Abstract
Despite the central role of TATA-binding protein (TBP) in transcription, changes in cellular TBP concentration produce selective effects on gene expression. Moreover, TBP is up-regulated by oncogenic signaling pathways. These findings suggest that TBP could be a nexus in pathways that regulate cell proliferation and that genetic lesions that result in cellular transformation may produce their effects at least in part through TBP. We provide evidence consistent with this hypothesis, demonstrating that increases in TBP expression contribute to cellular transformation. A Ras-mediated increase in TBP expression is required for full Ras transforming activity. TBP overexpression induces cells to grow in an anchorage-independent manner and to form tumors in athymic mice. These effects on cellular transformation require changes in RNA polymerase II-dependent transcription and on the selective recruitment of TBP to promoters via its DNA binding activity. TBP expression is elevated in human colon carcinomas relative to normal colon epithelium. Both Ras-dependent and Ras-independent mechanisms mediate increases in TBP expression in colon carcinoma cell lines. We conclude that TBP may be a critical component in dysregulated signaling that occurs downstream of genetic lesions that cause tumors.
The TATA-binding protein (TBP) is a central eucaryotic transcription factor used by all three cellular RNA polymerases. TBP associates with additional polypeptides to form at least three unique complexes, SL1, TFIID, and TFIIIB, which specify its role in the transcription of RNA polymerase I, II, and III genes, respectively. Two general mechanisms are used in the recruitment of TBP to these promoters. For those promoters that contain a TATA element, TBP binds directly to this sequence via its DNA binding domain. For promoters that lack this element, TBP is recruited solely through its interactions with other proteins that are bound to the promoter. The intracellular levels of TBP have been shown to be limiting for the transcription of both RNA polymerase I-dependent rRNA promoters (19) and RNA polymerase III-dependent tRNA, 5S RNA, and U6 RNA promoters (16). RNA polymerase II-dependent promoters are affected by increases in cellular TBP levels differently, depending on their architecture (3, 12). Generally, promoters containing TATA elements can be stimulated by TBP overexpression (3, 8). In contrast, TATA-lacking promoters are either unaffected or repressed by TBP overexpression (3, 8). Increased cellular TBP amounts also potentiate the effects of certain activators, such as VP16, while inhibiting the effect of others, such as Sp1 or NF-1 (12). Thus, increasing the cellular amounts of TBP can have profound effects on cellular gene activity. Decreases in the amount of TBP have also been shown to produce specific changes in gene expression. Expression of a cell cycle regulatory protein, cdc25B phosphatase, was found to be reduced in a chicken B-cell line when one copy of the TBP gene was disrupted (17). Furthermore, heterozygous disruption of the TBP gene in these cells caused abnormalities in cell growth and size and delayed mitosis (17). Thus, small alterations in cellular TBP concentrations affect cellular gene activity with profound phenotypic consequences.
It has previously been shown that the activation of specific cellular signaling pathways increases cellular TBP levels. Cells treated with the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA), a potent activator of protein kinase C (4), or cells expressing oncogenic Ras (20) exhibit increased TBP concentrations. Expression of the hepatitis B virus X protein has also been shown to increase TBP concentrations via the activation of Ras cellular signaling (6, 20). This event is mediated at the transcriptional level, whereby the human TBP promoter is induced by at least two Ras-activated pathways that require activation of mitogen-activated protein kinase kinase (MEK) (6). Since the activation of these signaling cascades can be key steps leading to oncogenesis and the development of many human tumors (1), we investigated whether increased cellular concentrations of TBP produced by these signaling events could contribute to cellular transformation and tumorigenesis.
Using several model systems, we present evidence to support the idea that increased cellular TBP amounts produce changes in cellular gene expression that lead to oncogenesis. First, we show that Ras-mediated increases in cellular TBP are necessary for Ras transforming function. Inhibiting the ability of oncogenic Ras to overexpress TBP in NIH 3T3 cells reduces Ras-mediated focus-forming activity. Second, enhanced expression of TBP in Rat1A cells, while not changing cell proliferation rates, significantly increases the ability of the cells to grow in an anchorage-independent manner. Third, enhanced expression of TBP also substantially increases the ability of these cells to form tumors in mice. Importantly, these TBP-mediated transforming activities require changes in cellular gene expression, as TBP mutants that are specifically defective for RNA polymerase II-dependent transcription fail to exhibit transforming activity. To determine the significance of these findings to human cancer, we have further analyzed colon carcinomas from six patients. Both normal and tumor tissues were isolated from each individual, and microdissections of epithelium from these samples were performed. Quantitative real-time reverse transcription-PCR (RT-PCR) analysis revealed that TBP mRNA was increased in a clinically significant proportion of the cases. Analysis of human colon nontumor and tumor cell lines revealed that increases in cellular TBP in tumors occur via both Ras-dependent and Ras-independent mechanisms. Together, these results are the first to support the idea that changes in cellular TBP levels play a role in oncogenesis.
MATERIALS AND METHODS
Expression plasmids.
