins containing an amino-terminal zinc ribbon and two 80-amino-acid imperfect direct repeats (Fig. 1A), two structural features of B-type factors. Besides duplicated genes coding for TFIIB (exhibiting 51 to 62% similarity with the entire human TFIIB sequence) (A. thaliana TFIIB1, -2, and -3 [At2g41630, At3g10330, and At3g29380.1, respectively]) (Fig. 1A) (7) and Brf (exhibiting 45% similarity with the human TFIIB core domain sequence) (A. thaliana Brf1 and -2 [At2g45100 and At3g09360) (Fig. 1A) proteins, we identified one additional gene coding for a novel putative Arabidopsis TFIIB-related protein of 503 amino acids (exhibiting 55% similarity with the human TFIIB core domain sequence) (A. thaliana pBrp [At4g36655]). We named it A. thaliana pBrp for A. thaliana plant-specific TFIIB-related protein (Fig. 1A). Sequence analysis of the putative pBrp protein indicated that, except for the zinc ribbon, its amino-terminal domain shared no homology with the corresponding region of the TFIIB or Brf proteins (see Fig. 3B). This suggests that A. thaliana pBrp might represent a novel TFIIB-related protein in Arabidopsis. Its characterization is presented in this work.
FIG. 1.
Characterization of a novel TFIIB-related protein in A. thaliana. (A) Schematic representation of A. thaliana TFIIB (AtTFIIB)-related proteins. Percentages refer to identities between human TFIIB (HsTFIIB) and Arabidopsis TFIIB, Brf, and pBrp proteins. The putative zinc ribbon domain, the imperfect direct repeats, and the carboxy-terminal P/E/D/S/T-rich domain are indicated. (B) Primary sequence of A. thaliana pBrp. The metal-coordinating cysteine residues of the putative zinc ribbon domain are in boldface. The two imperfect direct repeats are underlined. (C) A. thaliana pBrp can substitute for human TFIIB or A. thaliana TFIIB2 in stabilizing the interaction between A. thaliana TBP2 and the adenovirus major late promoter in gel shift assay. Additions to each reaction mixture are listed above each lane. FP, free probe.
FIG. 3.
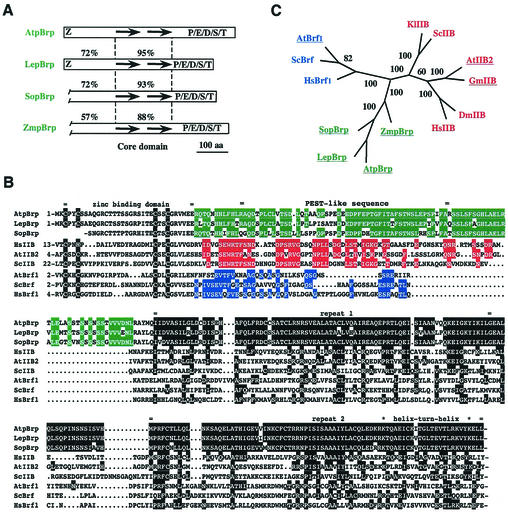
The pBrp proteins define a plant-specific TFIIB-related protein family. (A) Schematic representation of the pBrp protein family. Percentages refer to identities between A. thaliana pBrp (AtpBrp) and L. esculentum pBrp (LepBrp), S. oleracea pBrp (SopBrp), and Z. mays pBrp (ZmpBrp) in the TFIIB-like regions of these factors. The putative zinc ribbon domain, the imperfect direct repeats, and the carboxy-terminal P/E/D/S/T-rich domain are indicated. aa, amino acids. (B) Alignment of pBrps and related factors. Residues of related proteins that are identical to those of the A. thaliana pBrp core domain are highlighted in black. Conserved residues within the specific amino-terminal region are highlighted in green (pBrps), red (TFIIBs), and blue (Brfs). Locations of the zinc-binding domain, amino-terminal putative PEST-like sequence, direct repeats, and putative helix-turn-helix motif are indicated. ScIIB, Saccharomyces cerevisiae TFIIB; HsBrf1, Homo sapiens Brf1. (C) Evolutionary relationships between pBrps and related factors. The unrooted phylogenetic tree was inferred from the core domain alignment. Numbers refer to the score of bootstrap trials (out of 100) which support the branching. Abbreviations: DmIIB, Drosophila melanogaster TFIIB; GmIIB, Glycine max TFIIB; KlIIB, Kluyveromyces lactis TFIIB.
The corresponding A. thaliana PBRP annotated gene contains two introns and is located on the chromosome 4 (53). The annotation and expression of the A. thaliana PBRP gene was confirmed by the cloning and sequencing of its corresponding cDNA (Fig. 1B). Like other B-type factors, A. thaliana pBrp exhibits a putative amino-terminal metal-binding domain and a central core domain consisting of two imperfect direct repeats (Fig. 1B). However, in contrast to TFIIBs, A. thaliana pBrp harbors an additional carboxy-terminal domain that shows no significant homology to other known proteins, other than a noticeable high content of residues usually enriched in PEST-type sequences (P/E/D/S/T domain) (Fig. 1A).
One functional characteristic of B-type factors is their ability to bind TBP and form a stable ternary complex on TATA-containing promoters. We first tested whether A. thaliana pBrp could indeed bind A. thaliana TBP2 on promoter DNA. To do so, we performed gel shift experiments (49) using both human TFIIB (31) and A. thaliana TFIIB2 (7) recombinant proteins as positive controls. Consistent with previous reports, recombinant A. thaliana TBP2 bound very poorly to the promoter, while the further addition of either A. thaliana TFIIB2 or human TFIIB protein to the reaction mixture resulted in a stable ternary complex (Fig. 1C, compare lane 2 to lanes 5 and 7). Likewise, recombinant A. thaliana pBrp was able to form a stable complex with A. thaliana TBP2 (Fig. 1C, compare lane 2 to lane 3). Neither A. thaliana pBrp nor TFIIBs produced a stable shift in the absence of A. thaliana TBP2 (Fig. 1C, compare lanes 3, 5, and 7 to lanes 4, 6, and 8). This result is consistent with the sequence analysis of A. thaliana pBrp, which indicates the existence of a significant conservation at residues that are critical for binding to the A. thaliana TBP2-DNA binary complex in human TFIIB (of the 33 residues involved, 22 either are identical or represent conservative changes in A. thaliana pBrp) (56). In view of the similarity to B-type factors and the interaction with A. thaliana TBP2, we conclude that A. thaliana pBrp is a bona fide novel TFIIB-related protein in A. thaliana.
Arabidopsis PBRP is ubiquitously expressed.
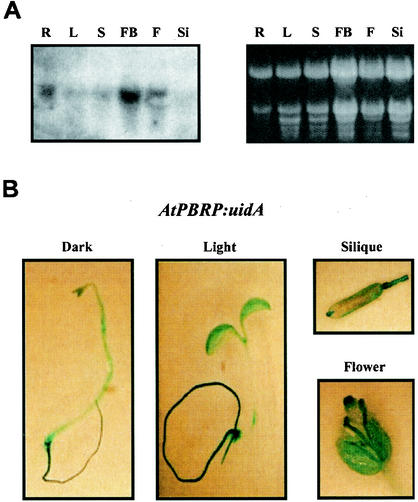
To determine the distribution pattern for A. thaliana PBRP messenger in Arabidopsis organs and tissues, we performed Northern blotting experiments with a cDNA probe that specifically hybridized to the coding region of the A. thaliana PBRP messenger. These analyses revealed a single transcript of 1.8 kb, which corresponds to the estimated size of the Arabidopsis cDNA clone. Northern analyses demonstrated that the A. thaliana PBRP messenger is ubiquitously expressed, although quantitative differences exist between different organs, with a higher level of expression in roots and a lower level in siliques (Fig. 2A). The expression of the A. thaliana PBRP gene was also found to be independent of the presence of light (data not shown). A. thaliana PBRP expression studies were extended by analyzing an A. thaliana PBRP-promoter:uidA (GUS) reporter gene fusion in Arabidopsis transgenic seedlings during plant development. Histochemical analysis of GUS expression in transgenic plants confirmed that A. thaliana PBRP is ubiquitously expressed in cotyledons, hypocotyls, and roots of both light- and dark-grown plants (Fig. 2B). Expression of the reporter gene was also observed in mature leaves (data not shown), flowers, and siliques (Fig. 2B). Taken together, these results indicated that A. thaliana PBRP is a constitutively expressed gene.
FIG. 2.
Arabidopsis PBRP is ubiquitously expressed. (A) Northern blot analysis of total RNA (∼20 μg) prepared from A. thaliana roots (lanes R), leaves (lanes L), stems (lanes S), flower buds (lanes FB), flowers (lanes F), and siliques (lanes Si) with PBRP cDNA as a probe. Ethidium bromide staining of the corresponding denaturing gel is shown as loading control. (B) Arabidopsis PBRP is ubiquitously expressed as revealed by GUS reporter expression. Transgenic seedlings expressing the GUS reporter gene under the control of A. thaliana PBRP (AtPBRP) promoter were grown for 1 week under light or dark conditions and then stained for GUS activity. Flowers and siliques from mature plants were also analyzed by histochemical assay.
pBrp polypeptides define a plant-specific TFIIB-related protein family.
To check whether pBrp-like proteins are present in other species, we performed a search of the nonredundant database with A. thaliana pBrp as the query sequence. Our screening identified a putative nuclear gene from tomato (Lycopersicum esculentum PBRP, gb/AAG01118.1) and several expressed sequence tags from diverse plant species, whose deduced protein sequences showed strong identity to A. thaliana pBrp. This result suggested that pBrp-like polypeptides might be widely expressed among plant species. Next, we isolated several partial clones from different cDNA libraries by low-stringency DNA hybridization. We identified A. thaliana pBrp orthologs from the dicotyledon Spinacia oleracea (accession number AJ295069) and the monocotyledon Zea mays (accession number AJ295070). As expected for bona fide orthologs, all identified pBrp proteins share a very high level of sequence identity throughout their TFIIB-like region, with values ranging from 60 to 95% (Fig. 3A). Although varying in size and primary sequence, the putative carboxy-terminal PEST-like domain was found to be present in all pBrp proteins identified so far (Fig. 2A), thus suggesting that the conservation of this domain might be functionally significant.
Sequence comparison of pBrps with representative members of the eukaryotic TFIIB and Brf families (Fig. 3B) indicates that pBrp proteins are more related to TFIIB (average of 34% identity) than to Brf (average of 25% identity) proteins in the core domain. This sequence conservation is significant and suggests that the A. thaliana pBrp core domain has the ability to fold into a structure similar to the cyclin box fold found in human TFIIB core domain (6, 56). This assumption is further supported by two other observations: (i) a significant conservation at nonpolar residues that are known to be responsible for the stability of the cyclin box fold domain in human TFIIB (56) and (ii) the α-helical predicted structure of the A. thaliana pBrp core domain, which harbors a putative helix-turn-helix motif (with a significant Dodd and Egan score of 4.72), two structural features of human TFIIB core domain (Fig. 3B) (20, 44, 56). To assess more precisely the relationship between these proteins, the core domain sequence comparison was used to build up an unrooted phylogenetic tree, from which three independent groups of B-type factors could be clearly discerned (Fig. 3C). Besides the expected Brf and TFIIB classes, pBrp factors form a novel group, indicating that plants harbor a third class of B-type factors which most probably arose from a TFIIB-like precursor. From these results, we conclude that pBrp proteins define a TFIIB-related protein family that is specific to plants. This statement is confirmed by the fact that no pBrp orthologs could be detected in the almost entirely sequenced genomes of yeast, C. elegans, Drosophila, and human.
pBrp proteins localize to plastids.
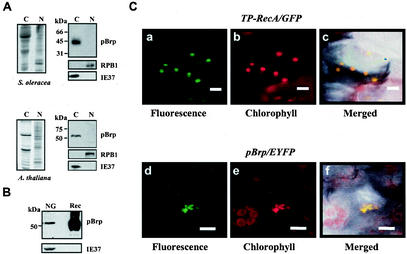
To determine whether pBrp proteins were expressed, a polyclonal rabbit antiserum was raised against the purified A. thaliana pBrp recombinant protein. Serum derived from rabbits immunized with the protein reacted with both Arabidopsis and spinach recombinant pBrp proteins. Preimmune serum demonstrated no reactivity toward these proteins (data not shown). No reactivity of the anti-pBrp serum to either human or Arabidopsis TFIIB recombinant protein was observed, confirming the specificity of the antibodies. The pBrp-specific antiserum was then tested for its ability to recognize a specific polypeptide in plants by Western blotting with protein extracts derived from highly purified spinach and Arabidopsis nuclei and chloroplasts (Fig. 4A). The organelles used in these experiments were purified on Percoll gradients and shown to be devoid of cross-contamination as measured by Western blotting with anti-RPB1 and anti-IE37 control antibodies (Fig. 4A). Moreover, these protein fractions were also found to be devoid of cytoplasmic or mitochondrial proteins (data not shown). In both cases, an immunoreactive protein with a molecular mass identical to that expected for S. oleracea pBrp (∼46 kDa) or A. thaliana pBrp (∼54 kDa) was detected exclusively in the chloroplast fraction (Fig. 4A). The preimmune serum did not detect any proteins of similar mobility either in chloroplasts or in nuclei (data not shown). Next, we tested whether pBrp protein was associated with nongreen amyloplasts purified from cauliflower buds (B. oleracea, a close relative of Arabidopsis) on Percoll gradients. We found that cauliflower bud amyloplasts harbor a ∼54-kDa protein doublet that was reactive to the anti-pBrp antibodies (Fig. 4B). Again, the preimmune serum did not recognize any polypeptides of similar mobility in extracts of nongreen plastids (data not shown). Taken together, these results demonstrate that pBrp proteins are expressed in plants and that they specifically localize to various types of plastids.
FIG. 4.
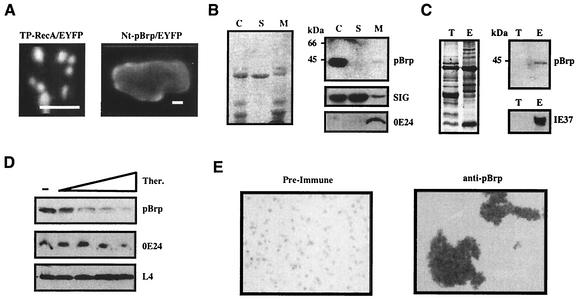
The pBrp proteins localize to plastids. (A) The pBrp proteins localize to leaf chloroplasts. Western analysis of protein extracts (∼20 μg) from S. oleracea (spinach) and A. thaliana (Arabidopsis) chloroplasts (lanes C) and nuclei (lanes N) probed with anti-pBrp, anti-RPB1, and anti-IE37 antibodies is shown on the right. Coomassie blue staining of the corresponding protein fractions is shown on the left. (B) PBrp proteins localize to nongreen plastids from cauliflower buds (NG). Rec, A. thaliana pBrp recombinant protein. (C) A. thaliana pBrp directs chimeric EYFP fusion protein to plastids. Transient expression in cotyledon cells from seedlings expressing either GFP fused to the transit peptide of plastid RecA protein as a plastid-targeted control (a, b, and c) or the A. thaliana pBrp/EYFP fusion protein (d, e, and f) is shown. a and d, GFP/EYFP fluorescence in two representative Arabidopsis cells monitored by confocal laser scanning microscopy; b and e, chlorophyll red autofluorescence; c and f, overlap of GFP/EYFP, chlorophyll, and bright-field images. Bars, 10 μm.
