Since the early work of Beadle and Tatum (6), fungi have been important model organisms for solving genetic problems, largely because of their ease of culture, their defined life cycles, and their short generation times. However, with the development of aggressive and successful medical practices and the rise in immunosuppressed patient populations, fungal infections have become an increasingly larger part of the infectious disease case load, and the etiologic agents have become independent objects of study.
In the last 15 years, the techniques that have been useful in model fungi have become important in studying the pathogens themselves, and the ease and power of these techniques have led to extremely rapid advances in the study of fungal disease. It seems useful to review the current molecular biological techniques employed to study fungal pathogenesis in order to demonstrate their usefulness and their possible problems.
Perhaps the tools most commonly used to study pathogenic fungi are gene isolation, gene expression analyses, and gene disruption by transformation. Gene isolation can be carried out in a variety of ways, including complementation of a mutant by transformation, hybridization with a homologous sequence, or, for those organisms with sequenced genomes, identification of the gene by bioinformatics followed by PCR amplification. Analysis of a cloned gene usually involves the study of its transcriptional regulation, most commonly by Northern blotting, although recently microarrays have become available for Candida albicans (61, 73). When transformation is available, reporter genes can be used to measure expression at the level of the protein product.
With transformation, presently available for C. albicans, Candida dubliniensis, Candida glabrata, Cryptococcus neoformans, Aspergillus fumigatus, Histoplasma capsulatum, Blastomyces dermatitidis, Coccidioides immitis, and Wangiella dermatitidis, the most frequently studied pathogens, targeted gene disruption is possible. For organisms with a high frequency of homologous recombination, disruption of a cloned gene is relatively simple, although in C. albicans the fact that the genome is diploid requires complicated strategies to inactivate both alleles. In H. capsulatum and B. dermatitidis, homologous recombination is infrequent, and identification of a transformant carrying the desired gene disruption requires a tedious search. In C. neoformans and A. fumigatus, the method of transformation used affects the level of homologous recombination.
The advent of genomics has provided the mycology community with complete or partial genome sequences of a number of pathogens, including C. albicans (http://www-sequence.stanford.edu/group/candida/), C. neoformans (http://www.tigr.org/tdb/e2k1/cna1/), A. fumigatus (http://www.tigr.org/tdb/e2k1/afu1/new.shtml), H. capsulatum (http://www.genome.wustl.edu/projects/hcapsulatum/index.php), and C. immitis (http://www.tigr.org/tdb/tgi/cigi/). The complete sequence of each of these organisms has necessarily to be determined from a single strain; however, the complex nature of virulence requires that different isolates be compared. Therefore, less complete sequencing of strains of equal or greater virulence is being carried out with H. capsulatum (G217B is the major strain, and G186AR will be sequenced to 2.5-fold coverage) and C. neoformans (serotype D will be completely sequenced, while an expressed sequence tag library and partial sequence [http://cneo.genetics.duke.edu/] are being derived from serotype A). A physical map of the C. neoformans genome has recently appeared (87). Genetic variation from the genome scale down to the sequence scale can complicate comparisons among isolates, and such variation needs to be taken into account in designing molecular experiments. In addition, experiments testing the contributions of single genes to pathogenesis are bound to be limited, since the complex nature of virulence suggests that pathogenesis is multifactorial. These considerations are likely to require the analysis of strains carrying several mutations in the future, especially as microarray experiments identify groups of interacting genes. Analyzing these strains will require a large number of controls and careful evaluation of the results.
Molecular Koch's postulates (40) have been proposed as a way to use molecular biology to study virulence. These postulates describe the kinds of experiments needed to firmly implicate a property in virulence. A major problem with implementing this kind of analysis in fungi is that the decrease in virulence in a mutant is often relative, not absolute, and ruling out extraneous factors, such as secondary mutations caused by transformation or haploinsufficiency (in diploids like C. albicans) is quite difficult. The effect of nutritional requirements on pathogenicity is another factor which complicates virulence determinations. For example, small changes in the activity of the URA3 (orotidine decarboxylase) gene are sufficient to alter in vivo phenotypes in C. albicans (96), and the site of integration of the URA3 gene affects its level of expression (62). Since URA3 is commonly used as a transformation marker, these findings make it imperative that mutants and reintegrants be as similar as possible with respect to the genomic location of URA3.
In this review we will discuss current work on the molecular analysis of medically important fungi, with an emphasis on the most extensively studied species, C. albicans. We will highlight the tools available and will point out pitfalls and problem areas that must be considered.
GENETIC TRANSFORMATION
The molecular genetics of fungi began with the demonstration of genetic transformation in Saccharomyces cerevisiae in 1977 (54), and it has grown with the development of many powerful tools for gene manipulation in vitro and in vivo. Success with model organisms has led to intense work on the major pathogenic fungi in order to utilize these powerful tools in the study of disease. An important advance was the ability to introduce new genetic material into an organism by transformation, first achieved in C. albicans in 1986 (58) and later made possible in C. neoformans (37) and H. capsulatum (114), followed by C, glabrata (69), A. fumigatus (97), W. dermatitidis (82), and B. dermatitidis (55). The transformation of C. immitis was reported most recently (1, 84). Although these transformation methods show some similarities, they differ in important aspects. Table 1 summarizes the similarities and differences in the transformation systems of the major fungal pathogens most prevalent in the United States.
TABLE 1.
Properties of transformation systems in pathogenic fungi
| Property | Valuea
|
|||||||
|---|---|---|---|---|---|---|---|---|
| C. albicans | C. glabrata | C. neoformans | H. capsulatum | A. fumigatus | W. dermatitidis | C. immitis | B. dermatitidis | |
| LiAc or spheroplasts | Y (58) | Y (69) | N | Y (115) | Y (98) | Y (83) | Y (85) | |
| Electroporation | Y (30) | Y (37) | Y (113) | Y (13, 105) | Y (83) | Y (10) | ||
| Biolistic | Y (100) | Y (A. nidulans) (44) | ||||||
| Other method | Y (1) | |||||||
| Linear/circular plasmid | C (59) | C (69) | L (36) | L (112) | ND | ND | ND | ND |
| ARS identified | Y (14, 59) | Y (69) | Y (103) | Y (112) | N (ARS-like sequence found in A. fumigatus [3]) | N | N | N |
| Drug resistance marker | Mycophenolic acid (7, 50, 56) | Neomycin (21); aureo basidin A (52) | Hygromycin (24); nurseothricin (68) | Hygromycin (113) | Hygromycin (98) | Hygromycin (83) | Hygromycin (1, 85) | Hygromycin (5); sulfonyl urea (10) |
| Homologous integration | High | >90% (21) | 2-50% (serotype A); 1-4% (serotype D) (28) | Rare (114) | 25% (98) to 50% (106) | 52% (116) | 10% (85) | Rare (55) |
ND, none detected; Y, yes; N, no; L, linear; C, circular. References are in parentheses.
Method of transformation.
Almost all of the fungal transformation protocols utilize spheroplasts or cells permeabilized with lithium acetate (LiAc) to promote uptake of DNA. C. albicans, H. capsulatum, C. glabrata, A. fumigatus, W. dermatitidis, and C. immitis can all be transformed in this way. In general, the generation of spheroplasts by treatment with cell wall-digesting enzymes is necessary for the filamentous fungi, while LiAc works with the yeasts. The major exception is C. neoformans, which, probably because of its thick polysaccharide capsule, resists LiAc and is usually transformed either by electroporation (37) or biolistically (DNA is dispersed on gold particles and propelled at high speed onto the cells spread on a grid) (99). Electroporation works with most fungi for which it has been tried and is the method used for B. dermatitidis (10). Electroporation also greatly increases the efficiency of transformation in H. capsulatum (113). The most unusual method of transformation is that developed by Abuodeh and coworkers, which employs Agrobacterium tumifaciens and its DNA transfer system to transform C. immitis (1).
State of the input DNA.
In A. fumigatus, B. dermatitidis, and C. immitis, the incoming DNA is unstable unless it integrates, by either homologous or illegitimate recombination, into the host genome. In C. albicans and C. glabrata, shuttle plasmids have been developed which contain autonomously replicating sequences (ARSs). In the case of C. albicans, several ARSs have been identified (14, 53, 59), and plasmids containing one or two ARSs have been constructed (83). C. glabrata is closely related to S. cerevisiae, and centromere-containing plasmids from S. cerevisiae will replicate in C. glabrata (69), although 2μ plasmids will not (117). In both H. capsulatum and C. neoformans, foreign DNA can be stabilized as linear plasmids by the addition of telomeres. In Histoplasma, the telomeres themselves serve as the ARS (111), while in Cryptococcus, stability is further enhanced by the addition of a 1.1-kb DNA sequence containing an ARS (102).
Dominant selectable markers.
The most frequently used auxotrophic marker for the selection of transformants in C. albicans is the gene encoding orotidine 5′ phosphate decarboxylase, URA3, since both selection for and counterselection against auxotrophy caused by loss of this enzyme is possible (9). Auxotrophy at URA5, which encodes orotate phosphoribosyltransferase, has also been used for positive and negative selection in C. neoformans (37, 60) and in H. capsulatum (88). Drug resistance markers are now available for seven of the major pathogenic fungi. Hygromycin resistance has been used to select transformants in C. neoformans (24), H. capsulatum (112), A. fumigatus (97), W. dermatitidis (82), C. immitis (1, 84), and B. dermatitidis (55). Selection for nurseothricin resistance has recently been reported for C. neoformans (68). For C. glabrata, both neomycin (21) and aureobasidin A (52) have been used. Sulfonyl urea resistance can be used for B. dermatitidis (10). The nonstandard codon usage in C. albicans (86) has precluded the use of heterologous drug resistance genes, although resistance to mycophenolic acid (50) has been used successfully, conferred either via overexpression of wild-type IMH3 (56) or by use of an IMH3 allele resistant to the drug (7, 93). Staib et al. have utilized the S. cerevisiae FLP recombinase engineered for C. albicans to make a useful gene disruption system for C. dubliniensis (94, 95). One possibility for a second marker is the hygromycin B resistance gene, HYG. A mutant form of HYG that corrects for the different codon usages in many Candida species was used successfully in Candida tropicalis (51).
Integration of incoming DNA.
Homologous integration of transforming DNA is important because it facilitates the disruption of a specific gene and therefore the construction of a specific mutant genotype. For all of the fungi but H. capsulatum and B. dermatiditis, integration at the homologous site can be achieved at relatively high frequencies among transformants, ranging from 10% (C. immitis) (84) to >90% (C. glabrata) (21). Notably, the method of DNA delivery affects the rate of production of desired integrants in C. neoformans: the use of biolistic transformation increases homologous recombination relative to that obtained with electroporation in both commonly used serotypes (A and D) (28). Recently, a method using PCR overlap has been devised to improve the frequency of targeting in C. neoformans by greatly increasing the amount of homology on the incoming DNA (27).
GENE CLONING AND DISRUPTION
A large number of C. albicans genes have been isolated by complementation for an analogous function in a known S. cerevisiae mutant. This approach to gene identification has become less necessary since the genome project has identified the homologues of S. cerevisiae genes that can be identified by sequence similarities. Nonetheless, if a gene does function in S. cerevisiae, then the effects of mutant alleles can be tested in S. cerevisiae prior to the more laborious process of testing them in C. albicans (35).
Some C. albicans genes have been cloned based upon their “gain-of-function” characteristics in S. cerevisiae. For example, CPH1, the C. albicans homologue of STE12, was isolated in a screen for genes that, when overexpressed, enhanced pseudohyphal formation in S. cerevisiae (66). An interesting approach was carried out by Clark et al., Csank et al., and Whiteway and coworkers; they identified Candida genes which interfered with mating factor arrest in S. cerevisiae (26, 107) or that disrupted normal control of the filamentation pathway (18). Other examples include the cloning of genes conferring the ability to make S. cerevisiae cells adhere to human cells (40, 42, 43, 46). Of these, two encode cell wall proteins that are important for adhesion in C. albicans, while one affects adhesion in S. cerevisiae cells through an indirect mechanism.
The ability to disrupt or delete genes efficiently in pathogenic fungi provides a powerful approach to study gene function. Gene disruption techniques have been explored most extensively in C. albicans, so we will focus on studies of that organism. Many of the concepts which apply to C. albicans are directly transferable to other pathogenic fungi. In order to delete a given gene in C. albicans, selectable markers, primarily genes that rescue auxotrophic requirements of the cell, are inserted into the gene of interest, resulting in an insertion-disruption mutant. To generate deletion strains, the marker is inserted using homologous sequences that flank the open reading frame (ORF) of interest. Since C. albicans is an asexual diploid, both alleles of a given gene must be inactivated. (Most other pathogenic fungi are haploids, and one transformation event is sufficient to prevent the gene function.) The diploidy of C. albicans complicates both the disruption and the interpretation of the results. Disruption can be accomplished in two ways. The first is by two independent integration events, usually achieved through two rounds of transformation. Since different constructs are often used to inactivate the two alleles, the resulting double disruptant is usually not truly homozygous at the disrupted locus. Alternatively, disruption of one allele can be accompanied by mitotic recombination, leading to homozygosis of the single knockout. In this case, mitotic recombination can lead to homozygosis of part of the chromosome carrying the gene of interest. In either case, restoration of the wild-type allele is necessary to ensure that the phenotype is due to the inactivation of the relevant gene (see below for more detail).
One of the C. albicans strains most commonly utilized for gene disruptions is CAI-4. This strain, derived from the clinical isolate SC5314, contains deletions of both copies of URA3. These deletions were constructed by homologous recombination and resulted in insertion of lambda bacteriophage DNA into both URA3 alleles (41). Thus, the genetic distance between pathogenic organism and useful laboratory strain is short. Since the ura3 deletion was introduced by recombination, this strain does not carry the plethora of secondary mutations which are likely to be introduced when mutagenesis strategies such as UV irradiation are used. Furthermore, URA3 can be used for both positive and negative selection. The Ura-Blaster cassette (the URA3 gene flanked by direct repeats that facilitate excision by mitotic recombination) (2) was adapted for use in C. albicans (41) and has been used successfully to make dozens of mutants. A potential problem with these studies is that CAI-4 carries a deletion of the 3′ untranslated region of the gene adjacent to the ura3 deletion and does not express this gene.
Recently, several new strains and gene disruption-deletion cassettes have been developed for the efficient construction of C. albicans mutant strains. BWP17 was derived from CAI-4 and is auxotrophic for URA3, HIS1, and ARG4 (110). Convenient cassettes that allow PCR-mediated insertion-deletion mutants to be constructed using this strain eliminate the need to recycle markers (109, 110). This system has benefited from the completion of the C. albicans genome-sequencing project because the time-consuming steps of cloning the gene of interest and constructing the disruption vectors has been eliminated. BWP17, however, carries a small deletion of the end of one homologue of chromosome 5 (B. Magee, personal communication).
A different approach has been pioneered by the Morschhauser laboratory. They altered the FLP gene from S. cerevisiae to fit C. albicans codon usage and added an appropriate promoter. This gene can be used to disrupt the gene of interest in a Ura strain using a cassette which includes the URA3 gene and the FLP gene flanked by the FLP target sequence, FRT. Activation of the FLP gene causes the cassette to be popped out, leaving the FRT sequence, and the disruption can be repeated on the second allele (70a).
As effective as these mutational strategies are, they do not allow easy identification of essential genes. Usually, initial evidence that suggests a gene is essential comes from the ability to generate a heterozygous, but not a homozygous, mutant strain. However, other factors could lead to the inability to disrupt both alleles. For example, heterozygosity at a given locus could dramatically affect the rate of homologous recombination and thus the integration of the transforming DNA (116). Another possibility is that the introduced mutation affects the expression of an essential neighboring gene. Thus, researchers must take other steps to demonstrate experimentally that a gene is essential. Often, a wild-type allele is introduced at an ectopic site in the heterozygous strain (effectively reconstructing a “diploid” strain). In this diploid strain, it should be possible to disrupt the remaining wild-type allele, demonstrating that the inability to construct a null strain is not due to inability to disrupt the second allele. In W. dermatitidis, a color-selectable system for specific ectopic integration of a gene has been devised (115). A polyketide synthesis gene that is required for pigment formation serves as a target for ectopic integration of a gene required for complementation. Transformants form albino colonies. Insertion into the locus selected does not affect the phenotype of the resulting strain. Similar ectopic integration sites for C. albicans include RP10 (ribosomal protein gene 10) (15).
Another approach to demonstrate that a gene is essential is to introduce a mutated second copy of the gene adjacent to the remaining wild-type allele in the heterozygous strain. Through negative selection, one copy of the gene is lost through recombination between the two adjacent alleles, and depending on the location of the recombination event, either the wild-type or the mutant copy will be retained. This system can be set up strategically so that most recombination events should lead to retention of the mutant copy. Failure to retain the mutant copy suggests that disruption or deletion of the gene is a lethal event and that the gene is essential for viability.
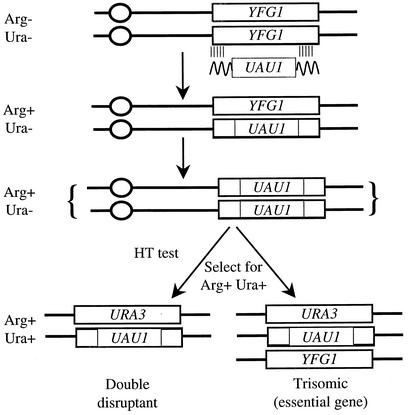
Recently, the identification of an essential gene was simplified by the introduction of a disruption cassette called UAU1 (39) (Fig. 1). This cassette consists of the ARG4 gene flanked by two incomplete (and nonfunctional) copies of the URA3 gene. The cassette includes ∼500 nucleotides of homology between the two ura3 copies so that recombination between the ura3 fragments results in an intact, functional URA3 gene and loss of the intervening ARG4 sequence. If UAU1 is integrated into a given gene, the resulting heterozygote will be Arg+ Ura−. However, mitotic crossing over or gene conversion, which occurs in all eukaryotic cells and is well documented in C. albicans (106), can result in homozygosis of the UAU1-marked allele, and in a second step, the cell can become Ura+ through recombination between the ura3 fragments on one chromosome while remaining Arg+ through retention of the UAU1 cassette on the other chromosome. Thus, both copies of a gene can be disrupted via a single transformation event followed by selection for the recombination events described above. Interestingly, selection for Arg+ Ura+ derivatives of strains transformed with the UAU1 cassette frequently gives rise to two populations of colonies. The first population lacks the wild-type allele of the given gene and contains one UAU1-marked allele and one URA3-marked allele, as expected. The second population also contains a UAU1-marked allele and a URA3-marked allele, but these colonies also retain the wild-type allele and thus are apparently triploid for the given locus. In the case of nonessential genes, both populations occur, but for known essential genes only the latter class is seen. Thus, the UAU1 cassette provides a rapid way to predict whether a gene is essential (19, 39).
FIG. 1.
Gene disruption with the UAU1 cassette. The UAU1 cassette, which contains ARG4 flanked by 5′ and 3′ sequences of URA3 with 530 bp of overlap, is amplified in a PCR using primers with 60-nucleotide tails and with homology to YFG1. These tails target integration to YFG1. Transforming a ura3/ura3 arg4/arg4 strain allows the selection of a heterozygote strain, YFG1/yfg1::UAU1, on synthetic medium lacking arginine. At some low frequency, the yfg1::UAU1 allele will become homozygous, one mechanism being mitotic recombination. The homozygous yfg1::UAU1/yfg1::UAU1 derivative is then able to generate an intact URA3 gene by recombination between the repeated URA3 sequences flanking the ARG4 gene. This results in a strain that is now Arg+ Ura+. Two distinct classes can be seen: double disruptants, which lack any wild-type YFG1, and trisomic strains, which contain both the yfg1::UAU1 and yfg1::URA3 alleles and retain a wild-type allele. Essential genes give rise only to trisomics, whereas nonessential genes give rise to both double disruptants and trisomics.
A strength of the UAU1 strategy is that since only one transformation step is required for strain analysis, it is possible to generate double disruptants that may not be recoverable by the sequential transformation steps. For example, null mutants in CNB1, a component of calcineurin, were constructed using the UAU1 technique but were not recovered using standard approaches (25). Probably this occurred because calcineurin is required for survival in the presence of lithium ions, which are used during transformation (70). It is important to note that UAU1 is useful in determining whether a gene is essential but is less useful as the only method for constructing double-disruption strains. This is because the mitotic recombination that leads to homozygosis of the inserted UAU1 cassette may also lead to homozygosis of genes centromere-distal to the UAU1 insertion site; this loss of heterozygosity could lead to unintended phenotypes that are not associated with disruption of the gene of interest.
In the construction of any mutant, steps should be taken to provide a level of confidence that the resulting mutation contributes to the phenotype; hence, analysis of independently derived mutants is critical. At least two independent heterozygous strains should be used to construct independent double disruptants (in the case of haploid fungi, at least two independent transformants should be tested), and two double knockouts from each heterozygous strain should be compared for initial analyses (a total of four double disruptants). If a given phenotype is due to the mutation, all four double disruptants should have identical phenotypes. At this point, more detailed studies can focus on a single pair of strains (a single disruptant and the double disruptant derived from it).
Once a given double disruptant is found to have a phenotype, regardless of the method used to construct it, a complementation test must be carried out. Complementation of the phenotype by the wild-type gene is the only way to ensure that the functions encoded by the gene of interest are responsible for the phenotype observed. For complementation, the wild-type allele is reintroduced into the double-knockout strain and should restore the wild-type phenotype at least to that of the heterozygous mutant. A major concern, however, is that ectopic insertion of any gene may affect its expression, so returning the wild-type allele and the selectable marker (if it is based on auxotrophy) can affect the phenotype in unexpected ways. For example, the presence or absence of URA3 can affect the adhesion of C. albicans to different substrates (5), and the level of URA3 expression affects virulence in a systemic mouse model (96). Thus, experiments must be designed to control for the location and expression of both the wild-type allele and the marker gene.
Reverse genetic approaches are very useful when a gene of interest is in hand, since the functions of that gene can be determined. However, forward genetic approaches are also valuable in the study of medically relevant fungi. For example, a collection of randomly mutagenized strains can be screened for a given phenotype, such as drug resistance. This approach differs from reverse genetics in that a given phenotype is in hand and the gene whose products confer that phenotype can be determined. Several forward genetic approaches have been successfully applied to C. albicans, C. glabrata, Aspergillus nidulans, and C. neoformans. One approach which has been successfully utilized is restriction enzyme-mediated integration (REMI). In REMI, random mutants are obtained by transforming target DNA with a selectable marker on a DNA fragment obtained by digestion with a given restriction enzyme. The same enzyme is included in the transformation mix and apparently enters the nucleus with the target DNA. where it cleaves the genomic DNA at random sites to direct DNA integration. REMI has been used to generate mutants in A. nidulans, A. fumigatus, and C. albicans (12, 13, 80, 81, 82). A limitation of this approach, in the case of C. albicans, is that only heterozygous mutations are generated.
Signature-tagged mutagenesis has been used to identify genes involved in important processes in fungal pathogens. In this system, a collection of mutants is generated, each containing a unique tag that can be readily identified via PCR and DNA hybridization. Pools of mutants are then passed through a selection against the phenotype of interest (persistence in an animal infection model, for example), and the strains that persist through the process are compared to the initial pool. Individual strains that are lost during the selection may carry mutations in genes that are important for the phenotype of interest. Using this approach, an adhesion factor in C. glabrata and several mutations that affect virulence in C. neoformans were identified (22, 79).
Recently, two forward genetic approaches which use transposon mutagenesis have been developed for C. albicans. One approach takes advantage of the fact that heterozygous mutants in C. albicans are often haploinsufficient. A collection of random heterozygous mutants was generated, and >140 genes that are important for the yeast-to-hyphal transition were identified (A. Uhl and A. Johnson, personal communication). The second approach utilized the UAU1 cassette, described above, which was inserted into a transposon, Tn7::UAU1. This modified transposon was then inserted randomly into a C. albicans genomic DNA library, and the sites of transposition were determined by high-throughput sequencing. Genomic-library inserts containing transposition events within likely ORFs were then transformed into C. albicans, and homozygous mutants were generated through the UAU1 system described above. Using this approach, >200 unique C. albicans homozygous mutants were generated, two of which affect pH responses, and >30 putative essential genes were identified (29).
CONDITIONAL MUTANTS
Conditional mutants in C. albicans have been generated primarily through the use of regulatable promoters. Promoters that regulate expression levels in response to nutritional conditions include GAL1 (49), MAL1 (4), MAL2 (101), PCK1 (64), and MET3 (15). Several of these promoters have been used to generate strains expressing very little or no gene product. For example, when the only copy of URA3 was expressed from the MAL2 promoter under repressing conditions (growth on glucose), the cells exhibited a Ura− phenotype: they required uridine for growth and were resistant to 5-fluoroorotic acid (4). In contrast, the MAL1 promoter was used, together with REMI, to generate strains expressing high levels of genes adjacent to the insert (12). This method identified CZF1, a gene encoding a transcription factor that is important for extensive hyphal growth in matrix-embedded cells (11). Another maltose-inducible, glucose-repressible promoter, MRP1, was used for conditional expression of the essential chitin synthase gene CaCHS1 (72).
The C. albicans MET3 promoter, like its S. cerevisiae counterpart, is induced when levels of methionine and/or cysteine drop below 1 mM. Care et al. (15) developed a vector for expressing any gene of interest from PMET3 in which URA3 and PMET3—linked to the gene of interest by conventional cloning methods—are expressed at the RP10 locus. Expression of green fluorescent protein (GFP) from PMET3 exhibits a wide range, with transcripts being undetectable under full repression conditions. The MET3 promoter has been used to generate “depletion” strains that lack the essential genes CaSEC20 (104), SRB1 (104), and CaVRG4 (80). This C. albicans MET3 promoter (as well as the PCK1 and GAL1 promoters) has been engineered into a vector that can be used for PCR-mediated insertion of a selectable marker sequence plus the promoter of interest into any genomic site (M. Gerami-Nejad, J. Berman, and C. Gale, unpublished data).
The PCK1 promoter, which drives PEP carboxykinase expression, is repressed in the presence of glucose and induced on gluconeogenic carbon sources (64). PPCK1 was used by Bockmuhl and coworkers (8) to study the functions of the C. albicans TPK1 and TPK2 genes, which encode two isoforms of protein kinase A. Strains lacking both copies of either TPK1 or TPK2 are viable, but no tpk1/tpk1 tpk2/tpk2 strains could be isolated, suggesting that the two genes encode a redundant, essential function. Expression of the only copy of TPK1 from the PCK1 promoter (in a PPCK1-TPK1/tpk1 tpk2/tpk2 strain) generated strains that grew normally under moderate induction conditions (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-Gal] medium) but that grew very slowly under repressing conditions (glucose). When high levels of expression were induced (on medium containing amino acids as the sole carbon source), the strain again grew slowly, presumably because excess Tpk is detrimental to cell growth. Interestingly, viable cells expressing the only copy of TPK1 from the MET3 promoter were not isolated, suggesting that the levels of expression from PMET3 did not produce sufficient levels of TPK1 mRNA for viability. Consistent with this result, repression of PPCK1-SEC20 resulted in slow growth, while repression of PMET3-SEC20 arrested growth. Together, these results suggest that repression of PPCK1 does not eliminate gene expression but rather reduces it to low levels that can maintain the viability of cells that would die if they expressed no gene product at all. In contrast, expression from repressed PMET3 is below the threshold of expression that is required for proteins such as Tpk1, Sec20, and Vrg4 to support cell viability.
In addition to the endogenous C. albicans promoters described above, a heterologous, tetracycline-regulatable expression system has been adapted for use in C. albicans (77). This system utilizes two components. One is a TR transactivator, which is a fusion protein between the Escherichiacoli tetracycline repressor protein and the activation domain of S. cerevisiae Hap4p. The other is a TR promoter, comprising a minimal promoter element with a tetracycline operator sequence (tetO). In the absence of tetracycline, tetR dimers specifically bind tetO, keeping it repressed. In the presence of tetracycline, tetR dimerization is inhibited and the genes are rapidly activated by the Hap4AD. This system has several advantages in that it is specific and, rather than requiring a change in the nutritional content of the medium, which can affect C. albicans physiology, it utilizes a nontoxic drug, tetracycline, which should not affect eukaryotic gene function appreciably. However, this system requires the use of a strain that expresses the TR transactivator. Researchers at Elitra, Inc., have been using a similar tetracycline-regulatable system to generate a collection of strains with specific C. albicans gene products depleted with the ultimate goal of generating a complete genomic set of strains that can be conditionally repressed by treatment with tetracycline (T. Roemer, personal communication).
Recently, “antisense” techniques have been used as a way to influence gene expression. De Backer and coworkers used antisense constructs coupled with promoter interference to identify unknown genes which were critical for growth (31). Antisense constructs have also been used in C. neoformans to inactivate the genes for laccase (LAC) and calcineurin (CNA1), either by the use of cDNA clones expressed in an antisense direction or, in the case of CNA1, by the use of 30-bp antisense oligonucleotides (48). This study demonstrated that the phenotypes of the antisense strains were similar to those of the null mutants, although the expression of the genes was only diminished, not abolished.
Liu and colleagues reported the use of RNA interference in C. neoformans (65). They transformed cells with a plasmid carrying inverted copies of the ADE2 and/or the CAP59 gene to generate double-stranded RNAs after expression of the inverted repeat from a single promoter. They were able to achieve mutant phenotypes, and when both genes were present in the inverted molecule, both mutant phenotypes were observed. This is a promising advance, as it is a relatively straightforward method, requiring a single transformation step, that should enable phenotype testing and, possibly, the analysis of gene function for genes present in multiple copies in the genome. It will be interesting to see how broadly it can be applied to pathogenic fungi.
The GAL7 promoter has been used in C. neoformans to regulate gene expression in both serotype D (16, 108) and serotype A (32) strains. The GAL7 promoter is regulated differently in the different serotypes; it was completely repressed in the presence of glucose in serotype D yet was only modestly regulated in serotype A cells.
REPORTERS OF GENE EXPRESSION
The β-galactosidase gene is the reporter gene most commonly used in microbiology, and the medically important fungi are no exception. The enzyme can be detected in colonies growing on X-Gal medium and, using o-nitrophenyl-β-d-galactopyranoside, in cell extracts. E. coli lacZ has been shown to be useful as a reporter of gene expression in C. glabrata (38), as well as in A. fumigatus (89), C. neoformans (98), and H. capsulatum (81), where codon usage is not an issue. C. albicans (as well as several other Candida species) decodes CUG as serine rather than leucine; thus, expression of a heterologous ORF often results in a nonfunctional protein. One of the earliest reporter systems used to study gene expression in C. albicans was Kluyveromyces lactis β-galactosidase encoded by the LAC4 gene, which contains two CUG codons (63). Limited usefulness was obtained with this reporter system: only a fraction of transformants had detectable expression of the reporter even when the gene was driven from the relatively strong ACT1 promoter. Recently, the Streptococcus thermophilus lacZ gene has been used to improve the utility of β-galactosidase as a reporter system (100). S. thermophilus lacZ contains only one CUG codon. The native enzyme, as well as a codon-optimized construct, gave similar and readily detectable levels of β-galactosidase activity when expressed from a constitutive (ACT1), hypha-specific (HWP1), or inducible (MAL2) promoter. The luciferase gene from the sea pansy Renilla reniformis (RLUC), which contains no CUG codons (92), and the firefly luciferase gene (FLUC), which contains nine CUG codons, have both been used in reporter gene constructs, although the latter is not expressed as a protein and must be assayed by Northern blotting (91). Luciferase acts on the substrate coelentrazine to form a bioluminescent product, the amount of which can be measured with a luminometer. When expressed from the WH11 promoter, RLUC activity was detected only in white-phase cells, whereas expression from the OP4 promoter was detected only in opaque cells. Importantly, this reporter system is useful in in vivo assays because the substrate is taken up by yeast cells and the resulting intracellular bioluminescence can be readily detected. β-Glucuronidase, encoded by the E. coli GUS gene, has been used as a reporter in C. neoformans to estimate the efficiency of regulation of the GAL7 promoter (108).
Genes complementing an enzymatic deficiency can also serve as reporters, but the host strain must contain the corresponding mutation. For example, C. albicans URA3 (75), XOG1 (β-glucanase) (17), and MET15 (O-acetylhomoserine O-acetylserine sulfhydrylase) (103) have been used as reporters of gene expression. The URA3 and XOG1 systems are quite sensitive and useful for determining quantitative levels of activity. The MET15 system utilizes a colony color change for qualitative screening of large numbers of colonies. met15 mutant colonies are dark brown when grown in the presence of lead, whereas expression of Met15p restores the normal white colony color.
An in vivo expression technology, based on genetic recombination, has recently been developed as a reporter of gene expression (93). This system utilizes the codon-optomized FLP recombinase sequence (ecaFLP) under the control of a specific promoter in a plasmid containing URA3. The cassette is then transformed into a Ura− C. albicans strain containing an integrated mycophenolic acid resistance (MPAR) marker flanked by FRT sequences (FRT-MPAR-FRT). Activity of the specific promoter sequence being studied can then be detected by a loss of MPAR. Because loss of the MPAR marker is irreversible, down-regulation of a gene after a previous induction cannot be detected with this particular in vivo system, and the sequence of gene regulation events must be inferred from a compilation of single data points from independent experiments. Staib et al. used this reporter system to look at differential expression of the secreted aspartyl proteinases in mouse infections differing in type (systemic versus mucosal) or in timing (early versus late) (93). The strength of this system is that it allows one to analyze the expression of a particular gene at different stages of the in vivo fungus-host interaction.
In the last several years, visually exciting gene reporter systems involving fluorescent-protein fusions have become available. The Aequorea victoria GFP reporter system offers the major advantage that both gene expression and protein localization can be monitored in living organisms. Both Cormack et al. (20) and Morschhauser et al. (71) described the usefulness of codon-optimized versions of GFP as a reporter of protein expression and localization in C. albicans. Both systems employed conventional cloning techniques to generate a promoter-GFP fusion cassette that could be transformed into C. albicans to give either a plasmid-based or a more stable chromosomally integrated expression construct. GFP has also been used in C. neoformans to monitor both in vitro and in vivo expression of the Mfα1 gene (33), as well as in H. capsulatum to examine the expression of the calcium binding protein, Cbp1p (57).
Recently, YFP (yellow fluorescent protein) and CFP (cyan fluorescent protein) variants of GFP have been generated by site-directed mutagenesis of Cormack's modified GFP (47). Differentially labeled proteins can be examined simultaneously by using multicolor imaging, facilitating protein coexpression and colocalization studies (45, 47). The fluorescent-protein cassettes are maintained in plasmids designed to allow PCR-mediated amplification of the fluorescent-protein sequence along with a selectable marker (URA3 or HIS1). The gene-specific sequence is incorporated into PCR primers to generate a cassette that can integrate, by homologous recombination, into a specific genomic location. This system allows one-step construction of full-length, C-terminally truncated, or promoter- fluorescent-protein fusions that are expressed from the native promoter. Thus, gene expression and protein localization can be analyzed in vivo in real time, an advantage over studies employing classical immunofluorescence techniques on fixed cells. A caveat to this system is that introduction of a fluorescent-protein tag may interfere with the expression, localization, and/or function of the fusion protein relative to the native protein. Thus, it is important to perform experimental controls, such as integration of the fluorescent-protein cassette into the remaining wild-type allele of the heterozygous strain, to determine whether the fluorescent-protein tag interferes with complementation. Other controls include using alternative epitope tags, such as hemagglutinin and myc (76; Gerami-Nejad et al., unpublished) or FLAG (101); placing the GFP tag in a different location (e.g., at the N terminus or in the middle of the gene) (47); or using classical antibody-based immunofluorescence techniques.
TRANSCRIPTION PROFILING USING DNA MICROARRAYS
With the completion of the C. albicans genome-sequencing project, several investigators are using DNA microarray technology to study gene expression on a genomewide scale. A key advantage of this technology is that patterns of expression involving many genes can be identified. One goal is to identify most or all of the targets of specific regulatory genes. In addition, microarrays may provide insight into unknown gene functions and provide information about how drugs achieve their therapeutic effects (mechanism-of-action studies).
Prior to the availability of C. albicans DNA arrays, Lorenz and Fink used S. cerevisiae microarrays to analyze the expression profiles of S. cerevisiae cells during their interactions with cultured macrophages (67). They found that genes encoding enzymes in the glyoxylate cycle are induced in live yeast cells isolated from the phagolysosome. Using this information, they determined that C. albicans cells also induce the principal enzymes of the glyoxylate cycle, isocitrate lyase (ICL1) and malate synthase (MLS1), during phagocytosis and that C. albicans cells lacking ICL1 are less virulent in mice (67). Experiments performed with C. albicans microarrays are consistent with these results (M. Lorenz, personal communication).
Also in 2001, Lane et al. developed DNA arrays containing 700 C. albicans ORFs and used them to study the differences in gene expression between wild-type C. albicans and the signaling mutants cph1/cph1, cph2/cph2, and efg1/efg1 during yeast form and hyphal-form growth (61). Based on their results, the authors hypothesized that several C. albicans key virulence factors are convergently regulated by distinct signaling pathways.
A European consortium led by Alistair Brown used DNA filter arrays containing 2,002 C. albicans ORFs to analyze expression profiles of cells lacking the transcriptional regulator TUP1, as well as NRG1 or MIG1. They found that a subset of Tup1p-regulated genes, which includes known hypha-specific genes and other virulence factors, is also repressed by Nrg1p (74). Furthermore, NRG1 and TUP1 repressed a different set of C. albicans genes than MIG1 and TUP1, supporting the idea that NRG1 and MIG1 target TUP1 to different promoters. In addition, they identified genes that were regulated by NRG1 or MIG1 in a TUP1-independent manner (73).
DNA microarrays developed by Incyte, Inc., contain 6,600 C. albicans ORFs, some identified from genomic DNA sequences and others identified from cDNA sequences. These ORFs were amplified by PCR, printed on glass slides, and used in differential hybridization experiments where mRNAs from different conditions or genetic strains were labeled with different fluors (most commonly Cy3 and Cy5), mixed, and hybridized to the same array (34). De Backer et al. used the Incyte arrays to study the response of C. albicans to a drug treatment (itraconazole) (31). A 24-h treatment of C. albicans with inhibitory concentrations of itraconazole affected the expression of 296 ORFS, including those expected because of their role in ergosterol biosynthesis. A newer technology is to print 70-mer oligonucleotides onto glass slides and to perform differential hybridizations using Cy3 and Cy5 fluors. A set of 6,266 70-mer probes available from Qiagen Operon (http://www.Operon.com) includes 5,921 probes designed from the genome sequence and another 345 probes designed from cloned genes found in GenBank.
Several other aspects of C. albicans gene expression have been examined using microarrays. Cowen et al. have used DNA microarrays to examine changes in gene expression during experimental acquisition of resistance to the antifungal drug fluconazole (23). They found two different terminal gene expression profiles among four different resistant populations. There was a reproducible sequence of changes as drug resistance increased in two of the four populations they examined. The final transcriptional profile common to three of the four populations was shared by some clinical isolates resistant to fluconazole. They suggest that there are several programs of adaptation to fluconazole, one of which is relatively frequent. Nantel et al. have used microarrays to profile the response of C. albicans to a shift to 37°C and serum and have shown that the genes which respond to serum are regulated by the two transcription factors EFG1 and CPH1 (78). Using a DNA microarray limited to genes involved in cell wall biosynthesis, Sohn and coworkers have found that EFG1 also plays a major role in regulating cell wall development in both the yeast and the hyphal phases (90).
For H. capsulatum, microarrays have been constructed by PCR amplification of 10,000 random 0.5- to 2-kb cloned genomic DNA fragments. These have been used to compare the profiles of cells growing in the mycelial form with those of cells growing in the pathogenic yeast form. In addition, the expression profiles of H. capsulatum cells after phagocytosis by resting macrophages and the profiles of cells responding to reactive nitrogen intermediates, like those produced by activated macrophages, have been analyzed (A. Sil, personal communication).
The different DNA array technologies should all be considered to provide qualitative, rather than quantitative, data regarding gene expression levels. It is important that experiments using differential hybridization include controls, such as labeling each sample with both Cy3 and Cy5 and performing reciprocal hybridization experiments to take into account differences in the efficiency of fluor labeling, as well as other fluor-related artifacts. Furthermore, each experiment should be repeated several times, as minor changes in cell growth and/or technical manipulations can influence the resulting expression data. The expression profiles of cells under different relevant environmental and pathogenesis conditions are likely to identify new genes and biochemical pathways involved in fungal pathogenesis.
CONCLUSIONS
During the past 10 years, genetic, molecular, and genomic approaches have been widely used to study a number of medically important fungi. The results have generated fundamental biological information about the organisms and have identified potential virulence factors and drug targets. Genetic transformation, gene disruption, generation and use of conditional mutants and reporter genes, and microarray analysis provide a powerful armamentarium with which to address important questions. However, effective use of these powerful techniques requires that careful control experiments be carried out and that hypotheses be critically tested rather than simply “confirmed.”
Acknowledgments
We thank B. B. Magee for sharing preliminary data and for reading the manuscript.
The work in P. T. Magee's laboratory was supported by NIAID grant AI16567 and NIAID contract AI05406. Work in J. Berman's laboratory was supported by Burroughs Wellcome Senior Scholar Award 0677 and NIH grant RO1-DE14666. The work in C. Gale's laboratory was supported by a March of Dimes Basil O'Connor Award (5-FY99-791) and NIAID grant AI01712-01.
Editor: D. A. Portnoy
REFERENCES
- 1.Abuodeh, R. O., M. J. Orbach, M. A. Mandel, A. Das, and J. N. Galgiani. 2000. Genetic transformation of Coccidioides immitis facilitated by Agrobacterium tumefaciens. J. Infect. Dis. 181:2106-2110. [DOI] [PubMed] [Google Scholar]
- 2.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aleksenko, A., and A. J. Clutterbuck. 1996. The plasmid replicator AMA1 in Aspergillus nidulans is an inverted duplication of a low-copy-number dispersed genomic repeat. Mol. Microbiol. 19:565-574. [DOI] [PubMed] [Google Scholar]
- 4.Backen, A. C., I. D. Broadbent, R. W. Fetherston, J. D. Rosamond, N. F. Schnell, and M. J. Stark. 2000. Evaluation of the CaMAL2 promoter for regulated expression of genes in Candida albicans. Yeast 16:1121-1129. [DOI] [PubMed] [Google Scholar]
- 5.Bain, J. M., C. Stubberfield, and N. A. Gow. 2001. Ura-status-dependent adhesion of Candida albicans mutants. FEMS Microbiol. Lett. 204:323-328. [DOI] [PubMed] [Google Scholar]
- 6.Beadle, G. W., and E. L. Tatum. 1941. Genetic control of biochemical reactions in Neurospora. Proc. Natl. Acad. Sci. USA 27:499-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beckerman, J., H. Chibana, J. Turner, and P. T. Magee. 2001. Single-copy IMH3 allele is sufficient to confer resistance to mycophenolic acid in Candida albicans and to mediate transformation of clinical Candida species. Infect. Immun. 69:108-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bockmuhl, D. P., S. Krishnamurthy, M. Gerads, A. Sonneborn, and J. F. Ernst. 2001. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol. Microbiol. 42:1243-1257. [DOI] [PubMed] [Google Scholar]
- 9.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 10.Brandhorst, T. T., M. Wuthrich, T. Warner, and B. Klein. 1999. Targeted gene disruption reveals an adhesin indispensable for pathogenicity of Blastomyces dermatitidis. J. Exp. Med. 189:1207-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, D. H., Jr., A. D. Giusani, X. Chen, and C. A. Kumamoto. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34:651-662. [DOI] [PubMed] [Google Scholar]
- 12.Brown, D. H., Jr., I. V. Slobodkin, and C. A. Kumamoto. 1996. Stable transformation and regulated expression of an inducible reporter construct in Candida albicans using restriction enzyme-mediated integration. Mol. Gen. Genet. 251:75-80. [DOI] [PubMed] [Google Scholar]
- 13.Brown, J. S., A. Aufauvre-Brown, and D. W. Holden. 1998. Insertional mutagenesis of Aspergillus fumigatus. Mol. Gen. Genet. 259:327-335. [DOI] [PubMed] [Google Scholar]
- 14.Cannon, R. D., H. F. Jenkinson, and M. G. Shepherd. 1990. Isolation and nucleotide sequence of an autonomously replicating sequence (ARS) element functional in Candida albicans and Saccharomyces cerevisiae. Mol. Gen. Genet. 221:210-218. [DOI] [PubMed] [Google Scholar]
- 15.Care, R. S., J. Trevethick, K. M. Binley, and P. E. Sudbery. 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34:792-798. [DOI] [PubMed] [Google Scholar]
- 16.Chang, Y. C., B. L. Wickes, and K. J. Kwon-Chung. 1995. Further analysis of the CAP59 locus of Cryptococcus neoformans: structure defined by forced expression and description of a new ribosomal protein-encoding gene. Gene 167:179-183. [DOI] [PubMed] [Google Scholar]
- 17.Cid, V. J., A. M. Alvarez, A. I. Santos, C. Nombela, and M. Sanchez. 1994. Yeast exo-beta-glucanases can be used as efficient and readily detectable reporter genes in Saccharomyces cerevisiae. Yeast 10:747-756. [DOI] [PubMed] [Google Scholar]
- 18.Clark, K. L., P. J. Feldmann, D. Dignard, R. Larocque, A. J. Brown, M. G. Lee, D. Y. Thomas, and M. Whiteway. 1995. Constitutive activation of the Saccharomyces cerevisiae mating response pathway by a MAP kinase kinase from Candida albicans. Mol. Gen. Genet. 249:609-621. [DOI] [PubMed] [Google Scholar]
- 19.Cognetti, D., D. Davis, and J. Sturtevant. 2002. The Candida albicans 14-3-3 gene, BMH1, is essential for growth. Yeast 19:55-67. [DOI] [PubMed] [Google Scholar]
- 20.Cormack, B. P., G. Bertram, M. Egerton, N. A. Gow, S. Falkow, and A. J. Brown. 1997. Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology 143:303-311. [DOI] [PubMed] [Google Scholar]
- 21.Cormack, B. P., and S. Falkow. 1999. Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics 151:979-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cormack, B. P., N. Ghori, and S. Falkow. 1999. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science 285:578-582. [DOI] [PubMed] [Google Scholar]
- 23.Cowen, L. E., A. Nantel, M. S. Whiteway, D. Y. Thomas, D. C. Tessier, L. M. Kohn, and J. B. Anderson. 2002. Population genomics of drug resistance in Candida albicans. Proc. Natl. Acad. Sci. USA 99:9284-9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox, G. M., D. L. Toffaletti, and J. R. Perfect. 1996. Dominant selection system for use in Cryptococcus neoformans. J. Med. Vet. Mycol. 34:385-391. [PubMed] [Google Scholar]
- 25.Cruz, M. C., A. L. Goldstein, J. R. Blankenship, M. Del Poeta, D. Davis, M. E. Cardenas, J. R. Perfect, J. H. McCusker, and J. Heitman. 2002. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 21:546-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Csank, C., C. Makris, S. Meloche, K. Schroppel, M. Rollinghoff, D. Dignard, D. Y. Thomas, and M. Whiteway. 1997. Derepressed hyphal growth and reduced virulence in a VH1 family-related protein phosphatase mutant of the human pathogen Candida albicans. Mol. Biol. Cell 8:2539-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. de Jesus Berrios, C. M. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607-2615. [DOI] [PubMed] [Google Scholar]
- 28.Davidson, R. C., M. C. Cruz, R. A. Sia, B. Allen, J. A. Alspaugh, and J. Heitman. 2000. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet. Biol. 29:38-48. [DOI] [PubMed] [Google Scholar]
- 29.Davis, D., V. Bruno, L. Loza, S. Filler, and A. Mitchell. 2002. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162:1573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Backer, M. D., D. Maes, S. Vandoninck, M. Logghe, R. Contreras, and W. H. Luyten. 1999. Transformation of Candida albicans by electroporation. Yeast 15:1609-1618. [DOI] [PubMed] [Google Scholar]
- 31.De Backer, M. D., B. Nelissen, M. Logghe, J. Viaene, I. Loonen, S. Vandoninck, R. de Hoogt, S. Dewaele, F. A. Simons, P. Verhasselt, G. Vanhoof, R. Contreras, and W. H. Luyten. 2001. An antisense-based functional genomics approach for identification of genes critical for growth of Candida albicans. Nat. Biotechnol. 19:235-241. [DOI] [PubMed] [Google Scholar]
- 32.Del Poeta, M., D. L. Toffaletti, T. H. Rude, C. C. Dykstra, J. Heitman, and J. R. Perfect. 1999. Topoisomerase I is essential in Cryptococcus neoformans: role in pathobiology and as an antifungal target. Genetics 152:167-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.del Poeta, M., D. L. Toffaletti, T. H. Rude, S. D. Sparks, J. Heitman, and J. R. Perfect. 1999. Cryptococcus neoformans differential gene expression detected in vitro and in vivo with green fluorescent protein. Infect. Immun. 67:1812-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 35.Devasahayam, G., V. Chaturvedi, and S. D. Hanes. 2002. The Ess1 prolyl isomerase is required for growth and morphogenetic switching in Candida albicans. Genetics 160:37-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edman, J. C. 1992. Isolation of telomerelike sequences from Cryptococcus neoformans and their use in high-efficiency transformation. Mol. Cell. Biol. 12:2777-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edman, J. C., and K. J. Kwon-Chung. 1990. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol. Cell. Biol. 10:4538-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El Barkani, A., K. Haynes, H. Mosch, M. Frosch, and F. A. Muhlschlegel. 2000. Candida glabrata shuttle vectors suitable for translational fusions to lacZ and use of beta-galactosidase as a reporter of gene expression. Gene 246:151-155. [DOI] [PubMed] [Google Scholar]
- 39.Enloe, B., A. Diamond, and A. P. Mitchell. 2000. A single-transformation gene function test in diploid Candida albicans. J. Bacteriol. 182:5730-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Falkow, S. 1988. Molecular Koch's postulates applied to microbial pathogenicity. Rev. Infect. Dis. 10(Suppl. 2):S274-S276. [DOI] [PubMed]
- 41.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu, Y., S. G. Filler, B. J. Spellberg, W. Fonzi, A. S. Ibrahim, T. Kanbe, M. A. Ghannoum, and J. E. Edwards, Jr. 1998. Cloning and characterization of CAD1/AAF1, a gene from Candida albicans that induces adherence to endothelial cells after expression in Saccharomyces cerevisiae. Infect. Immun. 66:2078-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu, Y., G. Rieg, W. A. Fonzi, P. H. Belanger, J. E. Edwards, Jr., and S. G. Filler. 1998. Expression of the Candida albicans gene ALS1 in Saccharomyces cerevisiae induces adherence to endothelial and epithelial cells. Infect. Immun. 66:1783-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fungaro, M. H., E. Rech, G. S. Muhlen, M. H. Vainstein, R. C. Pascon, M. V. de Queiroz, A. A. Pizzirani-Kleiner, and J. L. de Azevedo. 1995. Transformation of Aspergillus nidulans by microprojectile bombardment on intact conidia. FEMS Microbiol. Lett. 125:293-297. [DOI] [PubMed] [Google Scholar]
- 45.Gale, C., M. Gerami-Nejad, M. McClellan, S. Vandoninck, M. S. Longtine, and J. Berman. 2001. Candida albicans Int1p interacts with the septin ring in yeast and hyphal cells. Mol. Biol. Cell 12:3538-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaur, N. K., and S. A. Klotz. 1997. Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties uponSaccharomyces cerevisiae for extracellular matrix proteins, Infect. Immun. 65:5289-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerami-Nejad, M., J. Berman, and C. A. Gale. 2001. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18:859-864. [DOI] [PubMed] [Google Scholar]
- 48.Gorlach, J. M., H. C. McDade, J. R. Perfect, and G. M. Cox. 2002. Antisense repression in Cryptococcus neoformans as a laboratory tool and potential antifungal strategy. Microbiology 148:213-219. [DOI] [PubMed] [Google Scholar]
- 49.Gorman, J. A., W. Chan, and J. W. Gorman. 1991. Repeated use of GAL1 for gene disruption in Candida albicans. Genetics 129:19-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goshorn, A. K., and S. Scherer. 1989. Genetic analysis of prototrophic natural variants of Candida albicans. Genetics 123:667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hara, A., M. Arie, T. Kanai, T. Matsui, H. Matsuda, K. Furuhashi, M. Ueda, and A. Tanaka. 2001. Novel and convenient methods for Candida tropicalis gene disruption using a mutated hygromycin B resistance gene. Arch. Microbiol. 176:364-369. [DOI] [PubMed] [Google Scholar]
- 52.Hashida-Okado, T., A. Ogawa, I. Kato, and K. Takesako. 1998. Transformation system for prototrophic industrial yeasts using the AUR1 gene as a dominant selection marker. FEBS Lett. 425:117-122. [DOI] [PubMed] [Google Scholar]
- 53.Herreros, E., M. I. Garcia-Saez, C. Nombela, and M. Sanchez. 1992. A reorganized Candida albicans DNA sequence promoting homologous non-integrative genetic transformation. Mol. Microbiol. 6:3567-3574. [DOI] [PubMed] [Google Scholar]
- 54.Hinnen, A., J. B. Hicks, and G. R. Fink. 1978. Transformation of yeast. Proc. Natl. Acad. Sci. USA 75:1929-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hogan, L. H., and B. S. Klein. 1997. Transforming DNA integrates at multiple sites in the dimorphic fungal pathogen Blastomyces dermatitidis. Gene 186:219-226. [DOI] [PubMed] [Google Scholar]
- 56.Kohler, G. A., T. C. White, and N. Agabian. 1997. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 179:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kugler, S., B. Young, V. L. Miller, and W. E. Goldman. 2000. Monitoring phase-specific gene expression in Histoplasma capsulatum with telomeric GFP fusion plasmids. Cell Microbiol. 2:537-547. [DOI] [PubMed] [Google Scholar]
- 58.Kurtz, M. B., M. W. Cortelyou, and D. R. Kirsch. 1986. Integrative transformation of Candida albicans, using a cloned Candida ADE2 gene. Mol. Cell. Biol. 6:142-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kurtz, M. B., M. W. Cortelyou, S. M. Miller, M. Lai, and D. R. Kirsch. 1987. Development of autonomously replicating plasmids for Candida albicans. Mol. Cell. Biol. 7:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwon-Chung, K. J., A. Varma, J. C. Edman, and J. E. Bennett. 1992. Selection of ura5 and ura3 mutants from the two varieties of Cryptococcus neoformans on 5-fluoroorotic acid medium. J. Med. Vet. Mycol. 30:61-69. [PubMed] [Google Scholar]
- 61.Lane, S., C. Birse, S. Zhou, R. Matson, and H. Liu. 2001. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 276:48988-48996. [DOI] [PubMed] [Google Scholar]
- 62.Lay, J., L. K. Henry, J. Clifford, Y. Koltin, C. E. Bulawa, and J. M. Becker. 1998. Altered expression of selectable marker URA3 in gene-disrupted Candida albicans strains complicates interpretation of virulence studies. Infect. Immun. 66:5301-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leuker, C. E., A. M. Hahn, and J. F. Ernst. 1992. β-Galactosidase of Kluyveromyces lactis (Lac4p) as reporter of gene expression in Candida albicans and C. tropicalis. Mol. Gen. Genet. 235:235-241. [DOI] [PubMed] [Google Scholar]
- 64.Leuker, C. E., A. Sonneborn, S. Delbruck, and J. F. Ernst. 1997. Sequence and promoter regulation of the PCK1 gene encoding phosphoenolpyruvate carboxykinase of the fungal pathogen Candida albicans. Gene 192:235-240. [DOI] [PubMed] [Google Scholar]
- 65.Liu, H., T. R. Cottrell, L. M. Pierini, W. E. Goldman, and T. L. Doering. 2002. RNA interference in the pathogenic fungus Cryptococcus neoformans. Genetics 160:463-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 67.Lorenz, M. C., and G. R. Fink. 2001. The glyoxylate cycle is required for fungal virulence. Nature 412:83-86. [DOI] [PubMed] [Google Scholar]
- 68.McDade, H. C., and G. M. Cox. 2001. A new dominant selectable marker for use in Cryptococcus neoformans. Med. Mycol. 39:151-154. [DOI] [PubMed] [Google Scholar]
- 69.Mehra, R. K., J. L. Thorvaldsen, I. G. Macreadie, and D. R. Winge. 1992. Cloning system for Candida glabrata using elements from the metallothionein-IIa-encoding gene that confer autonomous replication. Gene 113:119-124. [DOI] [PubMed] [Google Scholar]
- 70.Mendoza, I., F. Rubio, A. Rodriguez-Navarro, and J. M. Pardo. 1994. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J. Biol. Chem. 269:8792-8796. [PubMed] [Google Scholar]
- 70a.Michel, S., S. Ushinsky, B. Klebl, E. Leberer, D. Thomas, M. Whiteway, and J. Morschhauser. 2002. Generation of conditional lethal Candida albicans mutants by inducible deletion of essential genes. Mol. Microbiol. 46:269-280. [DOI] [PubMed] [Google Scholar]
- 71.Morschhauser, J., S. Michel, and J. Hacker. 1998. Expression of a chromosomally integrated, single-copy GFP gene in Candida albicans, and its use as a reporter of gene regulation. Mol. Gen. Genet. 257:412-420. [DOI] [PubMed] [Google Scholar]
- 72.Munro, C. A., K. Winter, A. Buchan, K. Henry, J. M. Becker, A. J. Brown, C. E. Bulawa, and N. A. Gow. 2001. Chs1 of Candida albicans is an essential chitin synthase required for synthesis of the septum and for cell integrity. Mol. Microbiol. 39:1414-1426. [DOI] [PubMed] [Google Scholar]
- 73.Murad, A. M., C. d'Enfert, C. Gaillardin, H. Tournu, F. Tekaia, D. Talibi, D. Marechal, V. Marchais, J. Cottin, and A. J. Brown. 2001. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42:981-993. [DOI] [PubMed] [Google Scholar]
- 74.Murad, A. M., P. Leng, M. Straffon, J. Wishart, S. Macaskill, D. MacCallum, N. Schnell, D. Talibi, D. Marechal, F. Tekaia, C. d'Enfert, C. Gaillardin, F. C. Odds, and A. J. Brown. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20:4742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Myers, K. K., P. S. Sypherd, and W. A. Fonzi. 1995. Use of URA3 as a reporter of gene expression in C. albicans. Curr. Genet. 27:243-248. [DOI] [PubMed] [Google Scholar]
- 76.Nakayama, H., M. Izuta, S. Nagahashi, E. Y. Sihta, Y. Sato, T. Yamazaki, M. Arisawa, and K. Kitada. 1998. A controllable gene-expression system for the pathogenic fungus Candida glabrata. Microbiology 144:2407-2415. [DOI] [PubMed] [Google Scholar]
- 77.Nakayama, H., T. Mio, S. Nagahashi, M. Kokado, M. Arisawa, and Y. Aoki. 2000. Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus Candida albicans. Infect. Immun. 68:6712-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nantel, A., D. Dignard, C. Bachewich, D. Harcus, A. Marcil, A. P. Bouin, C. W. Sensen, H. Hogues, M. van het Hoog, P. Gordon, T. Rigby, F. Benoit, D. C. Tessier, D. Y. Thomas, and M. Whiteway. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nelson, R. T., J. Hua, B. Pryor, and J. K. Lodge. 2001. Identification of virulence mutants of the fungal pathogen Cryptococcus neoformans using signature-tagged mutagenesis. Genetics 157:935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nishikawa, A., J. B. Poster, Y. Jigami, and N. Dean. 2002. Molecular and phenotypic analysis of CaVRG4, encoding an essential Golgi apparatus GDP-mannose transporter. J. Bacteriol. 184:29-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Patel, J. B., J. W. Batanghari, and W. E. Goldman. 1998. Probing the yeast phase-specific expression of the CBP1 gene in Histoplasma capsulatum. J. Bacteriol. 180:1786-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peng, M., C. R. Cooper, Jr., and P. J. Szaniszlo. 1995. Genetic transformation of the pathogenic fungus Wangiella dermatitidis. Appl. Microbiol. Biotechnol. 44:444-450. [DOI] [PubMed] [Google Scholar]
- 83.Pla, J., R. M. Perez-Diaz, F. Navarro-Garcia, M. Sanchez, and C. Nombela. 1995. Cloning of the Candida albicans HIS1 gene by direct complementation of a C. albicans histidine auxotroph using an improved double-ARS shuttle vector. Gene 165:115-120. [DOI] [PubMed] [Google Scholar]
- 84.Reichard, U., C. Y. Hung, P. W. Thomas, and G. T. Cole. 2000. Disruption of the gene which encodes a serodiagnostic antigen and chitinase of the human fungal pathogen Coccidioides immitis. Infect. Immun. 68:5830-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Riggle, P. J., I. V. Slobodkin, D. H. Brown, Jr., M. P. Hanson, T. L. Volkert, and C. A. Kumamoto. 1997. Two transcripts, differing at their 3′ ends, are produced from the Candida albicans SEC14 gene. Microbiology 143:3527-3535. [DOI] [PubMed] [Google Scholar]
- 86.Santos, M. A., and M. F. Tuite. 1995. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 23:1481-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schein, J. E., K. L. Tangen, R. Chiu, H. Shin, K. B. Lengeler, W. K. MacDonald, I. Bosdet, J. Heitman, S. J. Jones, M. A. Marra, and J. W. Kronstad. 2002. Physical maps for genome analysis of serotype A and D strains of the fungal pathogen Cryptococcus neoformans. Genome Res. 12:1445-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sebghati, T. S., J. T. Engle, and W. E. Goldman. 2000. Intracellular parasitism by Histoplasma capsulatum: fungal virulence and calcium dependence. Science 290:1368-1372. [DOI] [PubMed] [Google Scholar]
- 89.Smith, J. M., C. M. Tang, S. Van Noorden, and D. W. Holden. 1994. Virulence of Aspergillus fumigatus double mutants lacking restriction and an alkaline protease in a low-dose model of invasive pulmonary aspergillosis. Infect. Immun. 62:5247-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sohn, K., C. Urban, H. Brunner, and S. Rupp. 2003. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol. Microbiol. 47:89-102. [DOI] [PubMed] [Google Scholar]
- 91.Srikantha, T., A. Chandrasekhar, and D. R. Soll. 1995. Functional analysis of the promoter of the phase-specific WH11 gene of Candida albicans. Mol. Cell. Biol. 15:1797-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Srikantha, T., A. Klapach, W. W. Lorenz, L. K. Tsai, L. A. Laughlin, J. A. Gorman, and D. R. Soll. 1996. The sea pansy Renilla reniformis luciferase serves as a sensitive bioluminescent reporter for differential gene expression in Candida albicans. J. Bacteriol. 178:121-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Staib, P., M. Kretschmar, T. Nichterlein, G. Kohler, S. Michel, H. Hof, J. Hacker, and J. Morschhauser. 1999. Host-induced, stage-specific virulence gene activation in Candida albicans during infection. Mol. Microbiol. 32:533-546. [DOI] [PubMed] [Google Scholar]
- 94.Staib, P., S. Michel, G. Kohler, and J. Morschhauser. 2000. A molecular genetic system for the pathogenic yeast Candida dubliniensis. Gene 242:393-398. [DOI] [PubMed] [Google Scholar]
- 95.Staib, P., G. P. Moran, D. J. Sullivan, D. C. Coleman, and J. Morschhauser. 2001. Isogenic strain construction and gene targeting in Candida dubliniensis. J. Bacteriol. 183:2859-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sundstrom, P., J. E. Cutler, and J. F. Staab. 2002. Reevaluation of the role of HWP1 in systemic candidiasis by use of Candida albicans strains with selectable marker URA3 targeted to the ENO1 locus. Infect. Immun. 70:3281-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tang, C. M., J. Cohen, and D. W. Holden. 1992. An Aspergillus fumigatus alkaline protease mutant constructed by gene disruption is deficient in extracellular elastase activity. Mol. Microbiol. 6:1663-1671. [DOI] [PubMed] [Google Scholar]
- 98.Toffaletti, D. L., and J. R. Perfect. 1997. Study of Cryptococcus neoformans actin gene regulation with a beta-galactosidase-actin fusion. J. Med. Vet. Mycol. 35:313-320. [PubMed] [Google Scholar]
- 99.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Uhl, M. A., and A. D. