Abstract
The hypoxic tumor microenvironment has been shown to contribute to genetic instability. As one possible mechanism for this effect, we report that expression of the DNA mismatch repair (MMR) gene Mlh1 is specifically reduced in mammalian cells under hypoxia, whereas expression of other MMR genes, including Msh2, Msh6, and Pms2, is not altered at the mRNA level. However, levels of the PMS2 protein are reduced, consistent with destabilization of PMS2 in the absence of its heterodimer partner, MLH1. The hypoxia-induced reduction in Mlh1 mRNA was prevented by the histone deacetylase inhibitor trichostatin A, suggesting that hypoxia causes decreased Mlh1 transcription via histone deacetylation. In addition, treatment of cells with the iron chelator desferrioxamine also reduced MLH1 and PMS2 levels, in keeping with low oxygen tension being the stress signal that provokes the altered MMR gene expression. Functional MMR deficiency under hypoxia was detected as induced instability of a (CA)29 dinucleotide repeat and by increased mutagenesis in a chromosomal reporter gene. These results identify a potential new pathway of genetic instability in cancer: hypoxia-induced reduction in the expression of key MMR proteins. In addition, this stress-induced genetic instability may represent a conceptual parallel to the pathway of stationary-phase mutagenesis seen in bacteria.
It has been argued that the large number of mutations found in malignant cells cannot be accounted for by the low rate of mutation generally found among somatic cells, leading to the suggestion that there is a mutator phenotype in cancer (26). The basis for this genetic instability has not been fully established. Much work has focused on the role of genetic defects in cancer cells affecting cell cycle regulation and DNA repair that could lead to genomic instability, such as in p53 (18). Hereditary nonpolyposis colon carcinoma is linked to inherited defects in several of the human DNA mismatch repair (MMR) genes, including Msh2, Mlh1, and Pms2 (29). Another cancer-prone syndrome, xeroderma pigmentosum, affects individuals with mutations in genes associated with the nucleotide excision repair (NER) pathway (56).
As an alternative mechanism by which genetic instability might arise in cancer, we and others have investigated the possible role of the tumor microenvironment (61). Developing tumors form a unique tissue environment because their growth outstrips their blood supply, leading to hypoxia, low pH, and nutrient deprivation (39, 55). Several studies have shown that hypoxia can alter chromosome metabolism, leading to gene amplification and fragile site induction (11, 37, 60). Our prior work found that cells exposed to hypoxia in culture have increased frequencies of point mutations at reporter gene loci (36). In addition, experimental tumors grown from cells implanted into mice show elevated levels of mutations compared to the same cells grown in parallel in normoxic culture, pointing to a deleterious effect of the tumor microenvironment on genome integrity in vivo (32, 36, 54).
We have hypothesized that hypoxia and associated physiologic changes could cause mutagenesis either by producing DNA damage or by hindering DNA repair. Previous experiments have shown that repair of UV-induced DNA damage is functionally diminished in hypoxic cells (62). However, in that work, we did not detect altered expression of factors in the NER pathway (which is responsible for UV damage repair), leading to the conclusion that the altered pH and other cellular changes related to hypoxia might serve to impair NER enzyme function rather than reduce NER gene expression.
In the present work, we have examined the status of the DNA MMR pathway under conditions of hypoxic stress. In mammalian cells, MMR is carried out by a series of proteins, including MSH2, MSH3, and MSH6 (homologues of the Escherichia coli MutS) and MLH1 and PMS2 (homologues of the E. coli MutL) (reviewed in reference 24). Heterodimers of the MutS homologues (either MSH2 and MSH3 or MSH2 and MSH6) act in concert with heterodimers of the MutL homologues (primarily MLH1 and PMS2) to mediate correction of replication errors as well as of DNA mismatches arising from other processes, such as recombination. We report here that levels of the MutL homologues MLH1 and PMS2 are reduced by hypoxia in mouse and human cells, whereas levels of the MutS homologues MSH2 and MSH6 are unaffected. We find that Mlh1 expression is specifically decreased at the mRNA level, and we provide evidence that this down-regulation can be attributed to diminished transcription due to histone deacetylation. In addition, a functional decrease in MMR activity under hypoxia was detected in both mouse and human cells by using two different reporter gene assays, including an assay for instability of a dinucleotide repeat sequence. These results identify a possible new pathway of acquired genetic instability in cancer.
MATERIALS AND METHODS
Cells.
Mouse 3340 cells are a fibroblast line described previously (13). HeLa cells (CCL-2) were obtained from the American Type Culture Collection (Manassas, Va.). EMT6 cells are derived from a mouse breast carcinoma (38). The 3340 and HeLa cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (Life Technologies, Rockville, Md.). EMT6 cells were grown in Waymouth medium supplemented with 10% fetal bovine serum (Life Technologies). HIF-1α wild-type and nullizygous mouse fibroblasts were described previously (42). Hypoxic cell culture conditions were established as described previously (36), using a continuous flow of a humidified mixture of 95% N2 and 5% CO2 gas certified to contain less than 10 ppm of O2 (Airgas Northeast, Cheshire, Conn.). Desferrioxamine mesylate (DFX) (Sigma, St. Louis, Mo.) treatment was carried out by supplementation of culture medium at a concentration of 250 μM under normoxic conditions. Cells were exposed to the cytosine methylation inhibitor 5-aza-2′-deoxycytidine (azaC) (Sigma) by addition to the culture medium at a concentration of 100 μM for 24 h prior to and during hypoxic exposure. Cells were treated with trichostatin A (TSA) (Sigma) by addition to the culture medium at a concentration of 300 nM immediately prior to and during hypoxic exposure.
Western blot analysis.
Western blotting was performed as previously described (34), using the following antibodies: MSH2 (clone Ab-2; Oncogene Research Products, San Diego, Calif.), MLH1 and PMS2 (clones A16-4 and G168-15, respectively; BD-PharMingen, Franklin Lakes, N.J.), MSH6 and HIF-1α (clones 44 and 54, respectively; Transduction Laboratories, Franklin Lakes, N.J.), and tubulin (clone B-512; Sigma). Proteins were visualized with horseradish peroxidase-conjugated anti-mouse immunoglobulin G and an ECL detection system (Amersham, Arlington Heights, Ill.), and bands were quantified by densitometry (UN-SCAN-IT software; Silk Scientific, Orem, Utah).
Northern blot analysis.