Wild-type human TBP cDNA was PCR amplified from pLTReTBP (22) and provided by A. J. Berk (University of California at Los Angeles). Oligonucleotide primers added EcoRI and AgeI sites 5′ of TBP and a HindIII site at the 3′ end. The PCR fragment was inserted into pBluescript SK at EcoRI and HindIII (TBP-psk). An oligonucleotide sequence containing a 5′ EcoRI site, the Kozak consensus translation initiation sequence 5′-CCA CCA TGG-3′, followed by two consecutive hemagglutinin (HA) epitope sequences (YPYDVPDYA) and a 3′ AgeI site, was PCR amplified and inserted 5′ of TBP-psk at EcoRI and AgeI sites amino terminal of the second codon (GAT) of TBP (E2TBP-psk). E2TBP-psk was mutagenized using the QuickChange Site-Directed Mutagenesis kit (Stratagene) to produce the RNA polymerase II-defective mutants E284R and L287E. E2TBP and mutant plasmids were sequenced and then subcloned into pSRαMSVtkneo expression vector (9) modified by site-directed mutagenesis to contain unique EcoRI and HindIII sites downstream of the 5′ long terminal repeat to make pLTR-E2TBP and its corresponding mutants. The TBP mutant with altered DNA binding specificity (pLTR-E2TBP-m3) has been described elsewhere (14). The TBP antisense expression construct was prepared by cloning a 991-bp XbaI/KpnI fragment from the 3′ end of the mouse TBP cDNA (15) into the KpnI and XbaI sites of the expression vector pcDNA3. A second construct expressing the same TBP cDNA fragment in the sense direction was prepared by subcloning the DNA sequence as a HindIII/KpnI fragment into pcDNA3. Orientation of the clones was confirmed by DNA sequencing. HA-tagged constitutively activated Ras (pDCR-RasV12) was described previously (6).
Cell lines.
Colon cell lines were maintained in Dulbecco modified Eagle medium (DMEM) with a high level of glucose (CCD 841 CoTr, HT-29, and NIH 3T3), McCoy's 5A medium (HCT116), or RPMI 1640 medium (COLO320) supplemented with 10% fetal calf serum (FCS). Cells were plated (106 cells/100-mm-diameter plates) and then serum starved in media with 0.5% FCS for 24 h, and whole-cell lysates were prepared. Where indicated, cells were treated with either 50 μM U0126, 200 nM FTI-277, or dimethyl sulfoxide vehicle at the time of serum starvation. NIH 3T3 cells were grown in high-glucose DMEM with 10% bovine calf serum. Rat1A cells were grown in low-glucose DMEM with 10% FCS.
Immunoblot analysis.
Subconfluent cell cultures were harvested from each of the cell lines, and whole-cell lysates were prepared. Lysates (100 μg of protein) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblot analysis. Membranes were probed with rabbit polyclonal anti-human TBP antibody (Geneka), rabbit polyclonal anti-HA antibody (Santa Cruz), goat polyclonal anti-eIF-2α antibody (Santa Cruz), or mouse monoclonal antiactin antibody (Chemicon) where indicated. Bound primary antibody was visualized using horseradish peroxidase-conjugated secondary antibody (Vector Laboratories) and enhanced chemiluminescence reagents (Amersham).
Focus formation assay.
NIH 3T3 cells were plated (2.5 × 105 cells/60-mm-diameter plate) and transfected with a total of 10 μg of DNA and 20 to 30 μl of Superfect reagent (Qiagen). After 24 h, the cells on each plate were split equally and transferred to four 100-mm-diameter plates, with the cells on a portion of these plates selected by the addition of 400 μg of G418 per ml. Fourteen days later, cells were fixed in methanol and treated with Giemsa stain to highlight foci, and the number of G418-resistant colonies was determined (10). To determine TBP mRNA levels in transfected NIH 3T3 cells, total RNA was purified using RNeasy Mini kit (Qiagen) and RT-PCR was performed with the ThermoScript RT-PCR system and Platinum Taq DNA polymerase (Invitrogen) using gene-specific primers. Mouse TBP mRNA was amplified using primers 5′-TCA CCA ATG ACT CCT ATG AC-3′ and 5′-GCC ACC TGT AAC TGA GTG T-3′, and actin primers were 5′-GAC AGG ATG CAG AAG GAG ATC AC-3′ and 5′-TCA GGA GGA GCA ATG ATC TTG A-3′.
Anchorage-independent growth assay.
Rat1A cells stably expressing c-myc were described previously (5). To generate pLTR-E2TBP or stable E2TBP mutant lines, Rat1A cells were plated (5 × 105 cells/100-mm-diameter plate) and transfected with 10 μg of plasmid DNA and Lipofectin (Invitrogen) (at a DNA/reagent ratio of 1:5). Cells were selected with 600 μg of G418 per ml, and after 4 weeks, G418-resistant cell populations were pooled. Resultant stable cell lines (105 cells/100-mm-diameter dish) were suspended in 0.4% low-melting-point agarose (Mallinckrodt) in media, over a bottom layer of media with 0.7% agarose (10). Cells were fed fresh complete media twice weekly. Colonies greater than 100 μm in diameter were counted 21 days after plating. Cell proliferation rates were determined by plating cell lines at 105 cells per 60-mm-diameter dish. Cells were harvested at 2-day intervals, and viable cells were counted.
Nude mouse tumorigenicity assay.
Stably transfected Rat1A cell lines (early passage number) expressing E2TBP or mutant E2TBP proteins (5 × 105 cells per animal) were injected subcutaneously into the groins of athymic nude (nu/nu) mice (four to six mice per group). Animals were monitored twice weekly for appearance and measurement of tumors. Tumor volumes were determined by measuring the three dimensions (height by weight by depth) of tumors (in millimeters) using vernier calipers. Depending on tumor burden, animals were sacrificed 4 to 6 weeks postinjection, and the tumors, where present, were weighed and analyzed histologically.
Laser capture microdissection of human colon tissue and real-time RT-PCR analysis.
Human colon tissues were obtained, analyzed, and prepared for laser capture microdissection by the University of Southern California (USC) Norris Translational Pathology Core Facility. Fresh surgical colon specimens from six patients were embedded in optimal-cutting-temperature compound and frozen at −80°C. Sections (6 μm thick) were cut, fixed, and stained with hematoxylin and eosin. Epithelial cells were isolated from matched normal and tumor sections using a PixCell II laser microdissection microscope (Arcturus, Mountain View, Calif.). Cells were digested and lysed from the membrane caps with GITC buffer (5.35 M guanidinium isothiocyanate, 50 mM Tris-Cl [pH 6.4], 20 mM EDTA, 1% [vol/vol] Triton X-100, 0.1 M 2-mercaptoethanol) and subjected to acid-phenol-chloroform extraction. Sodium acetate (2 M, 0.1× volume) was added to cell lysate, followed by 1× total volume water-saturated phenol and then 0.3× volume chloroform-isoamyl alcohol (24:1). Total RNA was precipitated from the aqueous layer with 1 μg of glycogen and isopropanol. RNA was collected by centrifugation. cDNA was prepared using Superscript First-Strand Synthesis system for RT-PCR (Invitrogen) with random primers. Sample cDNAs were cleaned up and concentrated with the MinElute Reaction Cleanup kit (Qiagen). Quantitative real-time PCR was performed by 5′ nuclease assay method with cDNA-specific fluorogenic probes on an Applied Biosystems 7700 Prism sequence detector (PE Applied Biosystems, Foster City, Calif.). Each reaction mixture contained cDNA, 200 nM fluorescent probe, primers (600 nM each), and Taqman Universal Master Mix (PE Applied Biosystems). TBP probe and primer sequences were as follows: probe, 5′-TGA CCC AGC AGC ATC ACT GTT TCT TGG-3′; forward primer, 5′-CTG GCC CAT AGT GAT CTT TGC-3′; and reverse primer, 5′-TCA ATT CCT TGG GTT ATC TTC ACA-3′. TBP probe sequence overlaps the junction between exons 1 and 2. Actin probe and primer sequences were as follows: probe, 5′-CCC TGG CAC CCA GCA CAA TGA AG-3′; primers as described above for TBP. Actin and TBP transcript concentrations were measured by RT-PCR from the same sample using separate tubes containing equal amounts of total cDNA. The relative amounts of TBP transcript were quantified by utilizing the comparative threshold cycle method with actin serving as the endogenous control reference (ABI Prism 7700 sequence detection system, Applied Biosystems user bulletin 2). cDNA (50 ng) reverse transcribed as described above and derived from adult human liver total RNA (Stratagene) was utilized as a calibrator to control for differences between experiments.
RESULTS
Increased cellular TBP is required for Ras transforming function.
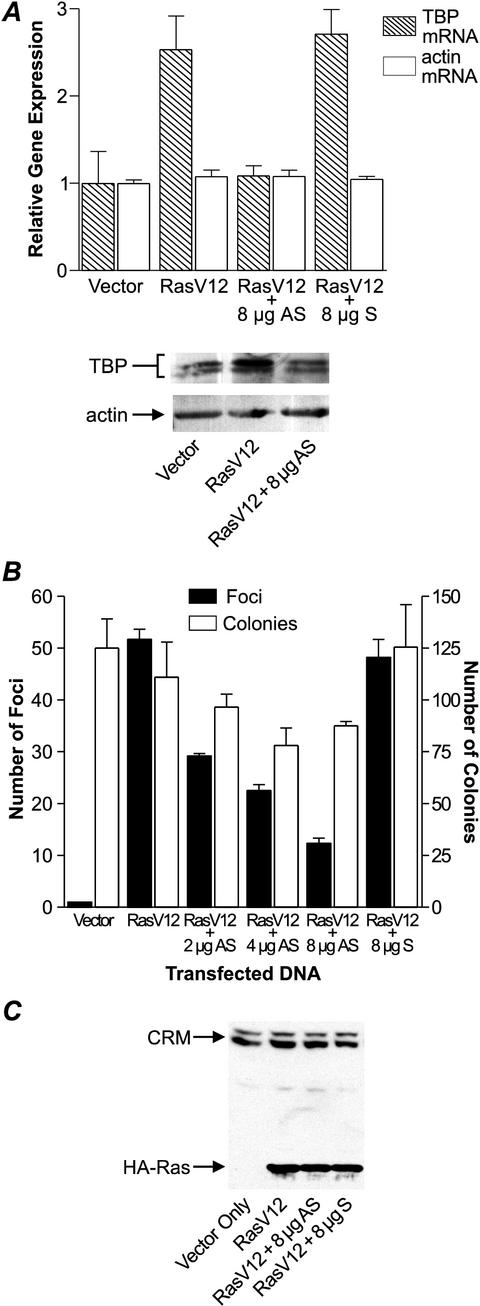
Previous studies have shown that oncogenic Ras increases cellular TBP levels (20). We therefore sought to determine whether this enhanced expression of TBP by Ras was necessary for Ras-induced transformation. To prevent the increased expression of TBP by oncogenic Ras, an antisense TBP expression plasmid (antisense TBP) was constructed that contained the 3′-terminal region of a mouse TBP cDNA in the reverse orientation. An expression plasmid containing the same 3′ region of the TBP cDNA in the sense orientation (sense TBP) was used as a control. Neither construct had a discernible open reading frame. An expression plasmid containing a cDNA encoding RasV12, a constitutively activated form of Ras, was transfected into NIH 3T3 cells. Consistent with previous studies, an approximately two- to threefold increase in cellular levels of TBP mRNA was observed in these RasV12-transfected cells (Fig. 1A, top). The RasV12-mediated increase in TBP mRNA was blocked by coexpression of the antisense TBP, but not by the sense TBP construct. This was compared to the levels of actin mRNA, which remained unchanged with the expression of RasV12 or antisense TBP. Analysis of TBP protein levels similarly showed an increase of approximately twofold in TBP protein upon expression of RasV12, which was blocked by coexpression of the antisense TBP (Fig. 1A, bottom). Since increasing the amount of transfected antisense TBP plasmid caused a corresponding decrease in the expression of TBP, in subsequent experiments we chose to use amounts of antisense plasmid that would not decrease the level of TBP below the basal level (i.e., the level of TBP observed in cells not expressing oncogenic Ras).
FIG.1.
Expression of an antisense TBP suppresses Ras transforming function. (A) Enhanced expression of TBP mRNA by oncogenic Ras is inhibited by the expression of antisense TBP. NIH 3T3 cells were cotransfected with 500 ng of HA-tagged constitutively activated human RasV12 (pDCR-RasV12) together with 8 μg of mouse TBP antisense (AS) or sense (S) expression plasmid or vector. TBP and actin levels were determined by RT-PCR (top) and immunoblot analysis (bottom). (B) Focus-forming activity of oncogenic Ras when coexpressed with TBP antisense expression plasmid. NIH 3T3 cells were transfected as described above. For each assay, a portion of the cells were selected with 400 μg of G418 per ml, beginning 24 h posttransfection. After 18 days, cells were fixed and then treated with Giemsa stain to highlight foci and count G418-resistant colonies (10). (C) The expression of oncogenic Ras is unchanged by altering TBP expression. NIH 3T3 cells were transiently transfected as described above with vector only or with pDCR-RasV12 with or without 8 μg of AS or S expression plasmid. Cell lysates (100 μg of protein) were subjected to immunoblot analysis with HA antibody. The CRM bands represent nonspecific, cross-reacting material in the lysates.
To determine whether changes in cellular TBP amounts contribute to Ras transforming function, the antisense TBP expression plasmid was coexpressed with RasV12 in NIH 3T3 cells, and the resultant abilities of the cells to grow in a density-independent manner and form foci were quantified (Fig. 1B). Expression of the antisense TBP produced a dose-dependent decrease in Ras-mediated focus formation. At the highest concentration of antisense TBP used, a fourfold reduction in foci was observed. In contrast, expression of the sense TBP had no effect on the ability of RasV12 to form foci. By comparison, the overall numbers of selected colonies were not affected as substantially by the expression of the antisense TBP, indicating that the transfection efficiencies were comparable. The decrease in Ras transforming activity cannot be attributed to changes in the expression of oncogenic Ras, as immunoblot analysis revealed that the levels of oncogenic Ras were comparable in cells cotransfected with either the antisense or sense TBP expression plasmids (Fig. 1C). These results support the idea that the increased expression of TBP by oncogenic Ras is required for full Ras transforming function.
Enhanced TBP expression promotes cellular transformation.
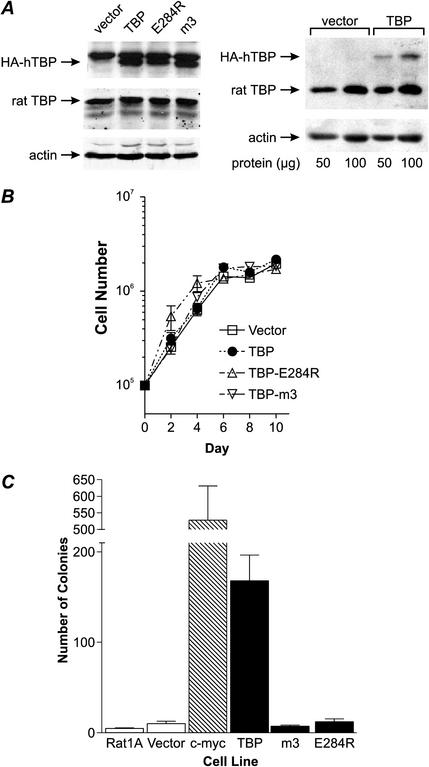
To further elucidate whether the increased levels of cellular TBP are a consequence of, or contribute to, the transformation process, we constructed stable Rat1A cell lines expressing HA-tagged human TBP (hTBP) or mutant hTBP proteins to examine the consequences of enhanced expression of TBP on the transformed phenotype. To determine the specificity of potential TBP-mediated transformation, we also used previously characterized hTBP mutants. Single amino acid mutations that alter the ability of the TBP protein to interact with specific transcription components have been characterized (2, 13). The mutant hTBP-E284R cannot associate with TFIIB, rendering it specifically defective for the transcription of RNA polymerase II-dependent genes. The hTBP-m3 mutant possesses three amino acid changes within the DNA binding domain, I292F, V301T, and L303V. These changes alter TBP DNA binding specificity, rendering hTBP-m3 able to bind and support both transcription from the nonconsensus TATAAA element, TGTAAA (14). Although hTBP-m3 is able to bind to the TATAAA consensus sequence in vitro, expression of the analogous yeast TBP mutations have been shown to confer a decrease in the ability of TBP-m3 to efficiently promote transcription from consensus TATAAA promoters. Moreover, TBP-m3 does not support normal growth rates in Saccharomyces cerevisiae (14). Using these two TBP mutants, we sought to determine whether direct effects on transcription were required to mediate TBP transforming activity.
Stable transformants of Rat1A cells (pooled, nonclonally selected) harboring the wild-type or mutant hTBP cDNAs were established. For each of the hTBP and mutant TBP cDNAs, four independent cell lines were generated, and representative results from each stable cell line are shown (Fig. 2). Lysates derived from each stable cell line were examined by immunoblot analysis to determine TBP levels. The endogenous rat TBP was expressed at similar levels in cells stably transfected with vector or cells with the wild-type or mutant hTBP genes (Fig. 2A, left). Each of the stable cell lines transfected with a wild-type or mutant hTBP expression plasmid expressed comparable amounts of the hTBP or mutant hTBPs (Fig. 2A, left). We have quantified the level of hTBP expression in the stable cell lines and estimate that it is approximately 30% above the endogenously expressed TBP (Fig. 2A, right). Examining the proliferation rates of each of the stable lines revealed no differences in the proliferation rate of the cells expressing hTBP or mutant hTBPs compared to those of the control (vector) and parental cell lines (Fig. 2B). However, the stable cell lines expressing hTBP exhibited a significant, approximately 20-fold increase in its ability to form colonies in soft agar compared to the control vector cell line or those expressing the mutant hTBPs (Fig. 2C). The TBP-mediated transforming activity was only threefold less than that of c-myc, previously shown to be strongly transforming in this assay (5). Analysis of an additional stable line of Rat1A cells expressing the TBP mutant, hTBP-L287E, specifically defective for RNA polymerase II-dependent transcription, revealed no change in proliferation rates or increased transforming activity, consistent with the results obtained for the hTBP-E284R mutant (data not shown). These results reveal that increased cellular TBP amounts significantly enhance the transformation potential of cells. The failure of the mutant TBPs to enhance transformation demonstrates that this event is specifically mediated through the ability of TBP to directly modulate gene expression. Furthermore, TBP-mediated transformation requires alterations in RNA polymerase II-dependent gene expression and its ability to selectively interact with TATA-containing promoters.
FIG. 2.
Increased cellular TBP levels promote anchorage-independent growth. (A) Stable cell lines expressing hTBP or mutant hTBP proteins. Rat1A cells were stably transfected with empty vector or expression plasmids containing HA epitope-tagged wild-type or mutant hTBP cDNA. Cells were selected for 4 weeks, and each resultant population of cells was pooled. Protein from total cell lysates was analyzed by immunoblot analysis to determine the relative levels of endogenous rat TBP, the HA-tagged hTBPs, and actin. (Left) The membrane was probed with anti-HA (top), anti-TBP (center), or antiactin (bottom) antibodies. (Right) The membrane was probed with anti-TBP (top) or antiactin (bottom) antibodies. (B) Proliferation rates of the cell lines expressing hTBP and mutant hTBP. Cells were plated and then harvested at 2-day intervals. Viable cells were determined and counted using trypan blue exclusion. Values are the means ± standard errors of the means of at least three independent determinations. (C) Enhanced expression of hTBP, but not mutant hTBP proteins, promotes cellular transformation. The stable Rat1A cell lines expressing hTBP, mutant hTBPs (E284R and m3), or c-myc were analyzed for growth in soft agar. Colonies greater than 100 μm in diameter were counted 21 days after plating. Values are the means ± standard errors of the means of four independent determinations per cell line.
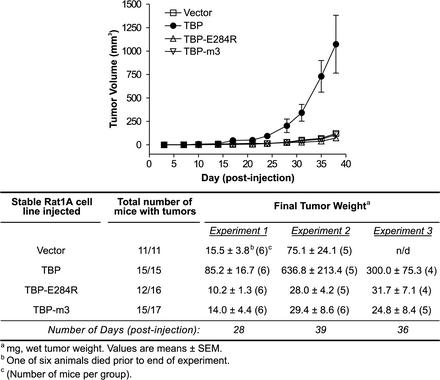
TBP overexpression enhances tumor formation.
An athymic (nude) mouse assay was used to further compare the transforming abilities of the wild-type TBP and the mutant TBP proteins. The stable cell lines expressing hTBP, hTBP-E284R, hTBP-m3, or vector alone were each injected subcutaneously into the groins of the mice. The tumor volume at the site of injection was measured twice weekly (Fig. 3, top). The stable hTBP cell lines showed a significant enhancement of tumor formation in the mice compared to the vector control line over a 5-week period. In contrast, the tumors formed by cells expressing either the RNA polymerase II-defective mutant, hTBP-E284R, or the altered DNA binding mutant, hTBP-m3, were comparable in size to tumors formed from the vector control cell line. Four to five weeks postinjection, the tumors were excised and analyzed (Fig. 3, bottom). All the mice inoculated with either the empty vector cell line or the wild-type TBP cell line formed tumors, and most, but not all, mice inoculated with the mutant TBP cell lines formed tumors. However, there was a marked difference in the weights of the tumors derived from the hTBP cell line compared to those of the tumors formed from the other stable cell lines. This result was reproduced with three additional stably transfected hTBP cell lines (data not shown). Histological analysis of the excised tumors confirmed that increased tumor volumes and weights were attributed to an increased number of tumor cells and not to infiltrating lymphotic cells or increased stromal mass. Thus, overexpression of TBP enhances the tumorigenicity of Rat1A cells. Consistent with the in vitro transformation results, this TBP-mediated process specifically requires alterations in RNA polymerase II-dependent gene expression.
FIG. 3.
Increased expression of cellular TBP enhances tumor formation in nude mice. Comparison of the rates of tumor formation in nude mice of cells stably expressing TBP or mutant TBP proteins. Rat1A cell lines expressing hTBP or mutant hTBPs were derived as described in the legend to Fig. 2. Early passage cells expressing hTBP, hTBP-E284R, hTBP-m3, or vector alone (5 × 105 cells per injection) were injected subcutaneously into the groins of athymic nude (nu/nu) mice. Animals were monitored at least twice a week for the appearance and measurement of tumors (top). The data shown are from one representative experiment of three independent experiments conducted. At the time indicated after injection, the mice were sacrificed, and the tumors, when present, were weighed and analyzed histologically to confirm that the isolated tissue was a tumor (bottom). In each experiment, four to six mice per group were injected with the indicated cell line. SEM, standard error of the mean; n/d, not determined.
TBP is up-regulated in human colon carcinomas.
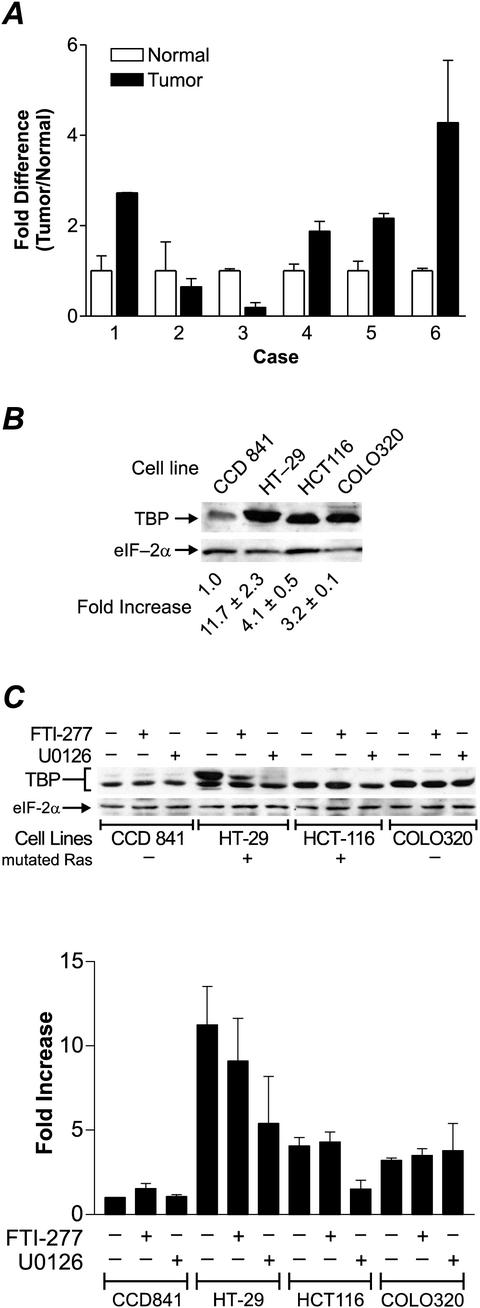
Our results above support that the overexpression of TBP contributes to cellular transformation. To determine the biological relevance of this finding to human cancer, we examined the expression of TBP in human colorectal tumors. To compare the expression of TBP in colon epithelium, matched normal and neoplastic cells were microdissected using laser capture methods from frozen biopsy specimens from six individuals. Quantitative RT-PCR was performed to determine the relative amounts of TBP mRNA relative to actin mRNA in each sample. Four of the six colon tumors exhibited an increase in TBP mRNA levels of approximately two- to fourfold compared to normal colon epithelium from the same individual (Fig. 4A). To further elucidate the mechanisms by which TBP may be up-regulated in colon carcinomas, TBP protein levels were further analyzed in human colon cell lines. Three tumorigenic cell lines (HT-29, HCT116, and COLO320) derived from human colon adenocarcinomas and a nontumorigenic cell line (CCD 841) derived from normal colon epithelium and immortalized were utilized. Lysates from each cell line was subjected to immunoblot analysis. We occasionally observed two cross-reacting polypeptides with apparent molecular masses of 42 and 40 kDa, the most prevalent being the 40-kDa band. The more variable 42-kDa band likely represents a phosphorylated form of TBP, although it is unknown how this modification may affect its function (21). A significant increase in TBP, from approximately 3- to 11-fold, was observed in the tumorigenic cell lines compared to the level in CCD 841 cells (Fig. 4B). The eIF-2α polypeptide, which has been previously shown to be unaffected by various oncogenic agents (17, 20) was used as an internal control.
FIG.4.
TBP is overexpressed in human colon carcinomas and colon carcinoma cell lines compared to nontumor controls. (A) Quantitative RT-PCR analysis of TBP mRNA in human colon tumors and normal colon epithelium. Human colon tissue samples were obtained, analyzed, and prepared for laser capture microdissection. Epithelial cells were microdissected from matched normal and tumor sections. Total isolated RNA was reverse transcribed, and sample cDNAs were subjected to quantitative real-time PCR as described in Materials and Methods. The TBP probe sequence overlaps the junction between exons 1 and 2. Actin and TBP mRNA concentrations were measured in separate tubes, and relative amounts of TBP transcript were determined. TBP mRNA levels were normalized to actin levels. The fold change of TBP mRNA levels in tumor are represented relative to TBP mRNA in matched normal tissue, with each value reflecting two sets of independently microdissected cells, each of which was subjected to independent RT-PCR. (B) Analysis of cellular TBP concentrations in human colon cell lines. Subconfluent cell cultures were harvested from each cell line indicated, and whole-cell lysates were prepared. Cell lysate (100 μg of protein) was subject to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblot analysis. Membranes were first probed with TBP antibody and then subsequently stripped and reprobed with eIF-2α antibody. Densitometry of the resulting autoradiographs was used to calculate the relative amount of each polypeptide. TBP amounts were normalized to the amount of eIF-2α polypeptide, and the fold increase was calculated relative to TBP amounts in the CCD 841 cell lysates. Values are the means ± standard errors of the means of at least four independent determinations. (C) Increased TBP levels in colon tumor cell lines are mediated via Ras-dependent and Ras-independent mechanisms. Subconfluent cell cultures were harvested from cell lines, and whole-cell lysates (100 μg of protein) were subjected to immunoblot analysis. Where indicated, cultures were incubated with either 50 μM U0126, 200 nM FTI-277, or dimethyl sulfoxide vehicle for 24 h. Membranes were probed with TBP antibody and then stripped and reprobed with eIF-2α antibody. Densitometry of the autoradiographs was used to calculate the relative amount of each polypeptide. TBP amounts were normalized to the amount of eIF-2α polypeptide, and the fold increase was calculated relative to the amount of TBP in the CCD 841 cell lysates. Values are the means ± standard errors of the means of at least four independent determinations.
Previous studies have demonstrated that activation of MEK, a downstream target of Ras, induces human TBP promoter activity (6). To determine whether the activation of Ras and/or MEK contributed to the increased TBP amounts observed in the colon carcinoma cell lines, the cells were incubated with either a Ras farnesylation inhibitor, FTI-277, or the MEK inhibitor, U0126. Both inhibitors consistently decreased TBP levels in the HT-29 carcinoma cells, while only the MEK inhibitor significantly decreased TBP in the HCT116 carcinoma cells (Fig. 4B). Both cell lines have been previously shown to possess activating ras mutations. Although cellular TBP concentrations in HCT116 cells are insensitive to FTI-277, we cannot exclude the possibility that the activated form of Ras in these cells is not inhibited by FTI-277. In contrast, treatment of the CCD 841 cells and COLO320 carcinoma cells with either of these inhibitors did not affect TBP levels, consistent with the fact that neither cell line contains activating mutations in ras. Together, the results of analyses of tumor tissues and cell lines support the idea that TBP may participate in human colorectal oncogenesis and that TBP levels may be increased through both Ras-dependent and Ras-independent mechanisms.
DISCUSSION
Previous studies have demonstrated that cellular TBP concentrations are increased by various oncogenic factors (4, 6, 20) and that increases in cellular TBP mediate selective alterations in gene expression (3, 8, 12, 16, 19). Our study provides several lines of evidence that the up-regulation of TBP contributes to the malignant transformation of cells. First, we find that the increase in TBP produced by the expression of oncogenic Ras is necessary for full Ras transforming function. Genetic lesions in ras, leading to its deregulation, are found in a wide variety of human cancers. While considerable efforts have been made to elucidate the signaling pathways that are activated by this important oncogene, little is still known about the downstream components of this pathway that directly alter gene expression. Thus, our results provide new evidence that the central transcription factor, TBP, is a key component of the Ras-induced transformation pathway.
Second, using another model system, our results demonstrate that Rat1A cells that are programmed to overexpress TBP exhibit a transformed phenotype. Increased TBP expression does not alter cellular proliferation rates, yet it significantly enhances the abilities of these cells to grow in an anchorage-independent manner and to form tumors in nude mice. The expression of mutant TBP proteins has further allowed us to investigate the mechanism for these TBP-mediated events. The hTBP-E284R mutant, while completely functional for RNA polymerase I and III transcription, is unable to support RNA polymerase II-dependent transcription, as it cannot form a complex with the RNA polymerase II-specific transcription factor, TFIIB (2, 13). In contrast to wild-type TBP, the expression of hTBP-E284R failed to promote either anchorage-independent growth or tumorigenesis, confirming that these events specifically require the ability of TBP to directly modulate RNA polymerase II-dependent transcription. The TBP-m3 mutant was used to assess its transformation function. TBP-m3 contains mutations within the DNA binding domain that have been shown to alter the DNA binding specificity of TBP (14). Thus, this mutant TBP, while affecting its recruitment to TATA-containing promoters, will functionally support transcription from promoters lacking a TATA element where TBP is recruited to the promoter exclusively through protein-protein interactions. The fact that the expression of TBP-m3 was unable to induce cellular transformation suggests that TBP transforming activity is dependent on the ability of TBP to change the expression of a subset of RNA polymerase II-dependent genes that possess a consensus TATAAA element. The assembly of TBP at eucaryotic promoters is one of a number of important regulatory steps in the transcription process that governs the magnitude by which a gene is expressed. The importance of TBP promoter interactions is supported by the many different proteins that directly regulate TBP, either positively, as in the case of TFIIA, or negatively, such as DR1 (11). Consistent with these results, the regulation of cellular TBP concentrations plays an important role in dictating cellular gene expression by all three RNA polymerases (3, 4, 12, 16, 19, 21). Interestingly, previous studies have shown that overexpression of TBP primarily induces TATA-containing promoters, but not TATA-less RNA polymerase II promoters, supporting the notion that recruitment of TBP to promoters through its DNA binding domain is a rate-limiting step for transcriptional activation of TATA-containing promoters but not TATA-lacking promoters (8). Together, these results support the view that specific changes in RNA polymerase II-dependent gene expression mediated by increases in cellular concentrations of TBP help to drive the cellular transformation process.
Third, as further support for the role of TBP in oncogenesis, we show that TBP expression is up-regulated in a number of human colon tumors. Isolation of epithelium from normal and tumor tissues from the same patients have allowed us to directly compare TBP expression. For these studies, we have used laser capture microdissection to isolate epithelium, avoiding potential artifacts that would have been measured with heterogeneous tissue biopsy specimens. We find that TBP expression is up-regulated in a clinically significant proportion of human colon tumors. This increase in TBP is recapitulated in colon tumor cell lines where cellular TBP levels are modulated by both Ras-dependent and Ras-independent mechanisms. Earlier studies suggested that TBP mRNA levels are also increased in several human breast and lung carcinoma tissues compared to those in normal tissue (18). These results indicate that enhanced TBP expression may occur in a variety of different types of human cancers. Together, our results support the idea that TBP overexpression may be associated with the development of certain human cancers that have acquired activating mutations in ras and that the overexpression of TBP contributes to Ras-mediated oncogenesis.
The results of the present study support the new concept that the regulation of a basal transcription factor impacts the oncogenic potential of cells. Since TBP is the transcription factor most common to all cellular genes, these results are perhaps unexpected. Yet mechanistically, the idea of alterations in a central transcription factor producing global but highly specific changes in gene expression leading to oncogenic transformation is intriguing. Increases in cellular TBP have been shown to up-regulate both RNA polymerase I- and III-dependent transcription (16, 19). Since the products of these genes, tRNAs and rRNAs, are tightly linked to cellular growth rates (7), increases in their production would serve to enhance the biosynthetic capacity of cells needed for cellular transformation. Since only a subset of RNA polymerase II promoters are responsive to changes in cellular TBP concentration (3, 8, 12), enhanced TBP expression could lead to qualitative and quantitative changes in cellular proteins that regulate growth control. Consistent with the idea that the overexpression of TBP contributes to cellular transformation, heterozygous disruption of a single TBP allele in chicken B cells resulted in significantly lower cell growth rates and delayed mitosis (8). Together, these studies reveal that small changes in the cellular concentrations of TBP produce specific changes in gene expression that govern cell proliferation rates and transforming potential. The discovery that TBP contributes to oncogenesis will now stimulate efforts to elucidate the specific changes in gene expression that lead to TBP-mediated oncogenesis and will further define this new role for the central transcription factor.
Acknowledgments
We thank our wonderful colleagues at the USC Norris Comprehensive Cancer Center Gene Regulation Group for many insightful discussions and Eileen Granada and Mo-Li Chen for technical expertise.
This work was supported in part by NIH grant CA74138 to D.L.J., grants CA79750 and CA51167 to L.D., and USC Cancer Center support grant 5P30CA14089.
REFERENCES
- 1.Bos, J. L. 1989. Ras oncogenes in human cancer, a review. Cancer Res. 49:4682-4689. [PubMed] [Google Scholar]
- 2.Bryant, G. O., L. S. Martel, S. K. Burley, and A. J. Berk. 1996. Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes Dev. 10:2491-2504. [DOI] [PubMed] [Google Scholar]
- 3.Colgan, J., and J. L. Manley. 1992. TFIID can be rate limiting in vivo for TATA-containing but not TATA-lacking, RNA polymerase II promoters. Genes Dev. 6:304-315. [DOI] [PubMed] [Google Scholar]
- 4.Garber, M., A. Vilalta, and D. L. Johnson. 1994. Induction of Drosophila RNA polymerase III gene expression by the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA) is mediated by transcription factor IIIB. Mol. Cell. Biol. 14:339-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoang, A. T., K. J. Cohen, J. F. Barrett, D. A. Bergstrom, and C. V. Dang. 1994. Participation of cyclin A in myc-induced apoptosis. Proc. Natl. Acad. Sci. USA 91:6875-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson, S. A. S., N. Mandavia, H.-D. Wang, and D. L. Johnson. 2000. Transcriptional regulation of the human TATA-binding protein by Ras cellular signaling. Mol. Cell. Biol. 20:5000-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larminie, C. G., H. M. Alzuherri, C. A. Cairns, A. McLees, and R. J. White. 1998. Transcription by RNA polymerases I and III, a potential link between cell growth, protein synthesis, and the retinoblastoma protein. J. Mol. Med. 76:94-103. [DOI] [PubMed] [Google Scholar]
- 8.Majello, B., G. Napolitano, P. De Luca, and L. Lania. 1998. Recruitment of human TBP selectively activates RNA polymerase II TATA-dependent promoters. J. Biol. Chem. 273:16509-16516. [DOI] [PubMed] [Google Scholar]
- 9.Muller, W. J., E. Sinn, P. K. Pattengale, R. Wallace, and P. Leder. 1991. BCR first exon sequence specifically active at the BCR/ABL tyrosine kinase oncogene of the Philadelphia chromosome-positive human leukemias. Mol. Cell. Biol. 11:1785-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pastan, I. 1979. Cell transformation. Methods Enzymol. 58:368-370. [DOI] [PubMed] [Google Scholar]
- 11.Pugh, B. F. 2000. Control of gene expression through regulation of the TATA-binding protein. Gene 255:1-14. [DOI] [PubMed] [Google Scholar]
- 12.Sadovsky, Y., P. Webb, G. Lopez, J. D. Baxter, P. M. Fitzpatrick, E. Gizang-Ginsberg, V. Cavailles, M. Parker, and P. J. Kushner. 1995. Transcriptional activators differ in their responses to overexpression of TATA-box-binding protein. Mol. Cell. Biol. 15:1554-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen, Y., G. A. Kassavetis, G. O. Bryant, and A. J. Berk. 1998. Polymerase (Pol) III TATA box-binding protein (TBP)-associated factor Brf binds to a surface on TBP also required for activated Pol II transcription. Mol. Cell. Biol. 18:1692-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strubin, M., and K. Struhl. 1992. Yeast and human TFIID with altered DNA-binding specificity for TATA elements. Cell 68:721-730. [DOI] [PubMed] [Google Scholar]
- 15.Trachtulec, Z., R. M. Hamvas, J. Forejt, H. R. Lehrach, V. Vincek, and J. Klein. 1997. Linkage of TATA-binding protein and proteasome subunit C5 genes in mice and humans reveals synteny conserved between mammals and invertebrates. Genomics 44:1-7. [DOI] [PubMed] [Google Scholar]
- 16.Trivedi, A., A. Vilalta, S. Gopalan, and D. L. Johnson. 1996. TATA-binding protein is limiting for both TATA-containing and TATA-lacking RNA polymerase III promoters in Drosophila cells. Mol. Cell. Biol. 16:6909-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Um, M., J. Yamauchi, S. Kato, and J. L. Manley. 2001. Heterozygous disruption of the TATA-binding protein gene in DT40 cells reduced cdc25B phosphatase expression and delayed mitosis. Mol. Cell. Biol. 21:2435-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wada, C., K. Kasai, T. Kameya, and H. Ohtani. 1992. A general transcription initiation factor, human transcription factor IID, overexpressed in human lung and breast carcinoma and rapidly induced with serum stimulation. Cancer Res. 52:307-313. [PubMed] [Google Scholar]
- 19.Wang, H.-D., A. Trivedi, and D. L. Johnson. 1998. Regulation of RNA polymerase I-dependent promoters by the hepatitis B virus X protein, activated Ras, and the TATA-binding protein. Mol. Cell. Biol. 18:7086-7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, H.-D., A. Trivedi, and D. L. Johnson. 1997. Hepatitis B virus X protein induces RNA polymerase III-dependent gene transcription and increases cellular TATA-binding protein by activating the Ras signaling pathway. Mol. Cell. Biol. 17:6838-6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White, R. J., T. M. Gottlieb, C. S. Downes, and S. P. Jackson. 1995. Mitotic regulation of a TATA-binding-protein-containing complex. Mol. Cell. Biol. 15:1983-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou, Q., P. M. Leiberman, T. G. Boyer, and A. J. Berk. 1992. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 6:1964-1974. [DOI] [PubMed] [Google Scholar]