Plastid-associated localization of pBrp proteins was confirmed by transient expression of an A. thaliana pBrp/EYFP fusion protein in Arabidopsis seedlings followed by confocal laser scanning microscopy. As illustrated for an epidermal transformed cell (Fig. 4C, panels d to f), a strong EYFP fluorescence was found to be associated with chlorophyll fluorescence. However, plastids appeared to be aggregated in all cases. This unexpected plastid profile was not due to the transient approach used in this experiment, since the control construct TP-RecA/GFP, in which GFP is fused to the transit peptide sequence of the plastid RecA protein (42), gave rise to fluorescence associated with individual plastids (Fig. 4C, panels a to c). The images shown are representative of those for many leaf cells expressing A. thaliana pBrp/EYFP. In no case was the EYFP fluorescence found to be associated with the nucleus or with any other compartment of the transformed cells. Together, these data demonstrate that pBrp proteins are associated with plastids rather than with nuclei and therefore give a rationale for the presence of this novel class of B-type factors solely in plant lineage.
The bulk of pBrp proteins are kept at the cytosolic face of plastid envelope.
A plastid location for the pBrp proteins was unexpected, since TFIIB-related proteins are anticipated to function in nuclei; indeed, Arabidopsis TBPs, which would be expected to function with A. thaliana pBrp, are detected only in nuclei by Western blotting (data not shown). Moreover, neither an amino-terminal extension nor a hydrophobic domain, which would be required to import these proteins into plastid stroma (40), is detectable in this novel class of TFIIB-related factors. Likewise, computer algorithms give ambiguous results for A. thaliana pBrp localization. While PREDOTAR version 0.5 (http://www.inra.fr/Predotar/) assigns a significant chloroplast targeting score, both CHLOROP (23) and TARGETP (24) do not predict a chloroplast location for A. thaliana pBrp. To address the question of whether A. thaliana pBrp harbors a targeting sequence, the amino-terminal domain of this protein was fused to EYFP protein (Nt-pBrp/EYFP) and its targeting properties were tested in transiently transformed Arabidopsis seedlings. As a positive control, similar transformation experiments were performed with the Tp-RecA/GFP control construct (42). In Arabidopsis cells transformed with the control construct, the RecA transit sequence directed GFP efficiently to plastids (Fig. 5A, left panel). In contrast, transformation with Nt-pBrp/EYFP resulted in a weaker fluorescence, which was located in the cytosol (Fig. 5A, right panel), therefore confirming the lack of targeting properties of the A. thaliana pBrp amino-terminal domain. How can we explain, then, the observed plastid association of pBrp proteins? To answer this question we have to consider two facts: (i) outer plastid envelope proteins are often devoid of targeting peptide sequence (40), and (ii) GFP-type proteins might dimerize when targeted to a membrane surface (82). Based on these observations, we infer that the observed plastid agglutination (Fig. 4C) could be due to the outer plastid envelope targeting and subsequent dimerization of A. thaliana pBrp/EYFP fusion protein in vivo. To test this hypothesis, we analyzed the suborganellar localization of pBrp proteins. Intact plastids were lysed in an hypotonic medium, and plastid membranes (containing both envelopes and thylakoids) were separated from the soluble stroma fraction by centrifugation. These subfractions were then tested by Western blotting with anti-pBrp antibodies or control antibodies directed against known plastid envelope (OE24 and IE37) and stroma (SIG3 and L4) proteins. Although a rapid decrease of pBrp protein was reproducibly observed upon plastid lysis, this experiment clearly indicated that pBrp proteins are associated with the membrane subfraction (Fig. 5B, compare lanes C and M). The decrease of the signal was not due to a loading artifact as judged by either Coomassie blue staining or Western blotting of the corresponding fractions with both anti-SIG3 and anti-OE24 control antibodies (Fig. 5B). It was rather due to a conditional instability of pBrp proteins revealed upon plastid lysis (see below). Further separation of the plastid membranes into envelopes and thylakoids followed by Western blotting analysis clearly indicated that pBrp proteins are indeed specifically associated with the plastid envelope (Fig. 5C, lanes E).
FIG. 5.
The pBrp proteins are kept in an inactive state at the cytoplasmic surface of plastid envelope. (A) The A. thaliana pBrp amino-terminal domain lacks chloroplast-targeting properties. Subcellular localization of the transiently expressed Tp-RecA/GFP and Nt-pBrp/EYFP fusion proteins was monitored by confocal laser scanning microscopy. Bars, 10 μm. (B) Western blot analysis of protein fractions (∼25 μg) from spinach chloroplasts (lanes C), stroma (lanes S), and membranes (lanes M) with anti-pBrp, anti-SIG3, and anti-OE24 antibodies is shown on the right. OE24 (an outer plastid envelope protein) and SIG (SIG3, a sigma-like transcription factor) were used as membrane and stroma control proteins, respectively. Coomassie blue staining of the corresponding fractions is shown on the left. (C) Western blot analysis of protein fractions (∼25 μg) from thylakoids (lanes T) and envelopes (lanes E) with anti-pBrp and anti-IE37 antibodies. IE37 (an inner plastid envelope protein) was used as an envelope control protein. Coomassie blue staining of the corresponding fractions is shown on the left. (D) Western blot analysis of protein fractions of thermolysin-treated, Percoll-purified, intact spinach plastids with anti-pBrp, anti-OE24, and anti-L4 antibodies. Plastid L4 protein (a stromal ribosomal protein) was used as an intraplastidic control protein. The different lanes correspond to nontreated plastids (−) and plastids treated with increasing concentrations of thermolysin (Ther.) (from 10 to 400 μg/ml). (E) Bright-field images of intact spinach chloroplasts incubated with either preimmune serum (left) or anti-pBrp antibodies (1/100 dilution) (right).
Association of pBrp with the plastid outer envelope was further confirmed by thermolysin digestion of isolated intact chloroplasts. After treatment with various concentrations of thermolysin, treated intact plastids were reisolated on Percoll gradients, and their constituent polypeptides were tested by Western blotting. As shown in Fig. 5D, both pBrp protein and OE24 (an outer envelope constituent 38) were hydrolyzed in a thermolysin-dependent manner. In contrast, the intraplastidic L4 ribosomal protein was not affected by the treatment. This result demonstrates that the bulk of pBrp proteins are located on the cytosolic side of the plastid envelope. The localization of pBrp proteins was further confirmed by an in vitro plastid agglutination test performed as described previously (38). Isolated intact chloroplasts were incubated with either anti-pBrp antibodies or preimmune serum and visualized by using bright-field microscopy. A strong and specific agglutination occurred with anti-pBrp antibodies, thus confirming that antigenic domains of these proteins are exposed to the cytosolic face of the outer plastid envelope (Fig. 5E). Taken together, our results demonstrate that the bulk of pBrp proteins are present at the cytosolic face of the plastid envelope.
pBrp proteins harbor conditional signals for rapid degradation.
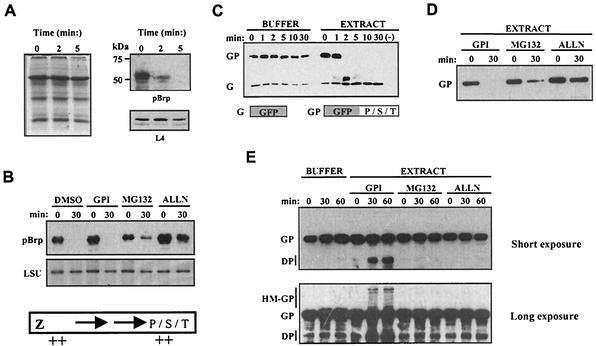
The observed instability of pBrp proteins after plastid lysis raises the question of whether the subcellular concentration of these factors may be regulated at the posttranslational level by controlled proteolysis. To test this hypothesis, we analyzed the intrinsic stability of A. thaliana pBrp by using an Arabidopsis cell-free degradation assay based on leaf cell extract (59). The levels of endogenous A. thaliana pBrp and of other cellular proteins were analyzed at various times after grinding of the leaf tissues in the degradation buffer by means of Western blotting and Coomassie blue staining. As shown in Fig. 6A, we observed a rapid degradation of the endogenous pool of plastid-localized A. thaliana pBrp protein in the extract. This degradation was specific to A. thaliana pBrp, as the endogenous L4 protein and numerous other cellular proteins showed no degradation in the extract over the same time period (Fig. 6A) and even after a much longer reaction time (data not shown). This experiment shows that the pBrp proteins are rapidly degraded if the plant cell integrity is disrupted. In order to characterize the proteolytic activity involved in A. thaliana pBrp degradation, we assayed several general classes of protease inhibitors in the degradation assay. While the addition of DMSO (the solvent for the inhibitors) or a general protease inhibitor cocktail (containing serine, aspartate, and cysteine protease inhibitors) had no effect, the addition of two proteasome inhibitors (ALLN and MG132) could partially prevent the degradation of A. thaliana pBrp in our cell-free system (Fig. 6B).
FIG. 6.
pBrp proteins harbor conditional signals for rapid degradation. (A) Cell-free degradation assay of the endogenous A. thaliana pBrp protein. Leaf extracts were resuspended in buffer supporting the proteasome degradation pathway. After the indicated times, the assay was stopped by addition of protein loading buffer and the protein extracts were analyzed by Western blotting with anti-pBrp and anti-L4 antibodies. Coomassie blue staining showing the global cellular protein profile is shown on the left. (B) Cell-free degradation assay of A. thaliana pBrp in the presence of various protease inhibitors. Extracts were treated with 2% DMSO (solvent for the inhibitors), 2% protease inhibitor cocktail (GPI) (Sigma), or a 40 μM concentration of the proteasome inhibitor MG132 or ALLN. After the times indicated, A. thaliana pBrp levels were examined by Western blotting. The large subunit of Rubisco (LSU) that showed no degradation in our cell-free assay is shown as loading control. Bottom panel, schematic diagram of the pBrp proteins. Relative positions of the predicted PEST-like sequences are indicated by ++. (C) Sensitivity to cell-free degradation of both GFP (G) and GFP/PST (GP) fusion proteins. The P/S/T domain refers to A. thaliana pBrp amino acids 311 to 503. Both GFP and GFP/PST recombinant proteins were incubated in buffer alone or in leaf extract for the indicated times and analyzed by Western blotting with an anti-GFP monoclonal antibody. No recombinant proteins were added in the leaf extract (−). (D) Cell-free degradation of GFP/PST was performed in cell extracts that were pretreated with various types of protease inhibitors, including the general cocktail inhibitor (GPI) or the specific proteasome inhibitors (MG132 and ALLN), using the same concentration as in panel B. After 0 and 30 min of incubation, GFP/PST protein was analyzed by Western blotting with an anti-GFP monoclonal antibody. (E) Cell-free degradation in conditions under which protease activity is limiting revealed the presence of high-molecular-weight GFP/PST intermediates that are reminiscent of ubiquitin conjugates. GFP/PST recombinant protein was incubated in buffer alone or in leaf extracts pretreated with protease inhibitors as indicated. After 0, 30, and 60 min, the recombinant protein was analyzed by Western blotting with an anti-GFP monoclonal antibody. DP, degradation product intermediates; HM-GP, high-molecular-weight GFP/PST intermediates.
To identify features of pBrp proteins that might mediate their specific degradation, we applied their primary sequences to the PEST-Find algorithm (63), and we identified several evolutionarily conserved putative PEST-like sequences in these proteins (see Fig. 6B, bottom panel, for a schematic representation). A strong PEST-like sequence with a significant score of 8.12 (S. oleracea pBrp) or 5.17 (A. thaliana pBrp) was identified in the flexible linker region that separates the amino-terminal zinc ribbon domain from the central core domain (Fig. 3B). A second group of putative PEST-like sequences that score up to 9.5 was found in the carboxy-terminal domain. Thus, the carboxy-terminal domain of pBrp might be functionally relevant for protein degradation and/or turnover. Due to their location in the protein, these sequences are likely to be solvent exposed. Interestingly, no PEST-like sequences could be detected in either human TFIIB or the plant TFIIB factors, which are stable proteins; i.e., the existence of PEST sequences seems to correlate with conditional instability in pBrp proteins.
To ascertain whether the carboxy-terminal extension is a bona fide PEST region that has the ability to promote pBrp destruction, we took advantage of the fact that these degradation signals are usually transplantable modules (63). We generated a chimeric protein in which the A. thaliana pBrp carboxy-terminal domain was fused to GFP, and its relative stability compared to that of GFP alone was tested in the cell-free degradation system (Fig. 6C). While the GFP showed no degradation, the GFP/PST fusion protein displayed a rapid degradation profile similar to that of endogenous A. thaliana pBrp in terms of kinetics (compare Fig. 6A and C). Protein degradation could be inhibited by the addition of proteasome inhibitors such as MG132 and ALLN, while the general protease inhibitor cocktail had no effect on degradation (Fig. 6D). Interestingly, the high-molecular-weight bands that are visible in extracts treated with the general protease inhibitor cocktail after longer exposure might correspond to GFP/PST intermediates, reminiscent of ubiquitin-protein conjugates (Fig. 6E, bottom panel). The appearance of these high-molecular-weight species seems to be correlated with the appearance of GFP/PST degradation products. They are not detectable in extracts treated with MG132 and ALLN proteasome inhibitors (Fig. 6E). Although the exact mechanism of protein degradation has to be elucidated, these experiments demonstrate that pBrp proteins harbor a specific conditional degradation signal(s) that probably targets these proteins for degradation by the proteasome.
Cells altered in proteasome-dependent protein degradation show an accumulation of A. thaliana pBrp in the nucleus.
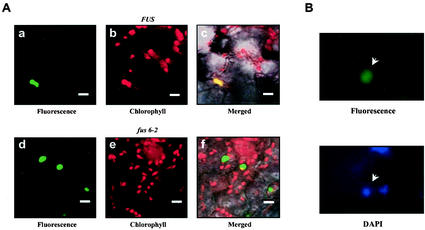
The fact that pBrp proteins harbor conditional proteolytic signals indicates a mechanism of action for these proteins which needs proteasome-controlled degradation. As an attempt to further examine this mechanism of action, we analyzed the intracellular expression level of pBrp proteins in a plant mutant background known to be deficient in regulated ubiquitin-dependent protein degradation. The Arabidopsis fus6 mutant (16, 81), which is known to be deficient in COP9 signalosome (CSN) (70), was transiently transformed by using the A. thaliana pBrp/EYFP fusion construct. CSN is an evolutionarily conserved nuclear-enriched protein complex that controls numerous physiological and developmental processes via the targeted destabilization of transcriptional regulators by the ubiquitin-dependent proteasome-mediated protein degradation pathway (33, 59, 68, 69). Surprisingly, while wild-type seedlings exhibited the plastid-localized fluorescence pattern (Fig. 7A), the fus6 mutant showed strong nuclear fluorescence as judged by DAPI (4′,6′-diamidino-2-phenylindole) staining (Fig. 7B). From these data, we conclude that under conditions of inhibition of proteasome-dependent degradation, pBrp proteins accumulate in the nucleus.
FIG. 7.
Nuclear localization of A. thaliana pBrp/EYFP fusion protein in a fus6 mutant. (A) Localization of the A. thaliana pBrp/EYFP fluorescence in wild-type (FUS6) or CSN mutant (fus6) seedlings. The mutant was selected based on its phenotype and transiently transformed with the A. thaliana PBRP/EYFP construct. Panels a and d show A. thaliana pBrp/EYFP fluorescence in either wild-type (a) or mutant (d) seedlings. Panels b and e show corresponding chlorophyll red autofluorescence. Panels c and f show the overlap of EYFP, chlorophyll, and bright-field confocal images. Bars, 10 μm. (B) Nuclear localization of the A. thaliana pBrp/EYFP fluorescence in the transformed fus6 mutant cells. The EYFP fluorescence was observed in the DAPI-stained nucleus.
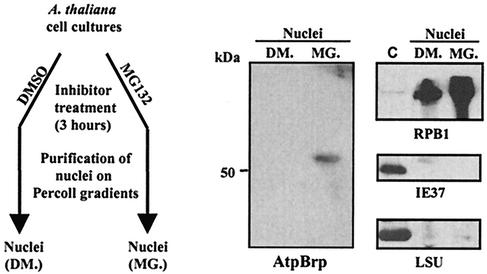
To confirm that the nuclear accumulation of A. thaliana pBrp/EYFP in CSN-deficient plants accurately reflects control of the endogenous factor, we assessed the level of A. thaliana pBrp in nuclei purified from Arabidopsis cells after treatment with an inhibitor of the proteasome or its solvent as control. Arabidopsis cells were preincubated with either MG132 or solvent (DMSO) for 3 h, and their nuclei were purified on Percoll gradients and analyzed by Western blotting with anti-pBrp and control antibodies (Fig. 8). While the level of A. thaliana pBrp protein in whole-cell extracts was not affected by the treatment (data not shown), a clear accumulation of A. thaliana pBrp could be detected in nuclei purified from MG132-treated cells (Fig. 8). No A. thaliana pBrp was present in the nuclei of DMSO-treated cells (Fig. 8). The presence of pBrp proteins in the nucleus was not due to cross-contamination of the nuclei with the plastid fraction as judged by control Western blottings performed with anti-RPB1, anti-IE37, and anti-LSU antibodies (Fig. 8). Taken together, these data suggest that pBrp function is controlled by nucleocytoplasmic partitioning, a process which is itself controlled by a proteasome-dependent protein degradation process.
FIG. 8.
Nuclear localization of endogenous A. thaliana pBrp protein in nuclei of Arabidopsis cells treated with proteasome inhibitor. Arabidopsis cells were preincubated with either 100 μM MG132 (MG.) or 2% DMSO (DM.) for 3h, and their nuclei were purified on Percoll gradients and analyzed by Western blotting with anti-pBrp, anti-RPB1, anti-IE37, and anti-LSU antibodies. A plastid fraction from leaves was used as a control (lane C).
DISCUSSION
The recent identification of tissue-specific and/or gene-selective GTFs and coactivators revealed variations in the components of the preinitiation complex as a means for more sophisticated metazoan gene regulation (11, 18, 19, 26, 35, 50, 60, 67, 74, 75, 77, 84). In the present study, we addressed the question of whether the plant lineage also evolved variants of the general transcription machinery to fulfill plant-specific functions. Searching of the A. thaliana genome with TBP and TFIIB queries showed that this plant does not express any TBP-related factors except the two previously characterized TBP proteins (TBP1 and TBP2) (28). The absence of additional TBP-related proteins in higher plants was further supported by a more systematic analysis of the available plant expressed sequence tag databases and the almost complete rice genome sequence. Hence, there is a strong probability that TBP may be a real universal GTF in higher plants.
In contrast to the case for TBP, we detected several genes coding for B-type GTFs in the Arabidopsis genome. We show that despite the apparent complexity of this gene family, plant B-type factors can be classified into three distinct groups. Besides the duplicated genes coding for the expected TFIIB- and Brf-type factors, whose presence emphasizes the high conservation of the RNAP II and III transcription systems in Arabidopsis (3), Arabidopsis expresses a novel TFIIB-related protein (pBrp) that is encoded by a single gene. A. thaliana pBrp contains sequence features that are characteristic of all members of the TFIIB/Brf/TFB protein families, including an amino-terminal metal-binding domain and a central core domain consisting of two imperfect direct repeats. In particular, Arabidopsis pBrp exhibits 35% identity with the conserved bipartite core domain of human TFIIB, which is known to be important for TBP binding (56) and interaction with DNA (44, 56). Likewise, a helix-turn-helix motif is also present at the carboxy terminus of the core domain region of pBrp proteins, suggesting that these proteins can engage in sequence-specific contact with the core promoter region. Consistent with the observation that TBP-binding residues are well conserved in A. thaliana pBrp, we show that this protein can form a stable complex with A. thaliana TBP2 on promoter DNA. Together, these data suggest that pBrp proteins probably interact with TBP and DNA in transcription initiation, thus representing the first example of the existence of a GTF variant in plants. Our results indicate that the strategy of duplicating GTFs had been independently invented in plants and metazoans to fulfill requirements specific to each lineage.
In this work, we provide molecular and phylogenetic evidence that the pBrp proteins represent a TFIIB-related protein family that is specific to plants. From an evolutionary standpoint, two hypothesis concerning the origin of these proteins can be formulated: (i) the pBrp proteins might have evolved early during eukaryotic cell evolution and were kept for still-unknown reasons only in the plant lineage or (ii) the pBrp proteins evolved after or concomitant with the separation of the plant lineage. The intricate relationships between pBrp proteins and plastids, i.e., plant-specific organelles, shown in this paper led us to favor the latter hypothesis. Considering the known cyanobacterial sequence data, we can infer that pBrp proteins are not likely to have been acquired from the eubacterial endosymbiont. It is more likely that pBrp proteins arose as a consequence of plastid acquisition, from the duplication of a preexisting B-type factor already present in the eukaryotic host ancestor. The high identity of the pBrp core domain to the core region of TFIIB factors suggests that the PBRP gene originated from TFIIB or its ancestor gene. So far, pBrp-type proteins have been only found in higher plants, which actually represent the majority of the database plant entries. There is not sufficient sequence information available to conclude whether pBrp-type proteins are also present in lower plants, algae, or even apicomplexa, and the question of whether these proteins are present in all plastid-containing eukaryotes or whether they are specific to higher plants remains open.
The novel TFIIB-related protein family that we have identified is characterized by several intriguing features that suggest an original mode of action for these proteins. First, we describe that the bulk of pBrp proteins are anchored to the outer membrane of the plastid envelope, therefore demonstrating a unique subcellular distribution among the so-far-characterized GTFs. However, in support of a role in nuclear transcription, we have shown that A. thaliana pBrp can interact with plant TBP. From these results, we reason that the cellular partitioning of pBrp proteins must be somehow regulated to allow this functional interaction to happen in the nucleus. Signal-induced change of the location of transcription factors has been already reported: modulation of nuclear gene expression by the regulated activation of cytoplasmic membrane-targeted transcription factors represents an increasingly recognized mechanism of gene regulation in eukaryotes (4, 13, 36, 58, 66). The activation of such membrane-associated factors involves release from the membrane by various signal-induced processes as diverse as proteolysis (13, 36), protein posttranslational modification (4, 58), and membrane modifications (66). In this respect, the specific association of pBrp proteins with the plastid envelope strongly suggests that the activation of these proteins would require either a plastid-originated signal or a signal that is relayed by this organelle. Unfortunately, all of our attempts aimed at identifying a developmental stage or an environmental condition under which these proteins could be detected in the nucleus have failed so far, and therefore the exact nature of this hypothetical signal remains unknown. Independently of the release step, the apparent lack of pBrp in the nucleus might be due to an ongoing degradation of the factor in this organelle. The specific PEST-type conditional degradation sequences that are present at the flexible linker region and the carboxy-terminal end of the protein might be implicated in such a rapid degradation. It is worth noting that proteasome-dependent conditional instability is an hallmark of the activated forms of numerous signaling factors, such as STATs, HSF-2, progesterone receptor, and SREBP (41, 52, 55, 79). Moreover, the CSN has been shown to be involved in the stimulus-dependent proteolytic control of several plant transcriptional regulators, such as HY5, and IAA proteins (59, 68, 69). We have shown here that Arabidopsis cells impaired in proteasome-dependent protein degradation exhibit an accumulation of pBrp in the nucleus. Although the exact mechanism by which the proteasome would control pBrp partitioning remains unclear, the presence of PEST sequences in these proteins together with the fact that the CSN is known to be localized mostly in the nucleus (70) strongly support a model in which the apparent absence of pBrp proteins in the nucleus can be traced to rapid protein turnover. Taken together, our results concerning the molecular characteristics and the cellular compartmentalization of the pBrp proteins are consistent with an involvement of these proteins in signal-induced nuclear gene expression.
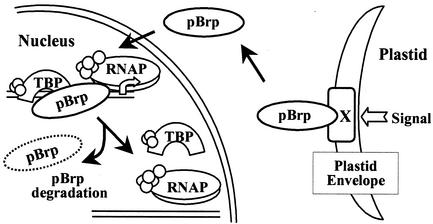
Although it is too early to conclude why plants evolved pBrp proteins, our results suggest that these factors might be involved in a novel intercompartmental regulatory pathway between plastids and the nucleus. In this respect, our results are in line with several reports showing that the expression of numerous nuclear genes encoding both plastid and nonplastid proteins is dependent on the functional status of plastids (64, 73). From these studies, it has been hypothesized that the chloroplast signals the nucleus by using various retrograde signaling pathways (64, 73). All approaches to identify putative components of these retrograde pathways have so far pointed towards plastid-localized proteins involved in porphyrin biosynthesis and transport and metabolites as signaling intermediates. The way by which these intermediates transmit the information on the nuclear transcription machinery remains unclear (64, 73). It is tempting to speculate that pBrp proteins represent one of the relays in the signal transduction pathways transmitting information from the outer plastid envelope to the nucleus by selectively recruiting the nuclear transcription machinery to particular gene promoters. To summarize our results, we present the hypothetical model for pBrp function shown in Fig. 9. In this model, pBrp is sequestered at the outer plastid envelope membrane by interaction either with a lipid that is highly enriched or specific within the plastid envelope membrane or by association with a specific protein partner. A plastid-derived signal(s) provokes the release of pBrp from the envelope membrane, enabling its translocation to the nucleus. In the nucleus, pBrp induces transcription of particular genes, and finally, rapid degradation of pBrp allows the nuclear transcription to be maintained under the control of the release process. Experiments to determine nuclear target genes of pBrp as well as the exact nature of the envelope-released signal are in progress.
FIG. 9.
Hypothetical model of intracellular signaling between plastids and the nucleus through pBrp protein action. The pBrp proteins are sequestered at the outer plastid envelope via interaction with lipid components or by association with an unknown protein (protein X). Interaction with the plastid envelope allows the pBrp proteins to act as sensor/regulators that monitor plastid status and transmit this information to the nucleus, where they can modulate nuclear gene expression by recruiting RNAP machinery. Finally, proteasome-dependent rapid degradation of pBrp may allow the nuclear transcription to be turned off rapidly once the release process is inhibited.
Acknowledgments
Thierry Lagrange and Mohamed-Ali Hakimi contributed equally to this work.
We thank Helene Pesey for excellent assistance, Jean-François Briat and Catherine Curie (Montpellier, France) for providing the maize library, Dia-Xao Zhou (Orsay, France) for providing the A. thaliana TBP2 vector, Danny Reinberg (Piscataway, N.J.) for providing anti-RPB1 monoclonal antibody and human TFIIB vector, R. E. Hanson for providing the plastid-targeted RecA/GFP vector, and Denis Falconet and Gerard Clabault for computer assistance.
This work was supported by the CNRS and by NSF grants to Eric Lam.
REFERENCES
- 1.Alban, C., J. Joyard, and R. Douce. 1988. Preparation and characterization of envelope membranes from nongreen plastids. Plant Physiol. 88:709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., L. D. Thomas, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipmann. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796-815. [DOI] [PubMed] [Google Scholar]
- 4.Attisano, L., and J. L. Wrana. 2002. Signal transduction by the TGF-β superfamily. Science 296:1646-1647. [DOI] [PubMed] [Google Scholar]
- 5.Baeza, L., A. Bertrand, R. Mache, and S. Lerbs-Mache. 1991. Characterization of a protein binding sequence in the promoter region of the 16S rRNA gene of the spinach chloroplast genome. Nucleic Acids Res. 19:3577-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagby, S., S. Kim, E. Maldonado, K. I. Tong, D. Reinberg, and M. Ikura. 1995. Solution structure of the C-terminal core domain of human TFIIB: similarity to cyclin A and interaction with TATA-binding protein. Cell 82:857-867. [DOI] [PubMed] [Google Scholar]
- 7.Baldwin, D. A., and W. B. Gurley. 1996. Isolation and characterization of cDNAs encoding transcription factor IIB from Arabidopsis and soybean. Plant J. 10:561-568. [DOI] [PubMed] [Google Scholar]
- 8.Barkan, A., and M. Goldschmidt-Clermont. 2000. Participation of nuclear genes in chloroplast gene expression. Biochimie 82:559-572. [DOI] [PubMed] [Google Scholar]
- 9.Bechtold, N., J. Ellis, and G. Pelletier. 1998. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82:259-266. [DOI] [PubMed] [Google Scholar]
- 10.Bell, S. D., and S. P. Jackson. 1998. Transcription in Archaea. Cold Spring Harbor Symp. Quant. Biol. 63:41-51. [DOI] [PubMed] [Google Scholar]
- 11.Berk, A. J. 2000. TBP-like factors come into focus. Cell 103:5-8. [DOI] [PubMed] [Google Scholar]
- 12.Blanc, G., A. Barakat, R. Guyot, R. Cooke, and M. Delseny. 2000. Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12:1093-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown, M. S., J. Ye, R. B. Rawson, and J. L. Goldstein. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100:391-398. [DOI] [PubMed] [Google Scholar]
- 14.Browning, K. S. 1996. The plant translational apparatus. Plant Mol. Biol. 32:107-144. [DOI] [PubMed] [Google Scholar]
- 15.Buratowski, S., and H. Zhou. 1992. A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell 71:221-230. [DOI] [PubMed] [Google Scholar]
- 16.Castle, L., and D. Meinke. 1994. A FUSCA gene of Arabidopsis encodes a novel protein essential for plant development. Plant Cell 6:25-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colbert, T., and S. Hahn. 1992. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev. 6:1940-1949. [DOI] [PubMed] [Google Scholar]
- 18.Dantonel, J. C., J. M. Wurtz, O. Poch, D. Moras, and L. Tora. 1999. The TBP-like factor: an alternative transcription factor in Metazoa? Trends Biochem. 24:335-339. [DOI] [PubMed] [Google Scholar]
- 19.Dikstein, R., S. Zhou, and R. Tjian. 1996. Human TAFII105 is a cell type-specific TFIID subunit related to hTAFII130. Cell 87:137-146. [DOI] [PubMed] [Google Scholar]
- 20.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douce, R., R. B. Holtz, and A. A. Benson. 1973. Isolation and properties of the envelope of spinach chloroplasts. J. Biol. Chem. 248:7215-7222. [PubMed] [Google Scholar]
- 22.Douce, R., and J. Joyard. 1982. Purification of the chloroplast, p. 239-256. In M. Edelman, R. B. Hallick, and N.-H. Chua (ed.), Methods in chloroplast molecular biology. Elsevier Biochemical Press, Amsterdam, The Netherlands.
- 23.Emanuelsson, O., H. Nielsen, and G. von Heijne. 1999. CHLOROP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emanuelsson, O., H. Nielsen, S. Brunak, and G. von Heijne. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005-1016. [DOI] [PubMed] [Google Scholar]
- 25.Felsenstein, J. 1984. Perspectives on the reconstruction of evolutionary history, p. 169-191. In T. Duncan and T. F. Stuessy (ed.), Cladistics. Columbia University Press, New York, N.Y.
- 26.Freiman, R. N., S. R. Albright, S. Zheng, W. C. Sha, R. E. Hammer, and R. Tjian. 2001. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science 293:2084-2087. [DOI] [PubMed] [Google Scholar]
- 27.Frishman, D., and P. Argos. 1996. Incorporation of non-local interactions in protein secondary structure prediction from the amino acid sequence. Protein Eng. 9:133-142. [DOI] [PubMed] [Google Scholar]
- 28.Gasch, A., A. Hoffmann, M. Horikoshi, R. G. Roeder, and N.-H. Chua. 1990. Arabidopsis thaliana contains two genes for TFIID. Nature 346:390-394. [DOI] [PubMed] [Google Scholar]
- 29.Gray, M. W. 1993. Origin and evolution of organelle genomes. Curr. Opin. Genet. Dev. 3:884-890. [DOI] [PubMed] [Google Scholar]
- 30.Gupta, R. S., and G. B. Golding. 1996. The origin of the eukaryotic cell. Trends Biochem. 21:166-171. [PubMed] [Google Scholar]
- 31.Ha, I., W. S. Lane, and D. Reinberg. 1991. Cloning of a human gene encoding the general transcription initiation factor IIB. Nature 352:689-695. [DOI] [PubMed] [Google Scholar]
- 32.Heard, D. J., T. Kiss, and W. Filipowicz. 1993. Both Arabidopsis TATA binding protein (TBP) isoforms are functionally identical in RNA polymerase II and III transcription in plant cells: evidence for gene-specific changes in DNA binding specificity of TBP. EMBO J. 12:3519-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hellmann, H., and M. Estelle. 2002. Plant development: regulation by protein degradation. Science 297:793-797. [DOI] [PubMed] [Google Scholar]
- 34.Hernandez, N. 1993. TBP, a universal eukaryotic transcription factor? Genes Dev. 7:1291-1308. [DOI] [PubMed] [Google Scholar]
- 35.Hiller, M. A., T.-Y. Lin, C. Wood, and M. T. Fuller. 2001. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 15:1021-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoppe, T., M. Rape, and S. Jentsch. 2001. Membrane-bound transcription factors: regulated release by RIP or RUP. Curr. Opin. Cell Biol. 13:344-348. [DOI] [PubMed] [Google Scholar]
- 37.Jefferson, R. A., T. A. Kavanagh, and M. W. Bevan. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joyard, J., A. Billecocq, S. G. Bartlett, M. Block, N.-H. Chua, and R. Douce. 1983. Localization of polypeptides to the cytosolic side of the outer envelope membrane of spinach chloroplasts. J. Biol. Chem. 258:10000-10006. [PubMed] [Google Scholar]
- 39.Kassavetis, G. A., G. L. Letts, and E. P. Geiduschek. 1999. A minimal RNA polymerase III transcription system. EMBO J. 18:5042-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keegstra, K., and K. Cline. 1999. Protein import and routing systems of chloroplasts. Plant Cell 11:557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim, T. K., and T. Maniatis. 1996. Regulation of interferon-γ-activated STAT1 by the ubiquitin-proteasome pathway. Science 273:1717-1719. [DOI] [PubMed] [Google Scholar]
- 42.Kohler, R. H., J. Cao, W. R. Zipfel, W. W. Webb, and M. R. Hanson. 1997. Exchange of protein molecules through connections between higher plant plastids. Science 276:2039-2042. [DOI] [PubMed] [Google Scholar]
- 43.Kunst, L. 1998. Preparation of physiologically active chloroplasts from Arabidopsis. Methods Mol. Biol. 82:43-48. [DOI] [PubMed] [Google Scholar]
- 44.Lagrange, T., H. Kapanidis, H. Tang, D. Reinberg, and R. Ebright. 1998. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 12:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larkin, R. M., and T. J. Guilfoyle. 1998. Two small subunits in Arabidopsis RNA polymerase II are related to yeast RPB4 and RPB7 and interact with one another. J. Biol. Chem. 273:5631-5637. [DOI] [PubMed] [Google Scholar]
- 46.Larkin, R. M., G. Hagen, and T. J. Guilfoyle. 1999. Arabidopsis thaliana RNA polymerase II subunits related to yeast and human RPB5. Gene 231:41-47. [DOI] [PubMed] [Google Scholar]
- 47.Li, Y.-F., J. Le Gourierrec, M. Torki, Y.-J. Kim, F. Guerineau, and D.-X. Zhou. 1999. Characterization and functional analysis of Arabidopsis TFIIA reveal that the evolutionarily unconserved region of the large subunit has a transcription activation domain. Plant Mol. Biol. 39:515-525. [DOI] [PubMed] [Google Scholar]
- 48.López-De-León, A., M. Librizzi, K. Puglia, and I. M. Willis. 1992. PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell 71:211-220. [DOI] [PubMed] [Google Scholar]
- 49.Maldonado, E., I. Ha, P. Cortes, L. Weis, and D. Reinberg. 1990. Factors involved in specific transcription by mammalian RNA polymerase II: role of transcription factors IIA, IID, and IIB during formation of a transcription-competent complex. Mol. Cell. Biol. 10:6335-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martianov, I., G.-M. Fimia, A. Dierich, M. Parvinen, P. Sassone-Corsi, and I. Davidson. 2001. Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol. Cell 7:509-515. [DOI] [PubMed] [Google Scholar]
- 51.Martin, W., and R. G. Hermann. 1998. Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol. 118:9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mathew, A., S. Mathur, and R. I. Morimoto. 1998. Heat shock response and protein degradation: regulation of HSF2 by the ubiquitin-proteasome pathway. Mol. Cell. Biol. 18:5091-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayer, K., et al. 1999. Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402:769-777. [DOI] [PubMed] [Google Scholar]
- 54.Nagy, F., M. Boutry, M.-Y. Hsu, M. Wong, and N.-H. Chua. 1987. The 5′-proximal region of the wheat Cab-1 gene contains a 268-bp enhancer-like sequence for phytochrome. EMBO J. 6:2537-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nawaz, Z., D. M. Lonard, A. P. Dennis, C. L. Smith, and B. W. O'Malley. 1999. Proteasome-dependent degradation of the human estrogen receptor. Proc. Natl. Acad. Sci. USA 96:1858-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nikolov, D., H. Chen, E. Halay, A. Usheva, K. Hisatake, D. Lee, R. G. Roeder, and S. Burley. 1995. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature 377:119-128. [DOI] [PubMed] [Google Scholar]
- 57.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657-2683. [DOI] [PubMed] [Google Scholar]
- 58.O'Shea, J. J., M. Gadina, and R. D. Schreiber. 2002. Cytokine signaling in 2002: new surprise in the Jak/Stat pathway. Cell 109:S121-S131. [DOI] [PubMed] [Google Scholar]
- 59.Osterlund, M. T., C. S. Hardtke, N. Wei, and X.-W. Deng. 2000. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405:113-118. [DOI] [PubMed] [Google Scholar]
- 60.Ozer, J., P. A. Moore, and P. M. Lieberman. 2000. A testis-specific transcription factor IIA (TFIIAτ) stimulates TATA-binding protein-DNA binding and transcription activation. J. Biol. Chem. 275:122-128. [DOI] [PubMed] [Google Scholar]
- 61.Parvin, J. D., and P. A. Sharp. 1993. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell 73:533-540. [DOI] [PubMed] [Google Scholar]
- 62.Privat, I., M.-A. Hakimi, L. Buhot, J. J. Favory, and S. Lerbs-Mache. 2003. Characterization of Arabidopsis plastid sigma-like transcription factors, SIG1, SIG2 and SIG3. Plant Mol. Biol. 55:385-399. [DOI] [PubMed]
- 63.Rechsteiner, M., and S. W. Rogers. 1996. PEST sequences and regulation by proteolysis. Trends Biochem. 21:267-271. [PubMed] [Google Scholar]
- 64.Rodermel, S. 2001. Pathways of plastid-to-nucleus signaling. Trends Plant Sci. 6:471-478. [DOI] [PubMed] [Google Scholar]
- 65.Roeder, R. G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. 21:327-335. [PubMed] [Google Scholar]
- 66.Santagata, S., T. J. Boggon, C. L. Baird, C. Gomez, J. Zhao, W. S. Shan, D. G. Myszka, and L. Shapiro. 2001. G-protein signaling through tubby proteins. Science 292:2041-2050. [DOI] [PubMed] [Google Scholar]
- 67.Schramm, L., P. S. Pendergrast, Y. Sun, and N. Hernandez. 2000. Different human TFIIIB activities direct RNA polymerase III transcription from TATA-containing and TATA-less promoters. Genes Dev. 14:2650-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwechheimer, C., G. Serino, J. Callis, W. L. Crosby, S. Lyapina, R. J. Deshaies, W. M. Gray, M. Estelle, and X.-W. Deng. 2001. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science 292:1379-1382. [DOI] [PubMed] [Google Scholar]
- 69.Schwechheimer, C., and X.-W. Deng. 2001. COP9 signalosome revisited: a novel mediator of protein degradation. Trends Cell Biol. 11:420-426. [DOI] [PubMed] [Google Scholar]
- 70.Staub, J. M., N. Wei, and X.-W. Deng. 1996. Evidence for FUS6 as a component of the nuclear-localized COP9 complex in Arabidopsis. Plant Cell 8:2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stern, D. B., D. C. Higgs, and J. Yang. 1997. Transcription and translation in chloroplasts. Trends Plant Sci. 2:285-323.
- 72.Sugiura, M. 1989. The chloroplast chromosomes in land plants. Annu. Rev. Cell. Biol. 5:51-70. [DOI] [PubMed] [Google Scholar]
- 73.Surpin, M., R. M. Larkin, and J. Chory. 2002. Signal transduction between the chloroplast and the nucleus. Plant Cell 14:327-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takada, S., J. T. Lis, S. Zhou, and R. Tjian. 2000. A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell 101:459-469. [DOI] [PubMed] [Google Scholar]
- 75.Teichmann, M., Z. Wang, and R. G. Roeder. 2000. A stable complex of a novel transcription factor IIB-related factor, human TFIIIB50, and associated proteins mediate selective transcription by RNA polymerase III of genes with upstream promoter elements. Proc. Natl. Acad. Sci. USA 97:14200-14205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Trifa, Y., I. Privat, J. Gagnon, L. Baeza, and S. Lerbs-Mache. 1998. The nuclear RPL4 gene encodes a chloroplast protein that co-purifies with the T7-like transcription complex as well as plastid ribosome. J. Biol. Chem. 273:3980-3985. [DOI] [PubMed] [Google Scholar]
- 77.Upadhyaya, A. B., S. H. Lee, and J. DeJong. 1999. Identification of a general transcription factor TFIIAα/β homolog selectively expressed in testis. J. Biol. Chem. 274:18040-18048. [DOI] [PubMed] [Google Scholar]
- 78.Vogel, J. M., B. Roth, M. Cigan, and M. Freeling. 1993. Expression of the two maize TATA binding protein genes and function of the encoded TBP proteins by complementation in yeast. Plant Cell 5:1627-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang, X., R. Sato, M. S. Brown, X. Hua, and J. L. Goldstein. 1994. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell 77:53-62. [DOI] [PubMed] [Google Scholar]
- 80.Watson, J. C., and W. F. Thompson. 1986. Purification and restriction endonuclease analysis of plant nuclear DNA. Methods Enzymol. 118:57-75. [Google Scholar]
- 81.Wei, N., S. F. Kwok, A. G. von Arnim, A. Lee, T. W. McNellis, B. Piekos, and X.-W. Deng. 1994. Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell 6:629-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zacharias, D. A., J. D. Violin, A. C. Newton, and R. Y. Tsien. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296:913-916. [DOI] [PubMed] [Google Scholar]
- 83.Zawel, L., and D. Reinberg. 1995. Common themes in assembly and function of eukaryotic transcription factors. Annu. Rev. Biochem. 64:533-561. [DOI] [PubMed] [Google Scholar]
- 84.Zhang, D., T. L. Penttila, P. L. Morris, M. Teichmann, and R. G. Roeder. 2001. Spermiogenesis deficiency in mice lacking the Trf2 gene. Science 292:1153-1155. [DOI] [PubMed] [Google Scholar]