Abstract
The control of the subcellular localization of cell cycle regulators has emerged as a crucial mechanism in the regulation of cell division. In the present work, we have characterized the function of the karyopherin Msn5p in the control of the cell cycle of Saccharomyces cerevisiae. Phenotypic analysis of the msn5 mutant revealed an increase in cell size and a functional interaction between Msn5p and the cell cycle transcription factor SBF (composed of the Swi4p and Swi6p proteins), indicating that Msn5p is involved in Start control. In fact, we have shown that the level of Cln2p protein is drastically reduced in an msn5 mutant. The effect on CLN2 expression is mediated at a transcriptional level, Msn5p being necessary for proper SBF-dependent transcription. On the contrary, loss of MSN5 has no effect on the closely related transcription factor MBF (composed of the Mbp1p and Swi6p proteins). Regulation of SBF by Msn5p is exerted by control of the localization of the regulatory subunit Swi6p. Swi6p shuttles between the nucleus and the cytoplasm during the cell cycle, and we have found that Msn5p is required for Swi6p export from the nucleus during the G2-M phase. What is more important, we have demonstrated that export of Swi6p to the cytoplasm is required for SBF activity, providing evidence for a functional switch of Swi6p linked to its nucleocytoplasmic shuttling during the cell cycle.
To maintain the temporal order of events, the cell cycle has to be tightly regulated. Progression through the cell cycle is governed primarily by the periodic activation and inactivation of different cyclin-dependent kinases (CDK). Multiple mechanisms have evolved to fine-tune CDK activity, including the phosphorylation of specific residues and control of the synthesis and destruction of their regulators (reviewed in reference 32). Recently, a new dimension was added to our understanding of the cell cycle in revealing the importance of the control of the subcellular localization of some cell cycle regulators (reviewed in references 35 and 42). Evidence has accumulated emphasizing that this level of spatial regulation, which includes the sequestration of those regulators in specific subcellular locations until they are required, or their dynamic trafficking through different compartments or structures is crucial for the proper temporal control of processes like the onset of DNA replication, mitosis, or cytokinesis.
In the G1 phase, in a process called Start in yeast or restriction point in mammals, cells decide whether or not to initiate a new round of cell division based on internal and external cues. The key process in Start is the triggering by specific CDK of a burst of new gene expression (reviewed in references 4 and 12). The cyclins and transcription factors involved in this process show specific patterns of subcellular localization, suggesting that mechanisms of spatial regulation are important for this transition of the cell cycle. In the yeast Saccharomyces cerevisiae, the execution of Start requires activation of the Cdc28p kinase through its association with the G1-phase-specific cyclin Cln3p when cells reach a critical size and environmental conditions are appropriate. This kinase induces, then, a transcription program (15, 38). More than 200 genes are periodically expressed at this stage (37). Among them, the cyclin CLN1 and CLN2 genes are transcribed, leading to an abrupt accumulation of Cln1p-Cdc28p and Cln2p-Cdc28p kinase activities, which in turn trigger all the early events of the cell cycle: initiation of DNA replication, spindle pole body duplication, and bud emergence.
The transcription factors mediating gene expression at Start are SBF and MBF (reviewed in reference 10). Both are heterodimers consisting of a common regulatory protein, Swi6p, and one of two homologous specific DNA-binding proteins: Swi4p in SBF and Mbp1 in MBF. SBF binds to the SCB element and controls the expression, among other things, of the CLN1 and CLN2 genes and genes encoding the proteins involved in morphogenesis. MBF recognizes the MCB element and regulates the periodic expression of many genes involved in DNA metabolism and the cyclin CLB5 and CLB6 genes. Considerable cross-talk between these factors is believed to occur. The importance of this transcription program in the late G1 phase is emphasized by the lethality of the swi4 swi6 and swi4 mbp1 double mutants. The molecular mechanism of the simultaneous activation of SBF- and MBF-dependent expression by Cln3p-Cdc28p is not yet completely understood, although, at least for SBF, it involves the binding of the transcription factor to its target promoters early in the G1 phase and the Cln3p-Cdc28p-mediated recruitment of RNA polymerase II onto SBF-containing promoters in the late G1 phase (11, 24). Both SBF- and MBF-dependent transcription events are silenced during the G2 phase. However, this inactivation is accomplished by different ways: SBF is displaced from the DNA by Clb-Cdc28p kinases, whereas MBF activity is repressed independently of Clb cyclins (2, 24).
The subcellular localization of several proteins involved in Start has been analyzed. Cln3p is primarily nuclear, whereas Cln2p is localized both in the cytoplasm and in the nucleus (16, 30, 31). Importantly, this different pattern of localization confers functional specificity to the respective Cdc28p kinase complexes. Regarding the SBF transcription factor, Swi4p is located in the nucleus during the entire cell cycle (3); in contrast, Swi6p shuttles between the nucleus and the cytoplasm, being nuclear for most of the cycle, aside from the period from G2 phase until the end of mitosis in which it is relocated to the cytoplasm (36, 39). Nuclear import of Swi6p is mediated by a nuclear localization signal that is negatively regulated by the phosphorylation of Ser160.
Active transport between the nucleus and the cytoplasm is mediated by the recognition of specific nuclear import and export signals by soluble transport receptors known as karyopherins (reviewed in reference 28). The Msn5p protein is one of the yeast karyopherins (17). It is involved in different cellular processes, like phosphate and carbon source metabolism, stress response, calcium signaling, mating response, pseudohyphal differentiation, and cell cycle control, Msn5p acting as a nuclear export receptor for several proteins, including the transcription factors Pho4p (23), Mig1p (14), Msn2p-Msn4p (18), Crz1p (9), Rtg1p-Rtg3p (25), and other proteins like the Cln-Cdc28p inhibitor Far1p (6) and the mitogen-activated protein kinase cascade scaffold protein Ste5p (29). Moreover, Msn5p is able to function bidirectionally since it acts as the importin of replication protein A (43). At least for Pho4p, Mig1p, and Crz1p, transport by Msn5p requires their phosphorylation, suggesting that Msn5p proteins specifically transport phosphoproteins.
In the present work, we studied the role of Msn5p in cell cycle control. We showed that Msn5p is required for the proper execution of Start, controlling the SBF (but not MBF) transcription factor. Regulation of SBF is mediated by the control of Swi6p subcellular localization, Msn5p acting as an export factor for Swi6p during the G2 and M phases of the cell cycle. Importantly, we demonstrated that shuttling of Swi6p between the nucleus and the cytoplasm during the cell cycle is necessary for SBF transcriptional activity.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The yeast strains used in this study are shown in Table 1. Strains containing the msn5Δ3::HIS3 or msn5Δ2::HIS3 disruption cassettes were obtained by using plasmid p335-Δ3::HIS3 or p335-Δ2::HIS3, respectively (1). The JCY167 (swi4::LEU2), JCY220 (swi6::TRP1), and JCY296 (cln2::LEU2) strains were derived from W303-1A by using the disruption cassette plasmids Bd194 and Bd197 (a gift from L. Breeden) and pcln2::LEU2 (gift from S. I. Reed), respectively. JCY450 was derived from PJ69-4A by disrupting the SWI4 gene. JCY324 was derived from the CLM240 strain (a gift from E. Herrero) by substituting the MSN5 promoter with the tetO7 promoter by integrating a DNA fragment amplified from plasmid pCM225 (a gift from E. Herrero). The JCY335 and JCY339 strains were derived from JCY324 by using the disruption cassette plasmids Bd197 and pswi4::ADE2 (a gift from L. H. Johnston). Tagging of the Swi6p protein at the C terminus with three copies of the hemagglutinin (HA) epitope in the W303-1A and CY19 (a gift from L. H. Johnston) strains to render the JCY114 and JCY355 strains, respectively, was achieved by integrating a DNA fragment amplified from plasmid pFA6a-3HA-kanMX6 (a gift from J. R. Pringle). The JCY287 and JCY353 strains were derived from JCY114 by disrupting the MSN5 or SWI4 genes as described above. Strains containing the Swi6p protein fused to an active (JCY438 and JCY442) or inactive (JCY440 and JCY445) version of the nuclear export signal (NES) from the PKI protein were obtained by including the appropriate sequence (30) in the 5′ primer used in the PCR amplification of the pFA6a-3HA-TRP1 template. Tagging of the Swi4p protein at the C terminus with three copies of the HA epitope in W303-1A and JCY196 to render the JCY331 and JCY333 strains, respectively, was achieved by integrating a DNA fragment amplified from plasmid pFA6a-3HA-TRP1.
TABLE 1.
Yeast strains
| Yeast | Phenotype |
|---|---|
| W303-1a | MATaade2 trp1 leu2 his3 ura3 can1 |
| JCY196 | MATaade2 trp1 leu2 his3 ura3 can1 msn5Δ3::HIS3 |
| JCY179 | MATaade2 trp1 leu2 his3 ura3 can1 msn5Δ2::HIS3 |
| BAM1-11Aa | MATα ade2 trp1 leu2 his3 ura3 met swi4ts |
| JCY047 | MATα ade2 trp1 leu2 his3 ura3 met swi4ts msn5Δ3::HIS3 |
| JCY167 | MATaade2 trp1 leu2 his3 ura3 can1 swi4::LEU2 |
| JCY220 | MATaade2 trp1 leu2 his3 ura3 can1 swi6::TRP1 |
| CML240b | MATaade2 trp1 leu2 his3 ura3 can1 CMV:tetR′SSN6-LEU2 |
| JCY324 | MATa ade2 trp1 leu2 his3 ura3 can1 CMV:tetR′SSN6-LEU2 tetO7:MSN5 |
| JCY335 | MATaade2 trp1 leu2 his3 ura3 can1 CMV:tetR′SSN6-LEU2 tetO7:MSN5 swi6::TRP1 |
| JCY339 | MATaade2 trp1 leu2 his3 ura3 can1 CMV:tetR′SSN6-LEU2 tetO7:MSN5 swi4::ADE2 |
| JCY114 | MATaade2 trp1 leu2 his3 ura3 can1 SWI6-3HA-kanMX6 |
| JCY287 | MATaade2 trp1 leu2 his3 ura3 can1 msn5Δ3::HIS3 SWI6-3HA-kanMX6 |
| CY19a | MATα ade2-1 trp1 leu2 his3 ura3 met can1 ho:lacZ mbp1::URA3 |
| JCY355 | MATα ade2-1 trp1 leu2 his3 ura3 met can1 ho:lacZ mbp1::URA3 SWI6-3HA-kanMX6 |
| JCY353 | MATα ade2 trp1 leu2 his3 ura3 met can1 swi4::LEU2 SWI6-3HA-kanMX6 |
| JCY438 | MATaade2 trp1 leu2 his3 ura3 can1 SWI6NES-3HA-TRP1 |
| JCY440 | MATaade2 trp1 leu2 his3 ura3 can1 SWI6nesi-3HA-TRP1 |
| JCY442 | MATaade2 trp1 leu2 his3 ura3 can1 msn5Δ3::HIS3 SWI6NES-3HA-TRP1 |
| JCY444 | MATaade2 trp1 leu2 his3 ura3 can1 msn5Δ3::HIS3 SWI6nesi-3HA-TRP1 |
| JCY331 | MATaade2 trp1 leu2 his3 ura3 can1 SWI4-3HA-TRP1 |
| JCY333 | MATaade2 trp1 leu2 his3 ura3 can1 msn5Δ3::HIS3 SWI4-3HA-TRP1 |
| MT244a | MATaade2 trp1 leu2 his3 ura3 can1 cln3::URA3 |
| JCY296 | MATaade2 trp1 leu2 his3 ura3 can1 cln2::LEU2 |
| PJ69-4Ac | MATatrp1 leu2 ura3 his3 gal4 gal80 GAL2:ADE2 GAL1:HIS3 GAL7:lacZ |
| JCY450 | MATatrp1 leu2 ura3 his3 gal4 gal80 GAL2:ADE2 GAL1:HIS3 GAL7:lacZ swi4::LEU2 |
A gift from L. H. Johnston.
A gift from E. Herrero.
A gift from E. A. Craig.
Centromeric plasmids pCM137, pCM239, and pCM194 containing HA-tagged versions of Cln1p, Cln2p, and Cln3p, respectively, and plasmids pCM166 (ptetO:CLN3 henceforth) and pCM250 (ptetO:CLN2 henceforth) overexpressing the CLN3 and CLN2 genes under the control of the tetO2 promoter were a gift from M. Aldea. Plasmids used in the expression analysis assays were derived from pLGΔ178, which contains a minimal promoter from the yeast cytochrome c gene fused to the Escherichia coli lacZ gene. pΔ5′728 (pCLN2:lacZ henceforth), which contains a fragment from positions −728 to −256 of the CLN2 promoter in front of the lacZ gene, was a gift from C. Wittenberg; pSCB:lacZ and pMCB:lacZ, which contain a fragment from positions −507 to −367 of the HO promoter bearing three SCB elements and a synthetic oligonucleotide with three MCB elements, respectively, were a gift from L. H. Johnston. Plasmid pBD-MSN5, used in the two-hybrid assay expressing Msn5p fused to the Gal4p DNA-binding domain, was a gift from M. Johnston.
Yeast cells were grown on standard yeast extract-peptone-dextrose (YPD) or YPGal rich medium or synthetic dextrose and SGal minimal media supplemented as required. To repress the tetO7 promoter, doxycycline was added to a concentration of 5 μg/ml.
Western blot analysis.
Approximately 108 cells were collected, incubated at 95°C for 5 min, resuspended in 15 μl of 5 M urea, and broken by vigorous shaking in the presence of glass beads. Extracts were supplemented with 50 μl of 2% sodium dodecyl sulfate (SDS)-0.125 M Tris-HCl (pH 6.8), incubated at 95°C for 2 min, and microcentrifuged. Protein concentration in the supernatant was determined by a Bio-Rad protein assay as indicated by the manufacturer. Approximately 25 μg of protein was resolved by SDS-polyacrylamide gel electrophoresis. After transfer to nitrocellulose filters, HA-tagged proteins were detected with the 12CA5 monoclonal antibody (Roche) and the enhanced chemiluminescence Western blotting analysis system (Amersham Biosciences) following the manufacturer's directions.
Indirect immunofluorescence.
Approximately 108 cells were fixed in growth medium with 37% formaldehyde (1:10 dilution) for 60 min with agitation. After mild sonication, cells were collected, washed with buffer A (1.2 M sorbitol, 0.1% β-mercaptoethanol, 50 mM KH2PO4 [pH 7.0]), and incubated at 37°C for 1 to 2 h in 0.5 ml of buffer A containing 100 μg of zimolyase 20-T/ml. Spheroplasts were collected, washed with buffer A, resuspended in 20 μl of a solution (8 μg of rat anti-HA antibody 3F10 [Roche]/ml in phosphate-buffered saline [PBS] containing 0.1% bovine serum albumin), and incubated overnight at 4°C. After washing with PBS, PBS containing 0.1% NP-40, and PBS, cells were incubated with the secondary antibody Alexa 546-labeled anti-rat antibody (Molecular Probes) as indicated for the primary antibody. DNA was stained at the end of the process by resuspending spheroplasts in PBS containing 1 μg of DAPI (4′,6′-diamidino-2-phenylindole; Sigma Inc.)/ml.
Chromatin immunoprecipitation analysis.
Approximately 5 × 108 cells were fixed in growth medium by adding formaldehyde to a final concentration of 1%, followed by a short incubation at room temperature for 20 min and overnight incubation with agitation at 4°C. Cells were washed four times with TBS (20 mM Tris [pH 7.5], 150 mM NaCl), resuspended in lysis buffer (50 mM HEPES [pH 7.5], 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride, complete protease inhibitors from Roche), and broken with glass beads. Lysis buffer was supplemented to a final volume of 500 μl, and samples were sonicated (four pulses of 1 min each) and microcentrifuged. Protein concentration was determined and adjusted if necessary in order to use the same amount of total protein in each sample. An aliquot of 50 μl was collected as a control of whole-cell extract and was supplemented with 200 μl of Tris-EDTA (TE)-1% SDS. HA-tagged proteins were immunoprecipitated by the addition of anti-HA Affinity Matrix (Roche) and incubation at 4°C for 7 h with agitation. Samples were successively washed twice for 5 min at 4°C with 1 ml of lysis buffer, lysis buffer containing 500 mM NaCl, LiCl buffer (10 mM Tris [pH 8.0], 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA), and TE. Elution of the retained proteins was done in two steps by adding 100 μl of TE-1% SDS or TE-0.67% SDS in each step and incubating at 65°C for 15 min. For the reversion of cross-linking, samples were incubated at 65°C for 15 h. DNA for PCRs was purified by proteinase K digestion, phenol extraction, and ethanol precipitation. The oligonucleotides used amplified DNA fragments from the URA3, MAT, RNR1 and CLN2 promoters (primer sequences are available upon request).
Miscellaneous.
Cell size was analyzed in exponentially growing cells after brief sonication in the Particle Count and Size Analyzer Z2 (Coulter Inc.). DNA content was determined by fluorescence-activated cell sorter (FACS) analysis with an EPICS XL (Coulter Inc.) flow cytometer as described previously (20). Northern analysis and β-galactosidase activity assays were performed as described previously (20).
RESULTS
Msn5p plays a role in Start.
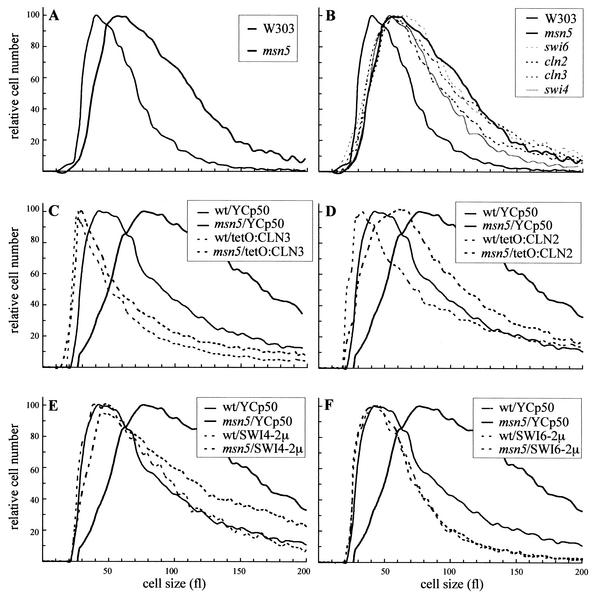
Previous work in our group has revealed a connection between the karyopherin Msn5p and cell cycle control at the G1-S phase transition (1). In order to obtain insights into the molecular function of Msn5p in cell cycle regulation, the phenotype of an msn5 mutant was analyzed. No important differences in the growth rate or budding index compared to that in the isogenic wild-type strain were detected. FACS analysis and microscopic observation of DAPI-stained cells showed no defects in DNA replication or mitosis. On the other hand, it is well established that the inactivation or overexpression of regulators of Start produces an altered cell size. Because of this, the size of msn5 cells in an exponentially growing culture was determined. As shown in Fig. 1A, inactivation of MSN5 caused a drastic increase in cell size. Strikingly, the alteration of cell size was very similar to that observed when Cln3p, Cln2p, Swi6p, or Swi4p was inactivated (Fig. 1B). Moreover, overexpression of the CLN3, CLN2, SWI6, and SWI4 genes rescued the cell size defect of the msn5 mutant (Fig. 1C through F). These results strongly suggest that Msn5p plays a role in the control of Start.
FIG. 1.
Analysis of the cell size in an msn5 mutant strain. (A and B) The sizes of the cells in exponentially growing cultures on YPD medium of wild-type (W303-1a), msn5 (JCY196), swi6 (JCY220), swi4 (JCY167), cln2 (JCY296), and cln3 (MT244) strains was determined in a Coulter cell counter. (C through F) Wild-type (wt) and msn5 mutant strains transformed with a control vector (YCp50), plasmids overexpressing CLN3 (ptetO:CLN3) or CLN2 (ptetO:CLN2), or multicopy plasmids containing the SWI6 or SWI4 gene were grown on synthetic medium, and cell sizes were determined. Graph are for the moving averages from at least four independent cultures of each of the strains.
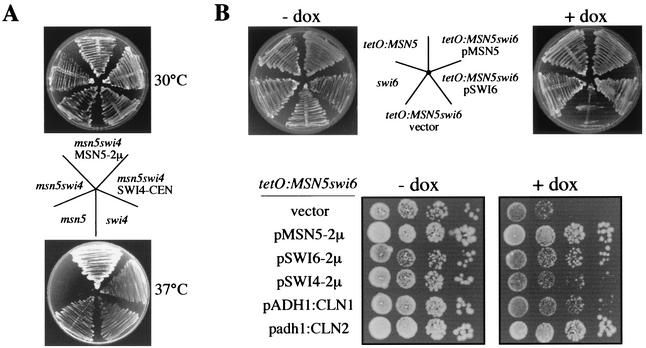
Based on tetrad analysis, a synthetic lethality between the msn5 and swi4 or swi6 mutations has been suggested previously (1). To confirm this interaction and go further in the characterization of the role of Msn5p in cell cycle control, strains combining the msn5 and swi6 or swi4 mutation were constructed and their phenotypes were analyzed. First, the MSN5 gene was deleted in a strain containing a thermosensitive allele of the SWI4 gene (swi4ts) and growth at 37°C was tested. In contrast to the swi4ts and msn5 single mutants, the msn5 swi4ts strain failed to grow (Fig. 2A). Lethality was suppressed by the reintroduction of a plasmid containing either the SWI4 or MSN5 gene. This result demonstrated a synthetic interaction between the msn5 and swi4 mutation. Analysis of the terminal phenotype of the msn5 swi4ts strain at the restrictive temperature indicated that cells were not blocked at a specific stage of the cell cycle but, however, that morphogenetic defects were evident. Thus, after 6 h at 37°C, more that 20% of particles corresponded to groups of cells that were not able to complete cytokinesis. Moreover, cell lysis was apparent with the accumulation of cell ghosts and debris. These phenotypes were also observed in the parental swi4ts strain, although to a much smaller extent (for instance, groups of cells represented only 4% of total cells), in agreement with the well-known function of SBF in both cytokinesis and the maintenance of cell integrity (20, 21). In conclusion, the results indicated that the inactivation of MSN5 aggravates the defects associated with SBF dysfunction.
FIG. 2.
Synthetic lethality between mutation in Msn5p and SBF components. (A) The swi4ts (BAM1-11A), msn5 (JCY196), and msn5 swi4ts (JCY047) strains, and the last transformed with a plasmid containing either the SWI4 or MSN5 gene, were streaked onto a YPD plate. Plates were incubated for 3 days at 30 or 37°C. (B) Upper panel, the swi6 (JCY220), tetO7:MSN5 (JCY324), and the tetO7:MSN5 swi6 (JCY335) strains, and the last transformed with a plasmid containing either the SWI6 or MSN5 gene, were streaked onto YPD plates with or without doxycycline and incubated at 28°C for 3 days. Lower panel, 10-fold serial dilutions from exponentially growing cultures of the tetO7:MSN5 swi6 strain transformed with a control vector, a multicopy plasmid containing the MSN5, SWI6, or SWI4 gene, a plasmid with the CLN1 gene under the control of the strong S. cerevisiae ADH1 promoter, or a plasmid bearing the CLN2 gene under the control of the mild Schizosaccharomyces pombe adh promoter were spotted onto medium with or without doxycycline (dox) and incubated at 30°C for 3 days.
To explore further the interaction between Msn5p and SBF, a strain containing a deletion of the SWI6 gene and the MSN5 gene expressed under the control of the doxycycline-repressed tetO7 promoter was constructed. When cells were transferred to a medium containing doxycycline, i.e., repressing expression of the MSN5 gene, the tetO7:MSN5 swi6 strain failed to grow, in contrast to the swi6 and tetO7:MSN5 strains (Fig. 2B). This growth defect was rescued by the reintroduction of a plasmid containing either the SWI6 or MSN5 gene. This observation demonstrated a synthetic lethal interaction between the msn5 and swi6 mutations. Investigation of the terminal phenotype of tetO7:MSN5 swi6 cells 10 h after the transfer to doxycycline-containing medium revealed an important increase in the percentage of unbudded cells, rising from 32 to 68% of the total number of cells. FACS analysis indicated that DNA replication was not blocked, and, in fact, cells with a 4C DNA content were detected (unpublished data). Observation of nuclei by DAPI staining and fluorescence microscopy showed the occurrence of binucleated unbudded cells (unpublished data). These phenotypic traits indicated a defect in the morphogenetic processes of Start that led to budding, a function that is dependent on SBF (5, 13). A similar result regarding the growth defect and terminal phenotype was obtained in a tetO7:MSN5 swi4 strain (unpublished data), consistent with the result described above and pointing to a functional link between Msn5p and SBF function.
SBF regulates transcription of the G1 cyclin genes CLN1 and CLN2, so it is possible that the phenotype of the strains described above could be related to a defect in Cln function. Neither the overexpression nor ectopic expression of CLN1, CLN2, or CLN3 suppressed the conditional lethal phenotype of the msn5 swi4ts and tetO7:MSN5 swi4 strains. On the contrary, ectopic expression of CLN2 rescued the growth defect of the tet07:MSN5 swi6 strain (Fig. 2B). Suppression of the lethality, although to a minor extent, was also observed when CLN1 or SWI4 genes were overexpressed. This result suggests that Msn5p could be controlling the level of Cln cyclins.
Msn5p controls SBF-dependent transcription.
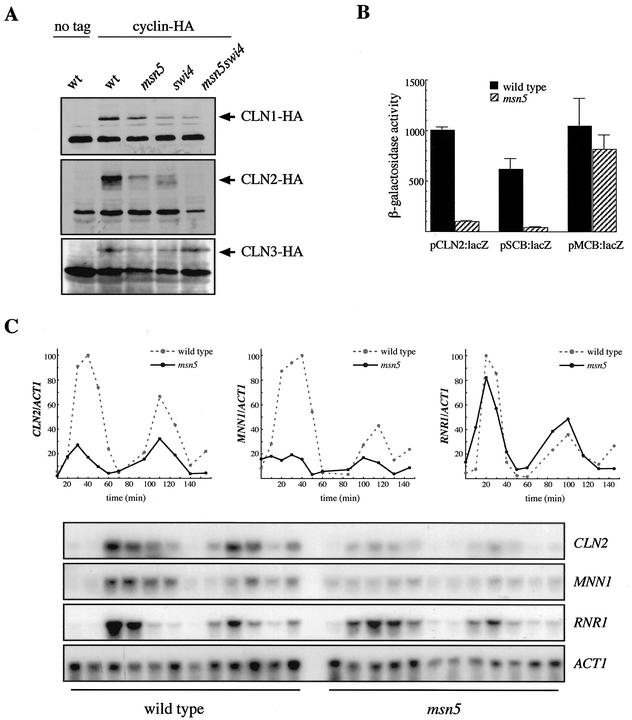
In order to check whether Msn5p regulates the expression of the G1 phase cyclins, Cln1p, Cln2p, and Cln3p were tagged with the HA epitope and the level of cyclins in msn5 mutant strains was determined by Western blot analysis. No significant differences in the levels of Cln1p or Cln3p were detected between the msn5 mutant strains and the respective wild-type and swi4ts parental strains (Fig. 3A). By contrast, inactivation of msn5 caused a severe reduction in Cln2p protein level in both the wild type and the swi4ts background. This result indicated that Msn5p is acting at Start at least by controlling the level of Cln2p cyclin.
FIG. 3.
Analysis of the regulation of gene expression by Msn5p. (A) Cln protein levels in extracts from wild-type (wt; W3031-a), msn5 (JCY196), swi4ts (BAM1-11A), and msn5 swi4ts (JCY047) strains bearing a centromeric plasmid containing the HA-tagged CLN1 (pCM137), CLN2 (pCM239), or CLN3 (pCM194) gene were examined by Western analysis. Control extracts from the wild-type strain transformed with an empty vector were included. A nonspecific band that cross-reacts with the 12CA5 antibody is shown as the loading control. (B) β-Galactosidase activity in extracts from wild-type (W303-1A) and msn5 (JCY179) strains transformed with pLGL-derived plasmids containing the lacZ reporter gene under the control of a fragment from the CLN2 promoter (pCLN2:lacZ), a fragment from the HO promoter with three SCB elements (pSCB:lacZ), or a synthetic oligonucleotide with three MCB elements (pMCB:lacZ) is shown. Values are derived from more than five extracts from at least three independent transformants. (C) Expression of the CLN2, MNN1, and RNR1 genes in α-factor-synchronized cultures of wild-type (W303-1A) and msn5 (JCY196) strains was studied by Northern analysis. Samples were collected at 0, 10, 20, 30, 40, 50, 60, 85, 100, 115, 130, and 145 min after release. ACT1 is shown as the loading control. Plots represent the amounts of CLN2, MNN1, and RNR1 mRNAs relative to that of ACT1 mRNA.
Regulation of Cln2p level could occur at the transcriptional or posttranscriptional level. To address this question, the expression of a fusion gene containing the β-galactosidase coding sequence under the control of the CLN2 promoter (plasmid pCLN2:lacZ) was studied. As shown in Fig. 3B, there is a 10-fold reduction in β-galactosidase activity in the msn5 mutant strain compared to that in the isogenic wild-type strain. A similar reduction was observed in mutants in the SWI4 or SWI6 genes (unpublished data). This indicates that Msn5p governs Cln2p production at a transcriptional level, probably by regulating SBF activity. This point was further characterized by Northern analysis, which confirms that CLN2 transcription was severely impaired in the msn5 mutant (Fig. 3C). In order to confirm whether Msn5p is controlling SBF function and that the effect of msn5 mutation is not specific for the CLN2 gene, the expression of another SBF-regulated gene, the MNN1 gene, was analyzed. As shown in Fig. 3C, MNN1 expression was also drastically reduced and periodic expression was altered in the msn5 mutant strain. In addition, the expression of the lacZ gene, driven by a fragment from the HO promoter that contains three SCB elements and whose function is totally dependent on SBF (plasmid pSCB:lacZ), was assayed. As observed for the pCLN2:lacZ plasmid, β-galactosidase expression from the pSCB:lacZ plasmid was also dramatically impaired in the msn5 mutant strain. These results confirm a control of SBF transcriptional activity by the Msn5p protein. It is interesting to point out that expression from the pSCB:lacZ is not totally abolished in the msn5 mutant, which suggests that SBF is not completely inactivated by the msn5 mutation.
Next, we explored the interaction between Msn5p and MBF transcription factor, closely related to SBF and also involved in transcriptional control at the G1-S phase boundary. In contrast to the lethality of the msn5 swi4 and msn5 swi6 double mutants, the msn5 mbp1 strain was viable without apparent defects. In addition, when a construct bearing a synthetic oligonucleotide containing MCB elements in front of the lacZ reporter gene (plasmid pMCB:lacZ) was used to test the functionality of MBF, no significant differences in β-galactosidase activities between the msn5 and wild-type strains were observed (Fig. 3B). Moreover, the expression of RNR1, a gene regulated by MBF, is not impaired in the msn5 mutant (Fig. 3C). Based on these results, it can be concluded that Msn5p is required specifically for proper SBF activity but does not affect MBF function.
Subcellular localization of Swi6p is controlled by Msn5p.
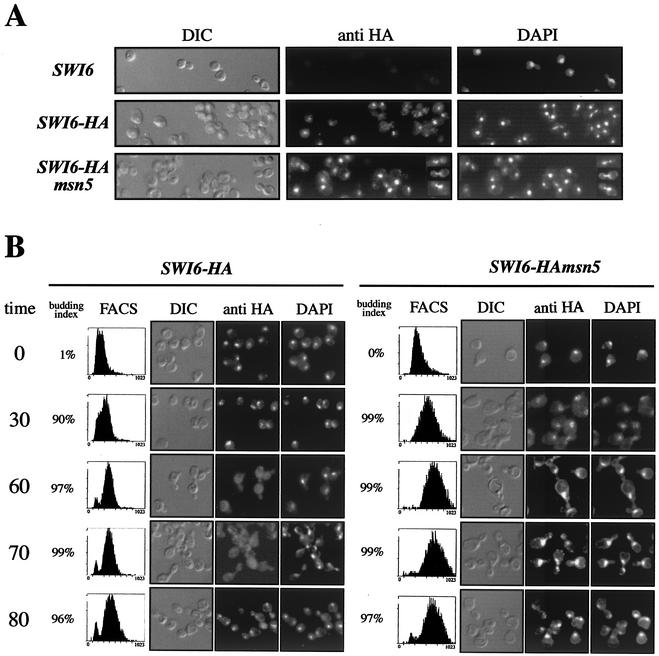
Msn5p is a karyopherin that has been implicated in the transport between the nucleus and the cytoplasm of different cargo proteins. On the other hand, it has been reported that Swi6p, the regulatory subunit of SBF, shuttles between the nucleus and the cytoplasm across the cell cycle. Because of this, we formulated the hypothesis that Msn5p could control SBF function by regulating the subcellular localization of Swi6p. To test this possibility, the Swi6p protein was tagged at the C terminus with the HA epitope. Confirmation of the tagging was obtained by Western analysis, which revealed no difference in the level of the Swi6p protein between the wild-type and msn5 strains (unpublished data). Tagging does not affect the functionality of the protein, as deduced from the expression of Swi6p-regulated reporter genes and cell size analysis (unpublished data). Location of Swi6p was determined by immunofluorescence in isogenic wild-type and msn5 mutant strains (Fig. 4A). In the case of wild-type cells, Swi6p protein was mainly nuclear in most but not all of the cells. As deduced from morphology and DAPI staining, cytoplasmic localization was observed in a period of the cell cycle between the G2 phase and the end of mitosis, in agreement with the reported cell-cycle-dependent regulation of the nuclear localization of Swi6p (36, 39). However, when msn5 cells were examined, Swi6p remained primarily nuclear over the entire cell cycle. The most dramatic example of this constitutive nuclear localization of Swi6p in the msn5 mutant was observed in cells undergoing anaphase. After a thorough screening, no anaphase cells in a wild-type culture showed a nuclear signal for Swi6p, as opposed to the strong nuclear signal observed in anaphase cells from the msn5 mutant (Fig. 4A).
FIG. 4.
Control of Swi6p localization by Msn5p. (A) Cells from asynchronous cultures of wild-type (JCY114) and msn5 (JCY287) strains expressing a HA-epitope-tagged Swi6p protein were assayed by indirect immunofluorescence as described in Materials and Methods. Differential interference-contrast (DIC) images, the Swi6p-HA indirect-fluorescence signal (anti-HA), and DAPI staining of DNA are shown. The first row corresponds to a control of the untagged wild-type strain. Several mitotic cells demonstrating the nuclear localization of Swi6p during mitosis in the msn5 mutant strain (see text) are included to the right of the corresponding pictures. (B) Exponentially growing cultures of the same strains were arrested with α-factor for 3 h and released. Samples were taken during the arrest and at 30, 60, 70, and 80 min after release and were analyzed for budding index, DNA content (by FACS analysis), and Swi6p localization (by indirect immunofluorescence). Representative cells are shown.
To gain more definitive evidence of the constitutive nuclear localization of Swi6p across the cell cycle in an msn5 mutant, an α-factor-synchronized culture was used (Fig. 4B). The α-factor-arrested cells showed nuclear localization in both the wild-type and msn5 strains. Thirty minutes after release, when cells were in S phase as deduced from FACS analysis, Swi6p was still nuclear in both strains. However, when cells were in the G2 phase (60 min after release), Swi6p remained mainly nuclear in the msn5 mutant, whereas in the wild-type cells, Swi6p was relocated to the cytoplasm. Ten minutes later, when cells were undergoing mitosis, Swi6p remained in the nucleus in msn5 cells in contrast to that observed in the wild-type strain, in which Swi6p was mainly cytoplasmic until reentering the nucleus at the end of mitosis (80 min after release). These results clearly demonstrate that the karyopherin Msn5p is involved in the regulation of the subcellular localization of the Swi6p transcription factor, Msn5p being required for the export of Swi6p to the cytoplasm during the G2-M phase.
Interaction between Msn5p and Swi6p.
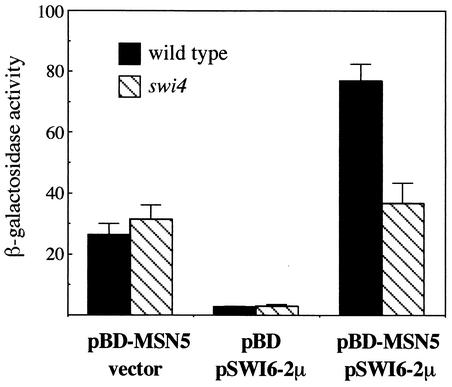
Our results strongly suggest that Msn5p is the exportin of Swi6p. To unequivocally confirm this, we tested a physical interaction between both proteins in vivo by two-hybrid assays. Considering that Swi6p has an intrinsic transcriptional activation function, the designed strategy involved overexpression of the Swi6p protein in the presence of Msn5p fused to the Gal4p DNA-binding domain and analysis of the expression of a GAL7:lacZ gene. As shown in Fig. 5, there was a significant increase in β-galactosidase activity when both the multicopy SWI6 and BD-MSN5 plasmids were present. This result demonstrates that Msn5p interacts with Swi6p in vivo, so the requirement for Msn5p in the export of Swi6p is not an indirect effect but Swi6p is the direct cargo of the karyopherin Msn5p.
FIG. 5.
Two-hydrid interaction assay between Msn5p and Swi6p. A plasmid expressing a fusion between Msn5p and the Gal4p DNA-binding domain (pBD-MSN5) or the corresponding empty vector (pBD) and a multicopy plasmid containing SWI6 (pSWI6-2μ) or the corresponding empty vector were introduced in the PJ69-4A strain and in JCY450 (swi4 mutant strain derived from PJ69-4A) in different combinations as indicated. The ability of the proteins to induce expression of a GAL7:lacZ gene was tested by measuring β-galactosidase activity in extracts from exponentially growing cells. Values are derived from more than eight extracts from at least three independent transformants.
Functionality of Swi6p depends on the proper control of its subcellular localization.
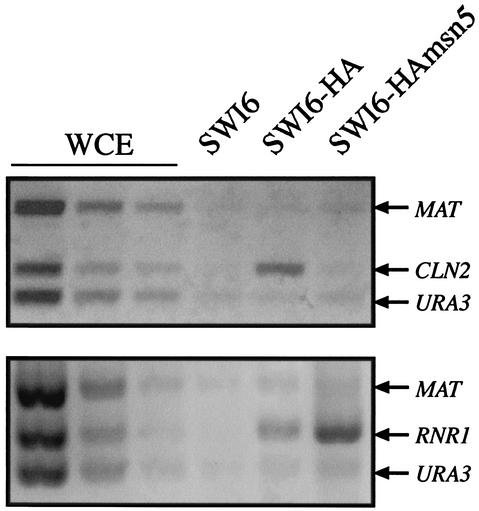
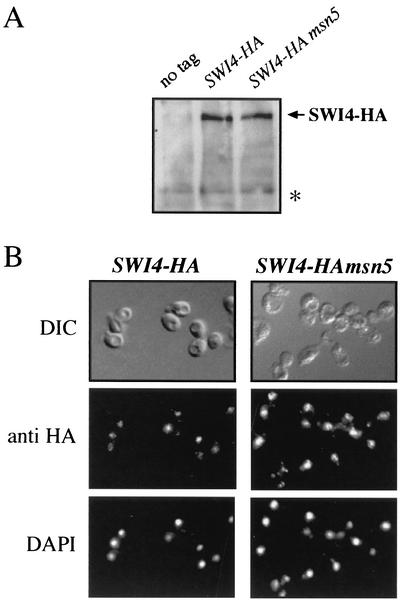
The result discussed above raises an apparent contradiction: how does one match the constitutive nuclear localization of Swi6p in the msn5 mutant with the impaired Swi6p-dependent transcription? One explanation is that Swi6p could be acting as a transcriptional repressor, but we were not able to obtain any data supporting this idea. A more likely explanation to this paradox is that the Swi6p protein that accumulated in the nucleus of msn5 mutant cells was not functional, being unable to activate transcription. To address this possibility, the binding capability of Swi6p to the CLN2 promoter was determined by chromatin immunoprecipitation assays. Because in wild-type cells, Swi6p is already bound to the promoter of its target genes through the G1 phase (24), cultures were synchronized with α-factor in order to arrive at conditions in which binding was optimized. As observed in Fig. 6, Swi6p immunoprecipitated from wild-type cells specifically purified a DNA fragment corresponding to the promoter of the CLN2 gene, indicating that, as expected, Swi6p is bound to the DNA. Remarkably, no specific purification of the DNA fragment containing the CLN2 promoter region compared to that of the control fragments was detected in Swi6p immunoprecipitates from msn5 cell extracts. The same result was observed by using asynchronous cultures (unpublished data). Thus, although in msn5 mutant cells, Swi6p was present in the nucleus, it was not bound to the CLN2 promoter. The lack of Swi6p binding to the CLN2 promoter could be due to a defect in the nuclear accumulation of Swi4p, the DNA-binding subunit of the SBF transcription factor. However, neither the protein level nor the import of Swi4p into the nucleus was affected in an msn5 mutant (Fig. 7), ruling out this possibility. In conclusion, these results strongly suggest that the export of Swi6p to the cytoplasm by Msn5p is essential for SBF activity.
FIG. 6.
Assay of the association of Swi6p with the CLN2 and RNR1 promoters. Cells from asynchronous cultures of wild-type (JCY114; lane SWI6-HA) and msn5 (JCY287; lane SWI6-HAmsn5) strains expressing a HA-epitope-tagged Swi6p protein were tested for Swi6p binding to the CLN2 and RNR1 promoters by chromatin immunoprecipitation assays. DNA samples were purified after cross-linking and immunoprecipitation and analyzed by PCR with primer pairs amplifying fragments from the CLN2, RNR1, MAT, or URA3 promoter. Control lanes show DNA amplified from different amounts of total DNA prior to purification (whole-cell extract [WCE]) and from the extract of a strain with untagged protein (W303-1a; lane SWI6)
FIG. 7.
Analysis of Swi4p protein level and localization in an msn5 mutant strain. (A) Swi4p protein levels in extracts from wild-type (JCY331; lane SWI4-HA) and msn5 (JCY333; lane SWI4-HAmsn5) strains expressing a HA-epitope-tagged Swi4p protein. Control extracts from the untagged wild-type strain were included. A nonspecific band labeled with an asterisk is shown as the loading control. (B) Cells from asynchronous cultures of the JCY331 (SWI4-HA) and JCY333 (SWI4-HAmsn5) strains were assayed by indirect immunofluorescence as described in the legend to Fig. 4. DIC, differential interference-contrast.
Binding of Swi6p to the RNR1 gene was also analyzed. Opposite to the result observed with the CLN2 gene, and consistent with the expression analysis reported above, Swi6p was detected bound to the RNR1 promoter in the msn5 mutant strain (Fig. 6). This result strongly reinforces the conclusion that Msn5p is controlling the SBF but not MBF transcription factor.
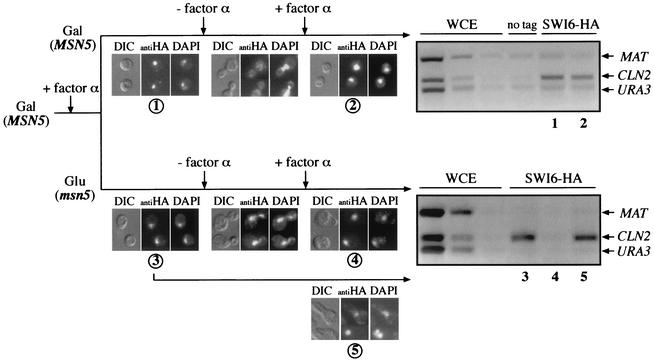
To investigate further the connection between the nucleocytoplasmic shuttling and functionality of Swi6p, a one-cycle experiment with a GAL:MSN5 gene was designed. This construct enables a rapid repression of the gene. Thus, 2 h after the addition of glucose, Msn5p protein is mostly depleted and the cells manifest defects associated with the loss of Msn5p (unpublished data). To carry out the experiment, galactose-grown cells were arrested by α-factor and, once blocked, half of the cells were transferred to glucose-containing medium. After two additional hours in the presence of α-factor, cells were washed and resuspended in galactose or glucose medium. Localization of Swi6p and binding of Swi6p to the CLN2 promoter was analyzed along a single cell cycle by immunofluorescence and chromatin immunoprecipitation, respectively. As expected, in control cells (cells grown on galactose), Swi6p was exported to the cytoplasm during the G2-M phase and the functionality of Swi6p was not affected, being bound to the CLN2 promoter in the next G1 phase (Fig. 8). On the contrary, when MSN5 was inactivated (cells grown on glucose), Swi6p remained nuclear during the whole cell cycle but, more important, it lost the capability of binding to the CLN2 promoter in the next G1 phase. Interestingly, in an aliquot of the cells shifted to glucose that was maintained in the presence of α-factor, Swi6p remained bound to the CLN2 promoter. These results indicate that loss of the functionality of Swi6p is not due to just the inactivation of Msn5p per se but that it requires progression through the cell cycle and, moreover, that this regulatory mechanism is acting in each cycle.
FIG. 8.
One cell cycle assay of the effect of blockage of nuclear export on Swi6p function. The JCY287 strain (SWI6-HAmsn5) transformed with the pGAL:MSN5 plasmid was grown on galactose medium and arrested by α-factor. Once blocked, culture was split and cells were transferred to YPGal or YPD medium containing α-factor. After two additional hours in the presence of α-factor, cells were released from the arrest, and after 40 min, α-factor was reinoculated to block progression in the next cycle. Samples were collected before the release and after the arrest in the next cycle (samples 1 and 2 for cells incubated on YPGal medium and samples 3 and 4 for cells grown on YPD medium). An aliquot of the cells transferred to YPD medium was maintained arrested during the whole experiment (sample 5). Samples were investigated for the subcellular localization of Swi6p by indirect immunofluorescence (as described in the legend to Fig. 4) and for association of Swi6p to the CLN2 promoter by chromatin immunoprecipitation (as described in the legend to Fig. 6). WCE, whole-cell extract; DIC, differential interference-contrast.
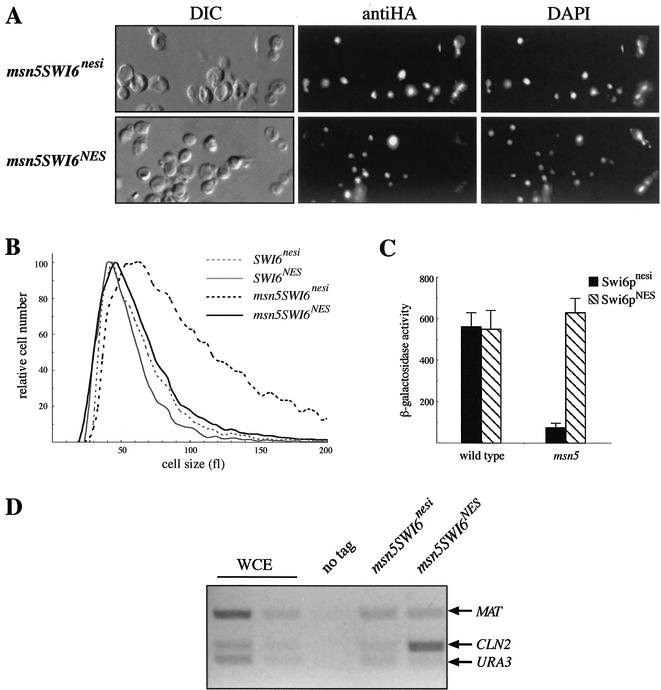
The previous results indicate that nuclear export is probably essential for Swi6p activity. If this is the case, then the forced nuclear export of Swi6p in an msn5 mutant should restore Swi6p activity. We tested this hypothesis by fusing a single copy of the PKI NES to the Swi6p protein. We hoped that, as described previously for Cln3p (30), this strategy would still allow nuclear accumulation of the Swi6p protein due to an intrinsic nuclear import signal (necessary to study Swi6p function). Activity of Swi6pNES was tested by analyzing the subcellular localization of the protein, the cell size, the expression of the SCB:lacZ gene, and the binding to the CLN2 promoter and comparing it with that of a control strain that contained the Swi6p protein fused to an inactive NES sequence (Swi6pnesi). In the wild-type strain, Swi6pNES was located mainly in the nucleus, unless in the G2-M period of the cell cycle (unpublished data), which indicates that the NES is not avoiding nuclear accumulation of the protein. Furthermore, analysis of the expression of the SCB:lacZ gene showed that the Swi6pNES protein is functional (Fig. 9C). In fact, binding of Swi6pNES to the CLN2 promoter was detected by chromatin immunoprecipitation assays (unpublished data). In the case of the msn5 mutant strain, the effect of the PKI NES in the localization of Swi6p was clearly observed: opposite to the control Swi6pnesi protein, which is mainly nuclear throughout the cell cycle, the Swi6pNES protein was relocated to the cytoplasm during the G2-M phase (Fig. 9A). This result confirmed that the NES sequence is forcing the nuclear export of Swi6p in the absence of MSN5. What it is most interesting is that this Msn5p-independent nuclear export was able to restore Swi6p function as tested by different methods: (i) msn5 mutant cells containing Swi6pNES have a normal cell size (Fig. 9B); (ii) whereas the SWI6nesi msn5 strain shows the low-level expression of the SCB:lacZ gene associated with the msn5 mutation, activity increases dramatically in the SWI6NES msn5 cells, reaching values similar to those observed in the wild-type strain (Fig. 9C); and (iii) Swi6pNES but not Swi6pnesi is bound to the CLN2 promoter in an msn5 mutant strain (Fig. 9D). All these results clearly demonstrate that inactivation of Swi6p in an msn5 mutant strain is due to the blockage of nucleocytoplasmic shuttling and that the nuclear export of Swi6p during the G2-M phase is essential for its functionality.
FIG. 9.
Effect of PKI NES-driven nuclear export on Swi6p function. The activity of a HA-epitope-tagged Swi6p protein fused to an active (Swi6pNES) or inactive (Swi6pnesi) version of the NES from the PKI protein was assayed in the wild type (JCY438 and JCY440 strains) and msn5 mutant (JCY442 and JCY444 strains) backgrounds. Subcellular localization of Swi6p (A), cell size (B), expression of the SCB:lacZ reporter gene (C), and binding of Swi6p to the CLN2 promoter (D) were analyzed as described previously. WCE, whole-cell extract; DIC, differential interference-contrast.
Nucleocytoplasmic shuttling of Swi6p requires the presence of Swi4p.
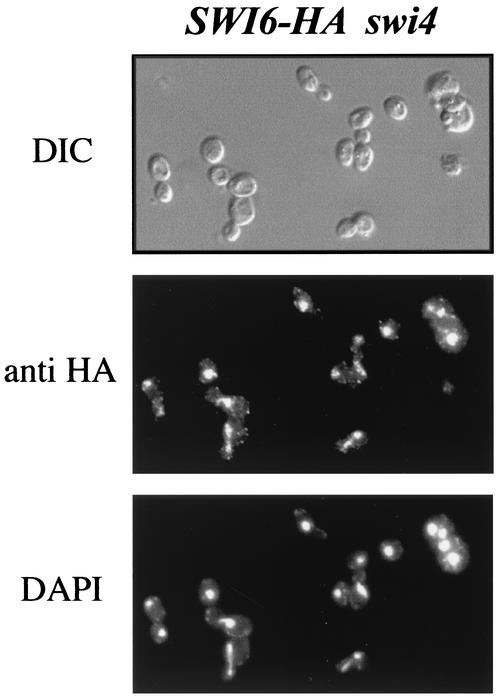
Swi6p forms, together with Swi4p or Mbp1p, the heterodimeric transcription factors SBF and MBF, respectively. We have shown that Msn5p is required for the nucleocytoplasmic shuttling of Swi6p and that this relocalization is crucial for SBF function but not for MBF function. The question arises whether the regulatory mechanism of Swi6p function described above depends on the existence of SBF or acts on Swi6p independent of the presence of Swi4p and, in this case, why Msn5p controls SBF but not MBF activity. To address these questions, the subcellular localization of Swi6p was analyzed in the mbp1 and swi4 mutant strains. In the case of the mbp1 mutant cells, no difference in Swi6p localization compared with that in wild-type cells was observed, that is, Swi6p was nuclear during most of the cell cycle, aside from a period in the G2-M phase (unpublished data). On the contrary, when swi4 cells were analyzed, Swi6p showed nuclear localization along the entire cell cycle, including cells in the G2-M phase (Fig. 10). This result is similar to that observed in the msn5 mutant and reflects the loss of Swi6p nucleocytoplasmic shuttling during the cell cycle when Swi4p was not present. In agreement with this, the interaction detected between Msn5p and Swi6p by the two-hybrid assay was lost in a swi4 mutant background (Fig. 5). These observations demonstrate that the described regulatory mechanism relies on the existence of the SBF complex and not on the monomeric Swi6p.
FIG. 10.
Subcellular localization of Swi6p in a swi4 mutant strain. Cells of a swi4 mutant strain expressing a HA-epitope-tagged Swi6p (JCY353) protein were fixed and assayed by indirect immunofluorescence as described in the legend to Fig. 4. DIC, differential interference-contrast.
DISCUSSION
Msn5p is involved in the control of the G1-S phase transition by regulating SBF.
The karyopherin Msn5p has pleiotropic functions controlling the subcellular localization of proteins involved in different processes such a catabolite repression, calcium signaling, phosphate metabolism, stress response, mating, DNA replication, and cell proliferation. We have investigated the connection between Msn5p and cell cycle control. The increased cell size of the msn5 mutant cells and the synthetic lethality between the msn5 and swi6 or swi4 mutations demonstrate that Msn5p is involved in the G1-S phase transition and suggest that the regulation of the subcellular localization of some protein involved in this transition is important for the proper execution of Start. The key process in Start is the activation of a transcriptional program, and most of the Msn5p-regulated proteins are transcription factors. Consistently, we have shown that transcriptional activation by SBF is severely impaired in an msn5 mutant. Furthermore, we have demonstrated that Msn5p controls the subcellular localization of the regulatory subunit of SBF, the Swi6p protein, and that a defect in the proper control of Swi6p location leads to a lack of SBF function.
A defective SBF can explain some of the phenotypes observed in msn5 mutant strains. Loss of SBF function causes an increase in cell size similar to that observed in the msn5 mutant strain. In addition, an impaired SBF activity leads to reduced transcription of the CLN2 gene, which explains the decrease in Cln2p protein levels observed in msn5 cells. It is striking that the inactivation of Msn5p has a strong effect on Cln2p levels but that it hardly affects the amount of Cln1p. This could be due to a different sensitivity of CLN1 and CLN2 gene expression to the loss of SBF function. Several reported observations support this possibility (see references 22 and 33). Interestingly, periodic expression of the CLN1 gene is mediated through MCB elements instead of SCB elements (34) so it could be that MBF substitutes for SBF more efficiently in the case of the CLN1 gene. On the other hand, the synthetic lethality between the msn5 and swi4 or swi6 mutations can be explained by an additive effect of both mutations on the expression of SBF-regulated genes and by the pleiotropic functions of both Msn5p and SBF.
It is interesting to point out that the inactivation of Msn5p reduces but does not completely abolish the expression of SBF-regulated genes. This is clearly observed in the Western analysis of the Cln2p protein level and in the expression assays, especially in the case of the SCB:lacZ gene, whose expression is totally dependent on SBF. One possible explanation is that the detected expression was due to MBF activity, but this is unlikely if we consider the viability of the msn5 mbp1 double mutant. Alternatively, and most probable, is that SBF is not completely inactivated by the msn5 mutation. This could otherwise explain the suppression of the cell size defect of the msn5 mutant by the overexpression of the CLN3, SWI4, and SWI6 genes. There are different possibilities to account for residual SBF activity in the msn5 mutant: first, newly synthesized Swi6p, which is functional upon entering the nucleus; second, export of Swi6p to the cytoplasm could occur at a very low rate in the absence of Msn5p; and third, SBF was not completely inactivated in the G2-M phase, allowing a basal level of transcription.
Msn5p is required for the export of Swi6p to the cytoplasm during the G2-M phase.
Analysis of the location of several proteins related to Start has enabled us to determine that Swi6p localization is regulated by Msn5p. Swi6p shows nucleocytoplasmic shuttling during the cell cycle, and our results indicate that Msn5p is required for the export of Swi6p to the cytoplasm during the G2-M phase. It cannot be ruled out that Msn5p could control other additional proteins involved in the G1-S phase transition, but we were not able to detect differences in the localization of Cln3p, Cln2p, and Swi4p between the wild-type and msn5 strains.
In the cell, Swi6p exists as a part of the transcription factors SBF and MBF, so the question arises whether the detected changes in Swi6p localization are related to both transcription factors. The fact that, in the swi4 mutant cells, which contain MBF but not SBF, the relocalization of Swi6p to the cytoplasm in the G2-M phase and the interaction between Mns5p and Swi6p were not observed demonstrates that the spatial regulation of Swi6p is affecting the subpopulation of the protein related to SBF specifically. In agreement with this, in mbp1 mutant cells, which contain SBF but no MBF, exportation of Swi6p to the cytoplasm in the G2-M phase is not affected. It cannot be inferred from the immunofluorescence assays whether all or only most of the Swi6p protein exits the nucleus in the G2-M phase. In fact, the assays show a shift from a mainly nuclear signal to an even staining of the whole cell, suggesting the persistence of a small amount of Swi6p inside the nucleus, probably the protein present in MBF. The observation that the changes in Swi6p localization are related to only SBF is in agreement with the results obtained in the analysis of gene expression and promoter-binding assays, which indicated that Msn5p is involved in the regulation of SBF function but, in contrast, is not required for transcriptional activation by MBF. Moreover, the lack of genetic interaction between the msn5 and mbp1 mutations, in contrast to the similarities in the phenotypes of msn5 and mutants in SBF, reinforces the idea that Msn5p is specifically required for the proper function of SBF but not MBF. Both SBF and MBF are activated at the same time by Cln3p-Cdc28p, but, strikingly, they are inactivated by a different mechanism in the G2-M phase since Clb2p-Cdc28p is required for switch-off SBF but not MBF-dependent transcriptional regulation (2). That the export mechanism would be linked to the inactivation mechanism acting in the G2 phase is an attractive hypothesis which otherwise could help to explain why the regulation of Swi6p by Msn5p affects SBF but not MBF.
Which is the molecular mechanism of export of Swi6p by Msn5p? We detected an in vivo interaction between Swi6p and Msn5p by two-hybrid assays, confirming that Msn5p is the exportin of Swi6p. Nevertheless, as reported for other Msn5p-regulated proteins (9, 14), we were unable to detect direct binding between Msn5p and Swi6p by different coimmunoprecipitation strategies, perhaps because it is a weak interaction or it requires conditions lost during the extraction. The nuclear export of Swi6p relies on the existence of SBF since it is abolished in a swi4 mutant. It has been described previously that Swi4p is a nuclear protein in all the phases of the cell cycle (3). This suggests a two-step mechanism based on the recognition or target of Swi6p in the SBF complex and then the export of Swi6p as a monomer. However, it cannot be ruled out that the whole SBF is the cargo of Msn5p but that the export of SBF does not produce detectable changes in Swi4p localization by using immunofluorescence assays. It has been suggested that Msn5p specifically exports phosphoproteins. Swi6p is a multiphoshorylated protein (36), so it is probable that export to the cytoplasm is regulated by the phosphorylation of specific residues. Interestingly, inactivation of SBF is dependent on Clb2p-Cdc28p kinase activity (24). The question arises whether phosphorylation by Clb2p-Cdc28p could be involved in targeting Swi6p for exportation.
Nucleocytoplasmic shuttling of Swi6p is essential for its functionality.
In the case of Pho4p, Mig1p, and Rtg1p/Rtg3p, their mislocalization in an msn5 mutant does not cause the misregulation of target genes, reflecting the existence of additional regulatory mechanisms. In other cases, like the Crz1p protein, its nuclear accumulation helps to explain the transcriptional activation of the Crz1p-regulated genes associated with msn5 mutation. However, the perturbed subcellular localization of Swi6p in the msn5 mutant raises an apparent contradiction, since the nuclear accumulation of the transcription factor correlated with an inactivation of target gene expression. The explanation is that Swi6p, although nuclear, is not functional, since it is not able to bind to the CLN2 promoter. Thus, our results have revealed a new mechanism regulating Swi6p/SBF: activation of Swi6p at Start is linked to its exit from the nucleus to the cytoplasm in the G2-M phase.
Reexamination of previous results in light of the present work provides more support for the functional regulation of Swi6p depending on its nucleocytoplasmic shuttling. A nuclear localization signal in Swi6p was identified, and the role of the phosphorylation of Ser160 in regulating this signal was characterized (36). By substituting this Ser residue with Ala, Swi6p (Swi6pAla160) becomes constitutively nuclear throughout the whole cell cycle, similar to that observed in an msn5 mutant. Nevertheless, Swi6pAla160 is functional since the transcriptional activation by SBF is not affected, as opposed to the impaired transcription detected in the msn5 mutant. This apparent contradiction could be explained if subcellular localization was analyzed as a balance between import and export. The Swi6pAla160 protein is nuclear because import remains active during the G2-M phase, whereas in the msn5 mutant, Swi6p is nuclear because it is not exported to the cytoplasm in the G2-M phase. Thus, Swi6pAla160, although mainly nuclear, is probably shuttling between the nucleus and cytoplasm, while in the msn5 mutant shuttling of Swi6p is abolished and function is lost. Supporting this interpretation, we have preliminary observations indicating that a Swi6p mutated in all putative Cdc28 phosphorylation sites including Ala160 (41)—which, like Swi6pAla160, is functional and is located in the nucleus for the entire cell cycle—lost its activity in an msn5 mutant strain. This observation further emphasizes the conclusion that export to the cytoplasm is required for Swi6p activation.
Licensing transcriptional activation in the G1-S phase transition.
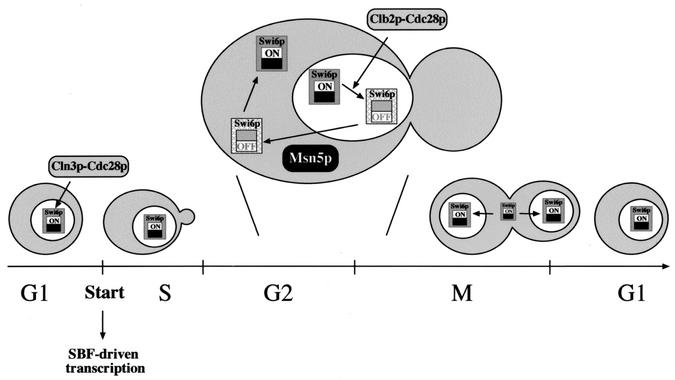
The picture emerging from our results is a functional switch of Swi6p linked to its nucleocytoplasmic shuttling during the cell cycle (Fig. 11). Swi6p goes into the nucleus at the end of mitosis and, as part of the SBF transcription factor, binds to the promoter of target genes. At Start, SBF is activated in situ by Cln3p-Cdc28p kinase, leading to the expression of corresponding genes. Later in the cycle, the Clb2p-Cdc28p kinase inactivates SBF, displacing it from DNA and thus switching off SBF-dependent transcription. This or an additional unknown molecular event then targets Swi6p (or SBF) for export to the cytoplasm by the karyopherin Msn5p. Nuclear export is essential for Swi6p/SBF functionality because, if this step is blocked, Swi6p remains nuclear but in a nonfunctional state even in the absence of Clb-Cdc28p kinase. Once in the cytosol, a functional re-setting of Swi6p reverses the inactivation, rendering a protein ready to activate transcription in the next cycle. Our results are a good example of the importance of spatial regulation in cell cycle control. Many proteins have to be properly located to carry out their function, and the number of cases in which this location is strictly controlled is expanding quickly. In the case of the Swi6p protein described here, the concept of spatial regulation goes a step further in the sense that a functional switch of the protein is linked to changes in subcellular localization.
FIG. 11.
Functional cycle of Swi6p. Different mechanisms affect Swi6p function and localization during the cell cycle. Swi6p is imported into the nucleus at the end of mitosis. As a part of the SBF transcription factor, it binds to the target promoter, although gene expression is not triggered until SBF is activated at Start by the Cln3p-Cdc28p kinase. This activation leads to progression through the cell cycle and subsequently to the accumulation of Clb-Cdc28p kinase activity. This kinase is responsible for the inactivation of SBF in the G2 phase, displacing it from the DNA. By this or another additional mechanism, Swi6p is targeted for being exported to the cytoplasm by the karyopherin Msn5p. Relocation of Swi6p to the cytoplasm enables a functional re-setting of Swi6p, rendering a protein able to activate transcription in the next cycle.
Why is there such a sophisticated regulation for Swi6p? Many cell cycle events have to be tightly regulated to guarantee that they happen once in each cycle or are restricted to a tight period of time. Perhaps the best example is DNA replication. The functional cycle depicted in Fig. 11 uses the licensing factor hypothesis raised to explain the control of DNA replication. As originally proposed, the initiation of DNA synthesis at S phase requires a previous step of licensing origins by binding during the mitosis of a cytosolic licensing factor, which would be destroyed in firing the origins (7; for a recent review on the real nature of the replication licensing system, see reference 8). Similarly, SBF transcriptional activation at Start requires, first, a re-setting of Swi6p functionality in the cytosol during mitosis, by which Swi6p gets licensed to activate transcription; firing of SBF-dependent transcription will lead to a loss of licensing by the inactivation of SBF by Clb-Cdc28p later on in the cell cycle. Restricting the functional re-setting of Swi6p to the cytoplasm helps to establish an alternation between the Start transcriptional program and mitosis. In cells with open mitosis, this re-setting would occur only after the disassembling of the nuclear envelope during mitosis; in the case of cells with closed mitosis, as in yeast cells, an active export mechanism during the G2-M phase is necessary, that being the role for Msn5p. It is not clear why yeast cells should avoid unscheduled transcription by SBF. However, in mammal cells, where a similar transcription program mediated by the E2F transcription factors is activated at the restriction point (19), inactivation of E2F-dependent transcription is essential for cell viability (26). There are several similarities between the function and regulation of SBF and E2F. Moreover, E2F shows changes in subcellular localization (27, 40). It will be interesting to test if the functional switch of a Start transcription factor associated with its nucleocytoplasmic shuttling described here also exists in higher eukaryotic cells.
.
Acknowledgments
We are very grateful to M. Aldea and L. H. Johnston for helpful discussions and the gift of reagents. We also thank L. Breeden, E. A. Craig, F. Estruch, B. Futcher, E. Herrero, M. Johnston, J. R. Pringle, S. I. Reed, and C. Wittenberg for kindly supplying plasmids and strains.
This work was supported by grant numbers PB97-1468-CO2-02 and BMC2001-0446-CO2-02 from the Spanish government and grant number GV98-5-86 from the Generalitat Valenciana. E.Q. is the recipient of an F.P.I. Fellowship from the Spanish government.
REFERENCES
- 1.Alepuz, P. M., D. Matheos, K. W. Cunningham, and F. Estruch. 1999. The Saccharomyces cerevisiae RanGTP-binding protein Msn5p is involved in different signal transduction pathways. Genetics 153:1219-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amon, A., M. Tyers, B. Futcher, and K. Nasmyth. 1993. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell 74:993-1007. [DOI] [PubMed] [Google Scholar]
- 3.Baetz, K., and B. Andrews. 1999. Regulation of cell cycle transcription factor Swi4 through auto-inhibition of DNA binding. Mol. Cell. Biol. 19:6729-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartek, J., J. Bartkova, and J. Lukas. 1996. The retinoblastoma protein pathway and the restriction point. Curr. Opin. Cell Biol. 8:805-814. [DOI] [PubMed] [Google Scholar]
- 5.Benton, B. K., A. H. Tinkelenberg, D. Jean, S. D. Plump, and F. R. Cross. 1993. Genetic analysis of Cln/Cdc28 regulation of cell morphogenesis in budding yeast. EMBO J. 12:5267-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blondel, M., P. M. Alepuz, L. S. Huang, S. Shaham, G. Ammerer, and M. Peter. 1999. Nuclear export of Far1p in response to pheromones requires the export receptor Msn5p/Ste21p. Genes Dev. 13:2284-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blow, J. J., and R. A. Laskey. 1988. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature 332:546-548. [DOI] [PubMed] [Google Scholar]
- 8.Blow, J. J., and B. Hodgson. 2002. Replication licensing—defining the proliferative state? Trends Cell Biol. 12:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boustany, L. M., and M. S. Cyert. 2002. Calcineurin-dependent regulation of Crz1p nuclear export requires Msn5p and a conserved calcineurin docking site. Genes Dev. 16:608-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breeden, L. 1995. Start-specific transcription in yeast. Curr. Top. Microbiol. Immunol. 208:93-125. [DOI] [PubMed] [Google Scholar]
- 11.Cosma, M. P., S. Panizza, and K. Nasmyth. 2001. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell 7:1213-1220. [DOI] [PubMed] [Google Scholar]
- 12.Cross, F. R. 1995. Starting the cell cycle: what's the point? Curr. Opin. Cell Biol. 7:790-797. [DOI] [PubMed] [Google Scholar]
- 13.Cvrckova, F., and K. Nasmyth. 1993. Yeast G1 cyclins CLN1 and CLN2 and a GAP-like protein have a role in bud formation. EMBO J. 12:5277-5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeVit, M. J., and M. Johnston. 1999. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr. Biol. 9:1231-1241. [DOI] [PubMed] [Google Scholar]
- 15.Dirick, L., T. Bohm, and K. Nasmyth. 1995. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 14:4803-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgington, N. P., and B. Futcher. 2001. Relationship between the function and the location of G1 cyclins in S. cerevisiae. J. Cell Sci. 114:4599-4611. [DOI] [PubMed] [Google Scholar]
- 17.Görlich, D., M. Dabrowski, F. R. Bischoff, U. Kutay, P. Bork, E. Hartmann, S. Prehn, and E. Izaurralde. 1997. A novel class of RanGTP binding proteins. J. Cell Biol. 138:65-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Görner, W., E. Durchschlag, J. Wolf, E. L. Brown, G. Ammerer, H. Ruis, and C. Schuller. 2002. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 21:135-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helin, K. 1998. Regulation of cell proliferation by the E2F transcription factor. Curr. Opin. Genet. Dev. 8:28-35. [DOI] [PubMed] [Google Scholar]
- 20.Igual, J., A. Johnson, and L. Johnston. 1996. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 15:5001-5013. [PMC free article] [PubMed] [Google Scholar]
- 21.Igual, J., W. Toone, and L. Johnston. 1997. A genetic screen reveals a role for the late G1-specific transcription factor Swi4p in diverse cellular functions including cytokinesis. J. Cell Sci. 110:1647-1654. [DOI] [PubMed] [Google Scholar]
- 22.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533-538. [DOI] [PubMed] [Google Scholar]
- 23.Kaffman, A., N. M. Rank, E. M. O'Neill, L. S. Huang, and E. K. O'Shea. 1998. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 396:482-486. [DOI] [PubMed] [Google Scholar]
- 24.Koch, C., A. Schleiffer, G. Ammerer, and K. Nasmyth. 1996. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at Start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 10:129-141. [DOI] [PubMed] [Google Scholar]
- 25.Komeili, A., K. P. Wedaman, E. K. O'Shea, and T. Powers. 2000. Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 151:863-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krek, W., M. E. Ewen, S. Shirodkar, Z. Arany, W. G. Kaelin, and D. M. Livingston. 1994. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell 78:161-172. [DOI] [PubMed] [Google Scholar]
- 27.Lindeman, G. J., S. Gaubatz, D. M. Livingston, and D. Ginsberg. 1997. The subcellular localization of E2F-4 is cell-cycle dependent. Proc. Natl. Acad. Sci. USA 94:5095-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macara, I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahanty, S. K., Y. Wang, F. W. Farley, and E. A. Elion. 1999. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell 98:501-512. [DOI] [PubMed] [Google Scholar]
- 30.Miller, M. E., and F. R. Cross. 2000. Distinct subcellular localization patterns contribute to functional specificity of the Cln2 and Cln3 cyclins of Saccharomyces cerevisiae. Mol. Cell. Biol. 20:542-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, M. E., and F. R. Cross. 2001. Mechanisms controlling subcellular localization of the G1 cyclins Cln2p and Cln3p in budding yeast. Mol. Cell. Biol. 21:6292-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgan, D. O. 1995. Principles of CDK regulation. Nature 374:131-134. [DOI] [PubMed] [Google Scholar]
- 33.Ogas, J., B. J. Andrews, and I. Herskowitz. 1991. Transcriptional activation of CLN1, CLN2, and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell 66:1015-1026. [DOI] [PubMed] [Google Scholar]
- 34.Partridge, J. F., G. E. Mikesell, and L. L. Breeden. 1997. Cell cycle-dependent transcription of CLN1 involves Swi4 binding to MCB-like elements. J. Biol. Chem. 272:9071-9077. [DOI] [PubMed] [Google Scholar]
- 35.Pines, J. 1999. Four-dimensional control of the cell cycle. Nat. Cell Biol. 1:E73-E79. [DOI] [PubMed] [Google Scholar]
- 36.Sidorova, J., G. Mikesell, and L. Breeden. 1995. Cell cycle-regulated phosphorylation of Swi6 controls its nuclear localization. Mol. Biol. Cell 6:1641-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stuart, D., and C. Wittenberg. 1995. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 9:2780-2794. [DOI] [PubMed] [Google Scholar]
- 39.Taba, M. R. M., I. Muroff, D. Lydall, G. Tebb, and K. Nasmyth. 1991. Changes in a SWI4,6-DNA-binding complex occur at the time of HO gene activation in yeast. Genes Dev. 5:2000-2013. [DOI] [PubMed] [Google Scholar]
- 40.Verona, R., K. Moberg, S. Estes, M. Starz, J. Vernon, and J. Lees. 1997. E2F activity is regulated by cell cycle-dependent changes in subcellular localization. Mol. Cell. Biol. 17:7268-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wijnen, H., A. Landman and B. Futcher. 2002. The G1 cyclin Cln3 promotes cell cycle entry via the transcription factor Swi6. Mol. Cell. Biol. 22:4402-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, J., and S. Kornbluth. 1999. All abroad the cyclin train: subcellular trafficking of cyclins and their CDK partners. Trends Cell Biol. 9:207-210. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida, K., and G. Blobel. 2001. The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J. Cell Biol. 152:729-740. [DOI] [PMC free article] [PubMed] [Google Scholar]