Total RNA was isolated by using Trizol (Life Technologies) followed by phenol-chloroform extraction. Equal amounts of RNA samples were fractionated by gel electrophoresis in 1% agarose with 6.6% formaldehyde in MOPS (morpholinepropanesulfonic acid) buffer at 60 V for 5 h. Gels were transferred to Hybond-N nylon membranes (Amersham) overnight in 20× SSPE buffer (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) and prehybridized in PerfectHyb Plus hybridization buffer (Sigma) for 3 h. Mouse probes were generated by reverse transcription-PCR amplification with RNA from 3340 cells (SuperScript one-step RT-PCR kit; Life Technologies) with the following primer pairs: for Mlh1, 5′-GCATAGCGGCGGGGGAAGTCAT-3′ (sense) and 5′-CGGTTGTGGCATTGGGCAGTGTT-3′ (antisense); for Pms2, 5′-CCAAGTGAGAAAAGGGGCGTGTTATCC-3′ (sense) and 5′-CTGTCTTGAAGCGCTTGGCATTTGTG-3′ (antisense); for Msh2, 5′-GCGGCCCGCGAGGTGTTCAA-3′ (sense) and 5′-TGCCTCAGTTTCCCCATGTCTCCAGTAGTC-3′ (antisense); for Msh6, 5′-CAGCCTAAGACACAAGGATCTAGGCGAAGTAGC-3′ (sense) and 5′-CTGTGTACCCTTGGTAATGATCCTACAGATCTCC-3′ (antisense); and for transketolase, 5′-GGAAGCCCCTCCCCAAAAACATGG-3′ (sense) and 5′-ATGGCGGCCATGCGAATCTGG-3′ (antisense). Probes were confirmed by DNA sequence analysis in each case. Probes were 32P labeled by using the Random Primers DNA Labeling System (Life Technologies) and purified by using NAP5 gel exclusion columns (Amersham). The human Mlh1 probe was 32P labeled from a 500-bp fragment corresponding to the 5′ end of the cloned Mlh1 cDNA (2). Blots were hybridized with the indicated probes overnight at 65°C and washed in 2× SSPE buffer-0.1% sodium dodecyl sulfate at 42°C, followed by visualization via autoradiography and quantification via phosphorimager analysis.
β-Galactosidase mutation assay.
The pCAR-OF plasmid (4 μg), in which the β-galactosidase gene contains a 58-bp out-of-frame (CA)29 insertion at the 5′ end of its coding region, was transfected by using cationic lipids (GenePorter; Gene Therapy Systems, San Diego, Calif.) into either HeLa cells or EMT6 cells along with the gWIZ luciferase vector (0.5 μg) as a normalization control. Three hours later, medium containing the transfection mixture was removed. Cells were replenished with fresh medium and were cultured either under normoxic or hypoxic conditions. After 48 h, cells were lysed, and the β-galactosidase and luciferase activities were measured under each condition. Values of β-galactosidase expression were normalized to the luciferase control and averaged over duplicate samples. In some cell samples, pCAR-OF plasmid DNA was rescued from the HeLa cells for genetic analysis of β-galactosidase gene function in indicator bacteria by an alkaline lysis procedure, as previously described (50).
supFG1 mutation assay.
3340 cells were treated with either hypoxia or normoxia for the 24 h. After a 3-day recovery period, the cells were harvested, and high-molecular-weight genomic DNA was prepared for lambda shuttle vector rescue from the chromosomal DNA and analysis of supFG1 gene function, as previously described (62).
RESULTS
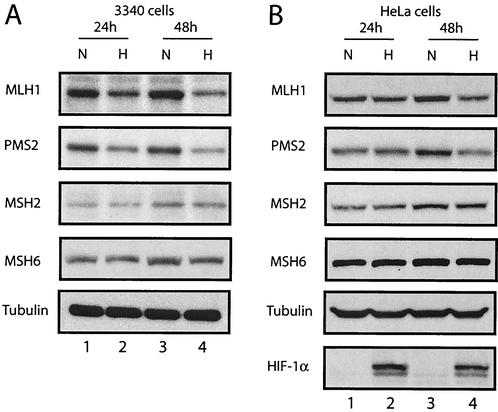
To determine whether the expression of the MMR proteins may be modulated by the conditions of the tumor microenvironment, a mouse fibroblast cell line, 3340, was placed under low-oxygen conditions, and samples of total cellular protein were obtained for Western blot analyses over a 2-day time period of exposure (Fig. 1A). After 48 h of hypoxic incubation, levels of the MutL homologues MLH1 and PMS2 were decreased in the hypoxic cells compared to normoxic cells, with relative decreases of seven- and fivefold, respectively. At the earlier 24-h time point, the differences were not as pronounced, but it was still possible to detect decreases in the ranges of twofold for both MLH1 and PMS2. In contrast, no hypoxia-related changes in the levels of either MSH2 or MSH6, two MutS homologues in the MMR pathway, were detected at any time during the 48-h period examined. We also saw no decrease in the levels of several NER proteins, including XPA, XPB, XPD, and XPG, in mouse cells under hypoxic conditions (reference 62 and data not shown). Levels of tubulin were also unchanged (Fig. 1A) and served as standards to confirm equal loading of cellular protein samples.
FIG. 1.
Decreased levels of MLH1 and PMS2 in mouse and human cells exposed to hypoxia. Western blot analyses were performed to determine the expression of the MMR proteins MLH1, PMS2, MSH2, and MSH6 in cells under normoxic (lanes N) or hypoxic (lanes H) conditions, as indicated. The period of time that the cells were maintained under hypoxic conditions (24 or 48 h) is given. Tubulin expression is presented to confirm equal sample loading. (A) Mouse 3340 cells. (B) HeLa cells. Expression of the hypoxia-inducible factor HIF-1α is also shown for comparison to verify that physiologically relevant levels of hypoxia were present in the treated cells.
A similar analysis was undertaken with human HeLa cells (Fig. 1B). Again, quantities of MLH1 and PMS2 were seen to drop with hypoxic incubation, especially at the 48-h time point, with decreases of six- and fourfold, respectively. Cellular levels of MSH2 and MSH6 were not altered. Note that in the same samples, the levels of the hypoxia-inducible transcription factor HIF-1α (44) were substantially increased by the hypoxic treatment, providing a clear contrast to the decreased MLH1 and PMS2 levels and verifying that the cells were exposed to a physiologically relevant degree of hypoxia.
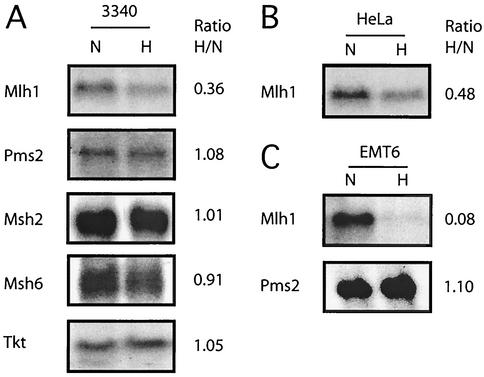
To determine if the reduced levels of MLH1 and PMS2 expression are the result of decreased mRNA levels, Northern blot analyses were performed (Fig. 2). In 3340 cells (Fig. 2A), Mlh1 mRNA levels were decreased upon hypoxic incubation (to 36% of the level in normoxia in the Northern blot shown), but those of Pms2 were essentially unchanged (108% of the level in normoxia). This experiment was repeated three times with similar results, with an average decrease in Mlh1 mRNA levels of approximately 3- to 3.5-fold and consistently with no change in Pms2 levels. Levels of Msh2 and Msh6 mRNAs were also unchanged (Fig. 2A).
FIG. 2.
Decreased levels of Mlh1 mRNA in mouse and human cells exposed to hypoxia. Northern blot analyses were performed on RNA samples obtained from cells grown under normoxic (lanes N) and hypoxic (lanes H) conditions for 24 h. (A) Mlh1, Pms2, Msh2, and Msh6 mRNA levels in mouse 3340 cells. To confirm equal sample loading, mRNA levels for transketolase (Tkt) were also determined. (B) Mlh1 mRNA levels in human HeLa cells under normoxic or hypoxic conditions. (C) Mlh1 and Pms2 mRNA levels in mouse EMT6 cells under normoxic or hypoxic conditions. Equal sample loading was verified in the cases of the HeLa and EMT6 cell blots by analysis of ethidium bromide-stained gels prior to transfer (not shown). Expression levels were quantified by phosphorimager analysis, and the ratio of expression under hypoxia to that under normoxia is listed to the right of each panel.
HeLa cells exposed to hypoxia also showed a decrease in Mlh1 mRNA levels (2- to 2.5-fold) (Fig. 2B). Analysis of a third cell line, EMT6, derived from a murine breast carcinoma (38), again showed a drop in Mlh1 mRNA levels in hypoxia (12-fold) (Fig. 2C) but no decrease in Pms2 mRNA levels. These results indicate that expression of Mlh1 is specifically reduced at the mRNA level in three different mouse and human cell lines exposed to hypoxia.
Note that Pms2 mRNA levels are not decreased by hypoxia, suggesting that the reduction in PMS2 protein levels seen in hypoxia may come about due to alterations in either translation efficiency or posttranslational stability. The latter is the more likely explanation, as destabilization of PMS2 in the absence of its heterodimer partner, MLH1, has been observed in the case of knockout mice deficient in MLH1 (58).
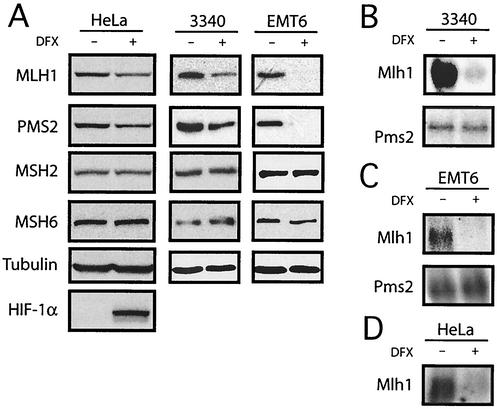
It is thought that cells detect low oxygen tension through a heme-containing sensor protein (14). The hypoxic state can be mimicked by using the iron chelator DFX, which is proposed to disrupt normal oxygen-sensing pathways in mammalian cells by inhibiting heme-Fe2+ interactions; DFX therefore has been used as a hypoxia mimetic in cell culture (52). Like hypoxia, DFX can stabilize HIF-1α (52), which stimulates the transcription of genes bearing the hypoxia response elements (28, 51, 52). The stabilization of HIF-1α by hypoxia and by DFX has been attributed to reduced proline hydroxylation (6, 20, 21). We exposed cells to DFX for 24 h and analyzed protein expression by Western blot analysis. Treatment of 3340, EMT6, and HeLa cells with DFX resulted in decreases in both MLH1 and PMS2 (Fig. 3A) but no changes in either MSH2 or MSH6. As expected, HIF-1α levels were increased in the DFX-treated HeLa cells. Northern blot analyses (Fig. 3B) revealed that Mlh1 mRNA was substantially decreased in the 3340 cells upon DFX exposure, but Pms2 mRNA levels were not affected. In EMT6 cells (Fig. 3C), Mlh1 mRNA was also reduced by DFX treatment, with no decrease in Pms2. Similarly, in HeLa cells treated with DFX (Fig. 3D), Mlh1 mRNA levels were decreased. These results show that Mlh1 expression is reduced not only in truly hypoxic cells but also in cells in which hypoxia is simulated by interfering with normal cellular oxygen-sensing and -signaling pathways.
FIG. 3.
Decreased MMR gene expression in mouse and human cells exposed to the hypoxia-mimetic drug DFX. (A) Western blot analyses of the expression of the MMR proteins MLH1, PMS2, MSH2, and MSH6 in human HeLa, mouse 3340, and mouse EMT6 cells treated with DFX. Tubulin expression is presented to confirm equal sample loading. Expression of the hypoxia-inducible factor HIF-1α in the HeLa cells is also shown for comparison to demonstrate that the DFX treatment provoked a hypoxia-like response in the cells. (B) Northern blot analyses of Mlh1 and Pms2 mRNA levels in mouse 3340 cells with and without DFX treatment. (C) Northern blot analysis of Mlh1 and Pms2 mRNA levels in mouse EMT6 cells with and without DFX treatment. (D) Northern blot analysis of Mlh1 mRNA levels in HeLa cells with and without DFX treatment.
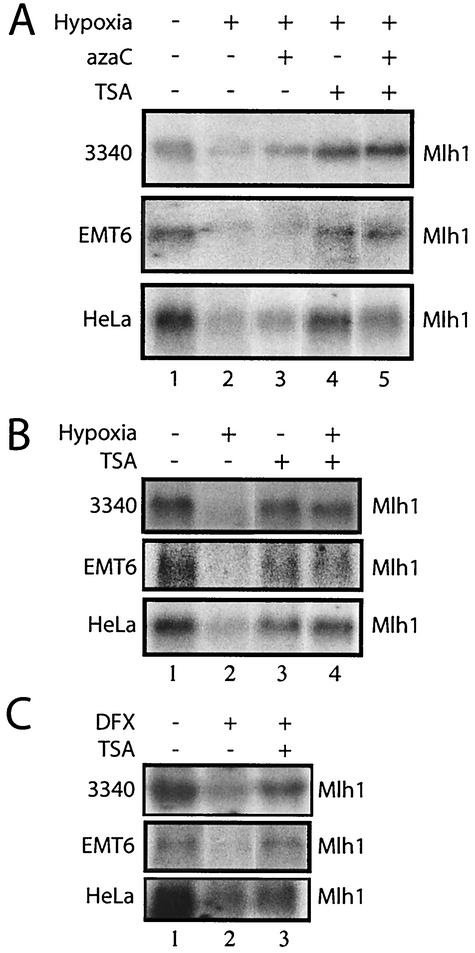
To investigate the mechanism by which hypoxia-induced signaling reduces Mlh1 gene expression, we considered the possibility that Mlh1 promoter hypermethylation might play some role. Mlh1 expression is reduced in many sporadic cancers, and this has been associated with methylation of CpG-rich promoter regions (16, 22). We used two sensitive assays for detection of methylation in genomic DNA, methylation-specific PCR (16) and combined bisulfite and restriction analysis (57). Neither assay revealed changes in Mlh1 promoter methylation status in the cells exposed to hypoxia for 24 or 48 h (data not shown). As a further test of a possible role for methylation, we pretreated either 3340, EMT6, or HeLa cells with the methylation inhibitor azaC (9) for 24 h and then exposed them to hypoxia, while still maintaining them in azaC. There was a minimal effect of azaC on Mlh1 mRNA levels in hypoxic cells (Fig. 4A).
FIG. 4.
The histone deacetylase inhibitor TSA prevents the down-regulation of Mlh1 expression by either hypoxia or treatment with the hypoxia mimetic drug DFX. (A) Northern blot analyses of Mlh1 expression levels were carried out on RNA samples from 3340, EMT6, and HeLa cells incubated under the following conditions as indicated: normoxia, hypoxia for 24 h, hypoxia plus the cytosine methylation inhibitor azaC, hypoxia plus TSA, or hypoxia plus both azaC and TSA. Equal sample loading was verified by analysis of ethidium bromide-stained gels prior to transfer (not shown). (B) Northern blot analyses of Mlh1 expression levels in 3340, EMT6, and HeLa cells under normoxic or hypoxic conditions and with or without TSA, as indicated. (C) Northern blot analyses of Mlh1 expression levels in 3340, EMT6, and HeLa cells with and without DFX treatment and with or without TSA, as indicated.
However, when the cells were incubated in the presence of TSA, a specific inhibitor of histone deacetylases (27, 59), the down-regulation of Mlh1 by hypoxia was prevented in all three cell lines (Fig. 4A [compare lanes 2 and 4 for each cell line] and B [compare lanes 2 and 4]). The combined effect of TSA and azaC was also effective in inhibiting the Mlh1 down-regulation. Hence, it appears that hypoxic stress reduces Mlh1 expression in a manner that is dependent upon histone deacetylase activity, since the decrease in Mlh1 expression is prevented by TSA-mediated inhibition of histone deacetylases. This finding is consistent with previous studies linking histone deacetylation with the down-regulation of gene expression by hypoxia (23). In the case of Mlh1, it is likely that the hypoxia-induced histone deacetylation occurs at the Mlh1 promoter locus, leading to a hypoacetylated state and thereby decreasing transcription of the Mlh1 gene. However, we cannot rule out the possibility that the reduced expression of Mlh1 in hypoxia is caused by a yet-to-be identified regulatory factor. The expression of this factor itself could be altered via changes in histone acetylation.
Interestingly, in normoxic cells, TSA did not cause an increase in Mlh1 mRNA and, in fact, appeared to cause a slight reduction in Mlh1 expression (Fig. 4B, compare lanes 1 and 3 for each cell line). That TSA might produce a decrease in Mlh1 gene expression in normoxic conditions is at first glance puzzling, since (i) the opposite effect is clearly seen under hypoxic conditions and (ii) TSA typically causes up-regulation of gene expression by altering the balance of histone acetylation and deacetylation to favor increased acetylation of chromatin (30). However, the ability of TSA to cause a reduction in the expression of certain genes under normoxic conditions (possibly via unexpected activation of histone acetyltransferase activity at certain loci) has been reported (30). Such genes include those for cdk-1, cyclin B1, and cyclin A (30). Hence, it is plausible that the expression of Mlh1, like that of these other genes, may be reduced by TSA under normoxic conditions but may be increased by TSA under hypoxia. Nonetheless, these results provide evidence for a specific hypoxia-induced pathway in which changes in histone deacetylase activity cause reduced Mlh1 expression.
Since DFX was found to produce a decrease in Mlh1 expression similar to that induced by hypoxia, we tested whether the down-regulation of Mlh1 caused by DFX could also be prevented by TSA treatment (Fig. 4C). Northern blot analyses of Mlh1 mRNA levels in 3340, EMT6, and HeLa cells revealed that TSA treatment does block Mlh1 down-regulation by DFX in all three cell lines. These results provide additional evidence for Mlh1 regulation at the level of histone acetylation, and they support the hypothesis that hypoxia and DFX cause decreased Mlh1 expression via similar mechanisms.
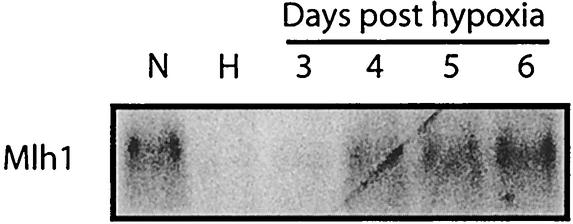
To test whether the hypoxia-induced Mlh1 down-regulation is stable or transient, we transferred 3340 mouse cells to normoxic conditions after 48 h of hypoxia, and we found that by 4 days the Mlh1 mRNA levels began to return to those of the normoxic controls (Fig. 5). This suggests that the down-regulation is transient and reversible, a result that is consistent with our model of regulation at the level of histone acetylation.
FIG. 5.
Reversibility of the hypoxia-induced down-regulation of Mlh1 after replacement of cells in normoxic conditions. Northern blot analysis of Mlh1 mRNA levels in 3340 mouse cells grown under normoxia (lane N), grown under hypoxia for 48 h (lane H), or grown under hypoxia for 48 h and then returned to normoxia for the indicated times is shown.
HIF-1α is a key factor that regulates gene expression in response to hypoxia (43). On examination of the Mlh1 promoter, we identified a putative hypoxia-responsive element at positions −84 to −80 (ACGTG) (19), suggesting that Mlh1 expression might be regulated by HIF-1α under hypoxia. To test this, we examined Mlh1 gene expression under normoxic and hypoxic conditions in otherwise isogenic mouse embryonic fibroblasts either wild type or nullizygous for HIF-1α (42). Down-regulation of Mlh1 was still detected in HIF-1α-deficient cells (data not shown), indicating that the down-regulation of Mlh1 expression in hypoxia does not require HIF-1α.
To examine whether MMR is functionally diminished in hypoxic cells, we examined mutagenesis at a chromosomal locus in the 3340 cells. 3340 cells are derived from transgenic mice that contain multiple stably integrated copies of a lambda vector DNA at a single locus on chromosome 7 (13). The mice and the mouse-derived 3340 cells were designed for mutation detection with the supFG1 reporter gene present in the lambda vector DNA (31). In experiments performed in parallel to the Western and Northern analyses described above, samples of 3340 cells grown under hypoxic or normoxic conditions were harvested for analysis of mutagenesis in the supFG1 gene via lambda phage vector DNA rescue from mouse cell genomic DNA by using lambda packaging extracts (15, 62). The phage vectors rescued from the hypoxic cells showed an approximately twofold increase in mutation frequency (Fig. 6A). Sequence analysis of the mutations obtained from the hypoxic cells revealed that a majority were 1-bp deletions or insertions within the G · C base pair repeats at positions 99 to 105 and 172 to 179 (data not shown). Using a second reporter gene in the lambda vector (the cII gene), we also found a twofold increase in mutation frequency under hypoxia (7.0 × 10−5 ± 1.0 × 10−5 [53 of 750,126] under normoxia and 14.4 × 10−5 ± 1.1 × 10−5 [79 of 549,297] under hypoxia).
FIG. 6.
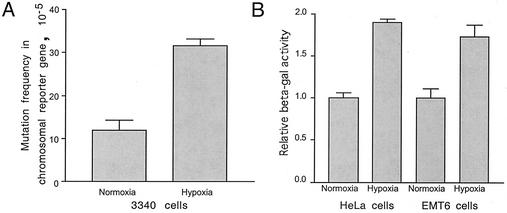
Induction of mutagenesis by hypoxia and association with MLH1 levels. (A) Frequencies of mutations in the chromosomal supFG1 reporter gene in 3340 cells exposed to hypoxia. Cells were maintained under normoxic or hypoxic conditions for 24 h. After 3 days of subsequent growth of the cells under normal culture conditions, mutagenesis in the supFG1 reporter gene was assayed by rescue of the chromosomally integrated λsupFG1 shuttle vector. Error bars indicate standard errors. (B) Restoration of β-galactosidase (beta-gal) activity via frameshift mutagenesis in a lacZ reporter gene construct in HeLa and EMT6 cells under normoxia or hypoxia. Cells were transfected with an episomal, replicative vector, pCAR-OF (33), containing the β-galactosidase gene interrupted by a 58-bp out-of-frame poly (CA)29 insertion tract at the 5′ end of its coding region. Restoration of the proper reading frame to generate a functional enzyme occurs when replication slippage errors within the repeat sequence tract are not corrected by MMR. This experiment was performed three times, and the relative β-galactosidase values were normalized to a value of 1 for the normoxic cells in each case. Standard errors are indicated.
As another approach to measure possible hypoxia-induced genetic instability, a mutation reporter construct was used to assay directly for the instability of simple repeated sequences that is the hallmark of MMR deficiency (48). HeLa and EMT6 cells were each transfected with an episomal, replicative reporter construct, pCAR-OF (33), containing the β-galactosidase gene interrupted by a 58-bp out-of-frame poly (CA)29 insertion tract at the 5′ end of its coding region. Restoration of the proper reading frame occurs when replication slippage errors within the (CA)29 tract escape correction by MMR. Hence, increased β-galactosidase activity is indicative of defective MMR. Following transfection (which included a luciferase vector as an internal control), the cells were placed under either hypoxic or normoxic conditions. After 48 h, cell lysates were assayed for β-galactosidase and luciferase activities (Fig. 6B). Under hypoxia, the HeLa and EMT6 cells were found to express 1.9- and 1.7-fold, respectively, higher levels of β-galactosidase (relative to luciferase) than the same cells under normoxic conditions. When an otherwise similar β-galactosidase expression vector lacking the out-of-frame 58-bp insertion was tested in the cells, no differences in the ratio of β-galactosidase to luciferase expression between hypoxia and normoxia were detected (data not shown), suggesting that hypoxia otherwise has no effect on the relative expression of the β-galactosidase and luciferase constructs.
Hence, in HeLa cells, for example, in which we detected a 6-fold decrease in MLH1 levels under hypoxia by Western blotting (Fig. 1), we measured a 1.9-fold increase in genetic instability with the pCAR-OF assay. For comparison, when the same pCAR-OF assay was used with colon cancer cells in which the Mlh1 genes had been completely inactivated by mutation, 100-fold increases in β-galactosidase expression relative to that in wild-type cells were observed (33). This comparison shows that there is still some residual MLH1 function and MMR activity in hypoxic cells and that hypoxic cells do not display the extreme genetic instability that is seen in completely MMR-deficient hereditary nonpolyposis colon carcinoma cells. However, the data do demonstrate that the reductions in MLH1 levels in HeLa and EMT6 cells under prolonged hypoxia are sufficient to produce measurable increases in genetic instability within a sequence context that is known to be unstable in the setting of MMR deficiency.
As yet another way to quantify the instability of the (CA)29 tract in hypoxic cells, the pCAR-OF plasmid vector DNA was rescued from the HeLa cells after 48 h under either normoxic or hypoxic conditions and used to transform indicator bacteria to assay for lacZ gene function (50). Enumeration of the resulting bacterial colonies revealed a 1.6-fold-higher frequency of β-galactosidase-positive colonies in the samples derived from the hypoxic versus the normoxic cells (5.2 × 10−4 [27 of 52,099] versus 3.3 × 10−4 [9 of 27,061]). This result is consistent with the measurements of β-galactosidase expression in HeLa cell lysates (see above) and again shows hypoxia-induced instability of the (CA)29 tract, as would be expected from a reduction in MMR activity. Hence, taken together, the experiments with the pCAR-OF assay in HeLa and EMT6 cells and with the supFG1 and cII chromosomal reporter gene assays in 3340 cells provide quantitative evidence for low but detectable levels of hypoxia-induced mutagenesis at repeated sequences, a characteristic of MMR deficiency.
DISCUSSION
To explore the mechanisms underlying tumor microenvironment-induced genetic instability, we investigated whether the hypoxic conditions found in solid tumors could influence the DNA MMR pathway. We found that in both human and mouse cells, prolonged exposure to hypoxia caused decreased levels of MLH1 and PMS2. Mlh1 expression, specifically, was found to be down-regulated at the mRNA level. Transcript levels from other MMR genes, including Pms2, Msh2, and Msh6, were unaffected. PMS2 protein levels were nonetheless reduced under hypoxia, in keeping with the known instability of PMS2 in the absence of its heterodimer partner, MLH1 (8, 58). Interference with oxygen-sensing and -signaling pathways by treatment of cells with the iron chelator DFX caused similar reductions in Mlh1 mRNA expression and in MLH1 and PMS2 protein levels, suggesting that the cellular perception of low oxygen tension is a key regulatory element. Mechanistically, the down-regulation was blocked by TSA, implicating histone deacetylation as a critical step in the process. Since transcription factors can alter histone acetylation, we tested for a possible role of the hypoxia-inducible transcription factor HIF-1α in the pathway, but it was not found to be required.
Consistent with a reduction in levels of MLH1 and PMS2, we detected increased frequencies of frameshift and slippage mutations in two different reporter constructs in hypoxic cells, the chromosomally based lambda shuttle vector and the episomal pCAR-OF vector. In the lambda vector system in the 3340 cells, mutagenesis in two reporter genes, supFG1 and cII, was examined. In the case of the pCAR-OF β-galactosidase assay, hypoxia-induced instability was detected in both HeLa and EMT6 cells. With the HeLa cells, two different methods of quantification were used. The observed genetic instability in hypoxic cells that was consistently detected in all of these assays is in keeping with diminished MMR activity. It should be noted that the reduced MMR activity could result both from the substantial decreases in MLH1 and PMS2 levels and from the suboptimal cellular conditions that are caused by hypoxia (such as low pH and decreased energy production), thereby also causing functional impairment of the remaining repair proteins.
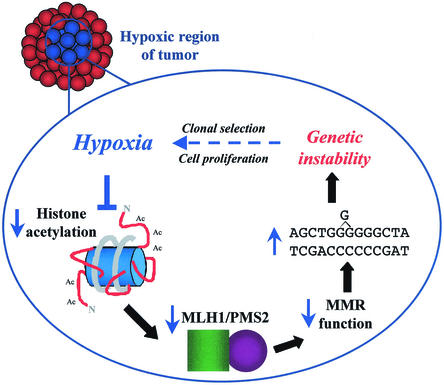
With respect to tumor biology, our results suggest that tumor cells that experience hypoxia may express transient mutator phenotypes (even in the absence of actual mutations in MMR genes) due to decreased Mlh1 expression, leading to acquired genetic instability and contributing to tumor progression (Fig. 7). Hypoxic regions in solid tumors vary over time and space, and there can be areas of both chronic and acute hypoxia. Hence, hypoxia-induced changes in Mlh1 gene expression may be variable and intermittent, being more pronounced in certain tumors (and certain regions within a tumor) and minimal in others. Overall, this process may help to explain why high levels of hypoxia in human tumors correlate with a worse prognosis and more aggressive behavior compared to the case for less hypoxic tumors (5, 17).
FIG. 7.
Model illustrating the pathway of hypoxia-induced genetic instability via down-regulation of the MMR gene Mlh1. The present data support a mechanism in which hypoxia causes histone deacetylation, leading to repression of Mlh1 transcription. Decreased MLH1 protein secondarily leads to destabilization of PMS2. In the setting of reduced levels of the MLH1/PMS2 heterodimer, cellular MMR activity is suboptimal, resulting in increased genetic instability.
In addition, the MMR pathway participates in the response of cells to certain types of DNA damage (13). Cells deficient in MMR factors, including MLH1, MSH2, and PMS2, show increased survival following exposure to alkylating agents and ionizing radiation (13). Hence, the down-regulation of Mlh1 under hypoxia in solid tumors may promote resistance to agents used in cancer therapy. This possibility is under active investigation.
The ability of hypoxia to alter gene expression in mammalian cells is well established (46). Genes induced by hypoxia include those for glycolytic enzymes, growth factors, and transcription factors, many of which have roles in tumor angiogenesis and metastasis (49, 55). Hypoxia has also been shown to decrease the expression of a number of genes, but without clear implications for DNA repair pathways (1, 4, 25, 35, 47, 53). Interestingly, the extended time course pattern of Mlh1 down-regulation that we observed may explain why previous surveys of the effect of hypoxia on gene expression profiles in mammalian cells via array technology have not identified Mlh1 as a hypoxia-responsive gene, as the prior studies were conducted over shorter time frames of hypoxia exposure (25).
Our studies provide some initial clues to the mechanism by which Mlh1 is down-regulated in hypoxia. It appears that Mlh1 expression is regulated, at least in part, at the level of transcription. Northern blot analysis revealed reductions in Mlh1 mRNA levels after 24 and 48 h of hypoxia in both mouse and human cells. In addition, the histone deacetylase inhibitor TSA was shown to prevent the hypoxia-induced down-regulation, suggesting that the reduction in Mlh1 mRNA results from changes in transcription initiation and that these changes are mediated by alterations in histone acetylation and chromatin structure.
We also considered the possibility that promoter methylation might play some role, prompted by reports that in many sporadic human cancers MLH1 is undetectable in histologic sections and that this can be associated with hypermethylation of cytosines at CpG sites within the Mlh1 promoter (16, 22). We did examine methylation of the Mlh1 promoter in genomic DNA from the cells under the conditions of our experiments, but we were unable to detect any changes in methylation by using either of two different assays (16, 57). In addition, the methylation inhibitor azaC had a minimal effect on the hypoxia-mediated down-regulation of Mlh1 mRNA (Fig. 4).
At this point, we cannot completely rule out a role for promoter methylation in Mlh1 down-regulation; however, we can conclude that extensive methylation is not required for the initial effect of hypoxia on Mlh1 expression. One possible mechanism is that Mlh1 down-regulation by hypoxia via histone deacetylation could serve as the initial event in a process of gene silencing that could eventually be augmented by hypermethylation. Such a pathway would be consistent with work pointing to a broad synergism between hypermethylation and histone deacetylation in the regulation of gene expression (3, 41).
While there appears to be a clear role for histone deacetylation in this pathway, the initiating signal that leads from hypoxia to Mlh1 down-regulation has not yet been elucidated. The fact that exposure to the iron chelator DFX could reproduce the same effects on MLH1 and PMS2 as caused by hypoxia is in keeping with a pathway in which an iron-containing protein responds to oxygen tension to initiate a signal pathway that can alter Mlh1 transcription (7, 45, 63). The similarity of the DFX-induced and the hypoxia-induced pathways of Mlh1 down-regulation is underscored by the finding that both are blocked by TSA treatment of the cells, indicating that both effects are mediated by histone deacetylation. In addition, while the effect on Mlh1 does not appear to require HIF-1α, some other HIF-1α-like transcription factor also subject to oxygen- and iron-dependent proline hydroxylation (or other modification) could still be involved (6, 20, 21, 45, 63).
Besides the hypoxia-induced repression of Mlh1 and the associated increase in repeat sequence instability reported here, other hypoxia-induced genome changes have been reported, including gene amplification and fragile site induction (11, 36, 37, 60). Interestingly, gene amplification has also been reported to arise in MLH1-deficient cells (10), but a direct connection between MLH1-induced and hypoxia-induced gene amplification has not been tested. Nonetheless, together these observations suggest that hypoxia can constitute a profound cellular stress that can promote genetic instability on several levels. This constellation of responses is reminiscent of the multiple genetic changes that have been associated with stationary-phase mutagenesis in E. coli (12, 40). In response to starvation conditions, several pathways of genome modification are activated in E. coli. Among these are break-induced recombination and a process of point mutagenesis that is proposed to result from an insufficiency of the MMR factor MutL (40), which is the Mlh1 homologue. It is tempting to draw a conceptual parallel between the hypoxia response reported here and the E. coli stress response. The response to hypoxia (and possibly to other stresses) may constitute an evolutionarily conserved mechanism by which mammalian cells, like E. coli, can increase their mutation rates under adverse conditions. Unlike the case of unicellular organisms, however, the expression of this pathway within a hypoxic tumor might benefit the individual cancer cells but would be disadvantageous to the organism as a whole.
Acknowledgments
Valia T. Mihaylova, Ranjit Bindra, and Jianling Yuan contributed equally to this work.
We thank M. Liskay, A. Giaccia, R. Hill, J. Sweasy, A. Perkins, and M. Snyder for helpful discussions and S. Peretz, R. Hickey, K. Wehner, L. Cabral, R. Franklin, and S. J. Baserga for their help. We also thank M. Liskay and B. Vogelstein for providing reagents.
This work was supported by grants from the NIH (ES05775 and CA16038) and the American Cancer Society (RPG-96-129-03-MGO) to P.M.G.
REFERENCES
- 1.Avila, M. A., M. V. Carretero, E. N. Rodriguez, and J. M. Mato. 1998. Regulation by hypoxia of methionine adenosyltransferase activity and gene expression in rat hepatocytes. Gastroenterology 114:364-371. [DOI] [PubMed] [Google Scholar]
- 2.Baker, S. M., A. W. Plug, T. A. Prolla, C. E. Bronner, A. C. Harris, X. Yao, D. M. Christie, C. Monell, N. Arnheim, A. Bradley, T. Ashley, and R. M. Liskay. 1996. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13:336-342. [DOI] [PubMed] [Google Scholar]
- 3.Baylin, S. B., M. Esteller, M. R. Rountree, K. E. Bachman, K. Schuebel, and J. G. Herman. 2001. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 10:687-692. [DOI] [PubMed] [Google Scholar]
- 4.Berk, J. L., N. Massoomi, C. Hatch, and R. H. Goldstein. 1999. Hypoxia downregulates tropoelastin gene expression in rat lung fibroblasts by pretranslational mechanisms. Am. J. Physiol. 277:L566-572. [DOI] [PubMed] [Google Scholar]
- 5.Brizel, D. M., S. P. Scully, J. M. Harrelson, L. J. Layfield, J. M. Bean, L. R. Prosnitz, and M. W. Dewhirst. 1996. Tumor oxygenation predicts for the likelihood of distant metastases in human soft tissue sarcoma. Cancer Res. 56:941-943. [PubMed] [Google Scholar]
- 6.Bruick, R. K., and S. L. McKnight. 2001. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294:1337-1340. [DOI] [PubMed] [Google Scholar]
- 7.Bunn, H. F., and R. O. Poyton. 1996. Oxygen sensing and molecular adaptation to hypoxia. Physiol. Rev. 76:839-885. [DOI] [PubMed] [Google Scholar]
- 8.Chang, D. K., L. Ricciardiello, A. Goel, C. L. Chang, and C. R. Boland. 2000. Steady-state regulation of the human DNA mismatch repair system. J. Biol. Chem. 275:18424-18431. [DOI] [PubMed] [Google Scholar]
- 9.Chen, R. Z., U. Pettersson, C. Beard, L. Jackson-Grusby, and R. Jaenisch. 1998. DNA hypomethylation leads to elevated mutation rates. Nature 395:89-93. [DOI] [PubMed] [Google Scholar]
- 10.Chen, S., S. H. Bigner, and P. Modrich. 2001. High rate of CAD gene amplification in human cells deficient in MLH1 or MSH6. Proc. Natl. Acad. Sci. USA 98:13802-13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coquelle, A., F. Toledo, S. Stern, A. Bieth, and M. Debatisse. 1998. A new role for hypoxia in tumor progression: induction of fragile site triggering genomic rearrangements and formation of complex DMs and HSRs. Mol. Cell 2:259-265. [DOI] [PubMed] [Google Scholar]
- 12.Foster, P. L., and J. Cairns. 1992. Mechanisms of directed mutation. Genetics 131:783-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fritzell, J. A., L. Narayanan, S. M. Baker, C. E. Bronner, S. E. Andrew, T. A. Prolla, A. Bradley, F. R. Jirik, R. M. Liskay, and P. M. Glazer. 1997. Role of DNA mismatch repair in the cytotoxicity of ionizing radiation. Cancer Res. 57:5143-5147. [PubMed] [Google Scholar]
- 14.Goldberg, M. A., S. P. Dunning, and H. F. Bunn. 1988. Regulation of the erythropoietin gene: evidence that the oxygen sensor is a heme protein. Science 242:1412-1415. [DOI] [PubMed] [Google Scholar]
- 15.Gunther, E. J., N. E. Murray, and P. M. Glazer. 1993. High efficiency, restriction-deficient in vitro packaging extracts for bacteriophage lambda DNA using a new E. coli lysogen. Nucleic Acids Res. 21:3903-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herman, J. G., A. Umar, K. Polyak, J. R. Graff, N. Ahuja, J. P. Issa, S. Markowitz, J. K. Willson, S. R. Hamilton, K. W. Kinzler, M. F. Kane, R. D. Kolodner, B. Vogelstein, T. A. Kunkel, and S. B. Baylin. 1998. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc. Natl. Acad. Sci. USA 95:6870-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hockel, M., K. Schlenger, B. Aral, M. Mitze, U. Schaffer, and P. Vaupel. 1996. Association between tumor hypoxia and malignant progression in advanced cancer of the uterine cervix. Cancer Res. 56:4509-4515. [PubMed] [Google Scholar]
- 18.Hollstein, M., D. Sidransky, B. Vogelstein, and C. C. Harris. 1991. p53 mutations in human cancers. Science 253:49-53. [DOI] [PubMed] [Google Scholar]
- 19.Ito, E., Y. Yanagisawa, Y. Iwahashi, Y. Suzuki, H. Nagasaki, Y. Akiyama, S. Sugano, Y. Yuasa, and K. Maruyama. 1999. A core promoter and a frequent single-nucleotide polymorphism of the mismatch repair gene hMLH1. Biochem. Biophys. Res. Commun. 256:488-494. [DOI] [PubMed] [Google Scholar]
- 20.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 21.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 22.Kane, M. F., M. Loda, G. M. Gaida, J. Lipman, R. Mishra, H. Goldman, J. M. Jessup, and R. Kolodner. 1997. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 57:808-811. [PubMed] [Google Scholar]
- 23.Kim, M. S., H. J. Kwon, Y. M. Lee, J. H. Baek, J. E. Jang, S. W. Lee, E. J. Moon, H. S. Kim, S. K. Lee, H. Y. Chung, C. W. Kim, and K. W. Kim. 2001. Histone deacetylases induce angiogenesis by negative regulation of tumor suppressor genes. Nat. Med. 7:437-443. [DOI] [PubMed] [Google Scholar]
- 24.Kolodner, R. 1996. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 10:1433-1442. [DOI] [PubMed] [Google Scholar]
- 25.Koong, A. C., N. C. Denko, K. M. Hudson, C. Schindler, L. Swiersz, C. Koch, S. Evans, H. Ibrahim, Q. T. Le, D. J. Terris, and A. J. Giaccia. 2000. Candidate genes for the hypoxic tumor phenotype. Cancer Res. 60:883-887. [PubMed] [Google Scholar]
- 26.Loeb, L. A. 1991. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 51:3075-3079. [PubMed] [Google Scholar]
- 27.Marks, P. A., V. M. Richon, and R. A. Rifkind. 2000. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J. Natl. Cancer Inst. 92:1210-1216. [DOI] [PubMed] [Google Scholar]
- 28.Minchenko, A., S. Salceda, T. Bauer, and J. Caro. 1994. Hypoxia regulatory elements of the human vascular endothelial growth factor gene. Cell. Mol. Biol. Res. 40:35-39. [PubMed] [Google Scholar]
- 29.Modrich, P., and R. Lahue. 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65:101-133. [DOI] [PubMed] [Google Scholar]
- 30.Nair, A. R., L. J. Boersma, L. Schiltz, M. A. Chaudhry, R. J. Muschel, and A. Chaudry. 2001. Paradoxical effects of trichostatin A: inhibition of NF-Y-associated histone acetyltransferase activity, phosphorylation of hGCN5 and downregulation of cyclin A and B1 mRNA. Cancer Lett. 166:55-64. [DOI] [PubMed] [Google Scholar]
- 31.Narayanan, L., J. A. Fritzell, S. M. Baker, R. M. Liskay, and P. M. Glazer. 1997. Elevated levels of mutation in multiple tissues of mice deficient in the DNA mismatch repair gene Pms2. Proc. Natl. Acad. Sci. USA 94:3122-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paquette, B., and J. B. Little. 1994. In vivo enhancement of genomic instability in minisatellite sequences of mouse C3H/10T1/2 cells transformed in vitro by X-rays. Cancer Res. 54:3173-3178. [PubMed] [Google Scholar]
- 33.Parsons, R., G. M. Li, M. J. Longley, W. H. Fang, N. Papadopoulos, J. Jen, A. de la Chapelle, K. W. Kinzler, B. Vogelstein, and P. Modrich. 1993. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell 75:1227-1236. [DOI] [PubMed] [Google Scholar]
- 34.Peretz, S., R. Jensen, R. Baserga, and P. M. Glazer. 2001. ATM-dependent expression of the insulin-like growth factor-I receptor in a pathway regulating radiation response. Proc. Natl. Acad. Sci. USA 98:1676-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reisdorph, R., and R. Lindahl. 1998. Hypoxia exerts cell-type-specific effects on expression of the class 3 aldehyde dehydrogenase gene. Biochem. Biophys. Res. Commun. 249:709-712. [DOI] [PubMed] [Google Scholar]
- 36.Reynolds, T. Y., S. Rockwell, and P. M. Glazer. 1996. Genetic instability induced by the tumor microenvironment. Cancer Res. 56:5754-5757. [PubMed] [Google Scholar]
- 37.Rice, G. C., C. Hoy, and R. T. Schimke. 1986. Transient hypoxia enhances the frequency of dihydrofolate reductase gene amplification in Chinese hamster ovary cells. Proc. Natl. Acad. Sci. USA 83:5978-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rockwell, S. 1977. In vivo-in vitro tumor systems: new models for studying the response of tumours to therapy. Lab. Anim. Sci. 27:831-851. [PubMed] [Google Scholar]
- 39.Rockwell, S. 1997. Oxygen delivery: implications for the biology and therapy of solid tumors. Oncol. Res. 9:383-390. [PubMed] [Google Scholar]
- 40.Rosenberg, S. M. 2001. Evolving responsively: adaptive mutation. Nat. Rev. Genet. 2:504-515. [DOI] [PubMed] [Google Scholar]
- 41.Rountree, M. R., K. E. Bachman, J. G. Herman, and S. B. Baylin. 2001. DNA methylation, chromatin inheritance, and cancer. Oncogene 20:3156-3165. [DOI] [PubMed] [Google Scholar]
- 42.Ryan, H. E., M. Poloni, W. McNulty, D. Elson, M. Gassmann, J. M. Arbeit, and R. S. Johnson. 2000. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 60:4010-4015. [PubMed] [Google Scholar]
- 43.Semenza, G. L. 2000. Expression of hypoxia-inducible factor 1: mechanisms and consequences. Biochem. Pharmacol. 59:47-53. [DOI] [PubMed] [Google Scholar]
- 44.Semenza, G. L. 2000. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J. Appl. Physiol. 88:1474-1480. [DOI] [PubMed] [Google Scholar]
- 45.Semenza, G. L. 2001. HIF-1, O(2), and the 3 PHDs: how animal cells signal hypoxia to the nucleus. Cell 107:1-3. [DOI] [PubMed] [Google Scholar]
- 46.Semenza, G. L. 2000. Hypoxia, clonal selection, and the role of HIF-1 in tumor progression. Crit. Rev. Biochem. Mol. Biol. 35:71-103. [DOI] [PubMed] [Google Scholar]
- 47.Sook Kim, M., J. Hyen Baek, M. K. Bae, and K. W. Kim. 2001. Human rad21 gene, hHR21(SP), is downregulated by hypoxia in human tumor cells. Biochem. Biophys. Res. Commun. 281:1106-1112. [DOI] [PubMed] [Google Scholar]
- 48.Strand, M., T. A. Prolla, R. M. Liskay, and T. D. Petes. 1993. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365:274-276. (Erratum, 368:569, 1994.) [DOI] [PubMed]
- 49.Sutherland, R. M. 1998. Tumor hypoxia and gene expression—implications for malignant progression and therapy. Acta Oncol. 37:567-574. [DOI] [PubMed] [Google Scholar]
- 50.Wang, G., D. D. Levy, M. M. Seidman, and P. M. Glazer. 1995. Targeted mutagenesis in mammalian cells mediated by intracellular triple helix formation. Mol. Cell. Biol. 15:1759-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, G. L., and G. L. Semenza. 1993. Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J. Biol. Chem. 268:21513-21518. [PubMed] [Google Scholar]
- 52.Wang, G. L., and G. L. Semenza. 1993. Desferrioxamine induces erythropoietin gene expression and hypoxia-inducible factor 1 DNA-binding activity: implications for models of hypoxia signal transduction. Blood 82:3610-3615. [PubMed] [Google Scholar]
- 53.Warren, S. M., D. S. Steinbrech, B. J. Mehrara, P. B. Saadeh, J. A. Greenwald, J. A. Spector, P. J. Bouletreau, and M. T. Longaker. 2001. Hypoxia regulates osteoblast gene expression. J. Surg. Res. 99:147-155. [DOI] [PubMed] [Google Scholar]
- 54.Wilkinson, D., J. K. Sandhu, J. W. Breneman, J. D. Tucker, and H. C. Birnboim. 1995. Hprt mutants in a transplantable murine tumour arise more frequently in vivo than in vitro. Br. J. Cancer 72:1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams, K. J., R. L. Cowen, and I. J. Stratford. 2001. Hypoxia and oxidative stress. Tumour hypoxia—therapeutic considerations. Breast Cancer Res. 3:328-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood, R. D. 1996. DNA repair in eukaryotes. Annu. Rev. Biochem. 65:135-167. [DOI] [PubMed] [Google Scholar]
- 57.Xiong, Z., and P. W. Laird. 1997. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 25:2532-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yao, X., A. B. Buermeyer, L. Narayanan, D. Tran, S. M. Baker, T. A. Prolla, P. M. Glazer, R. M. Liskay, and N. Arnheim. 1999. Different mutator phenotypes in Mlh1- versus Pms2-deficient mice. Proc. Natl. Acad. Sci. USA 96:6850-6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshida, M., M. Kijima, M. Akita, and T. Beppu. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265:17174-17179. [PubMed] [Google Scholar]
- 60.Young, S. D., R. S. Marshall, and R. P. Hill. 1988. Hypoxia induces DNA overreplication and enhances metastatic potential of murine tumor cells. Proc. Natl. Acad. Sci. USA 85:9533-9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuan, J., and P. M. Glazer. 1998. Mutagenesis induced by the tumor microenvironment. Mutat. Res. 400:439-446. [DOI] [PubMed] [Google Scholar]
- 62.Yuan, J., L. Narayanan, S. Rockwell, and P. M. Glazer. 2000. Diminished DNA repair and elevated mutagenesis in mammalian cells exposed to hypoxia and low pH. Cancer Res. 60:4372-4376. [PubMed] [Google Scholar]
- 63.Zhu, H., and H. F. Bunn. 2001. Signal transduction. How do cells sense oxygen? Science 292:449-451. [DOI] [PMC free article] [PubMed] [Google Scholar]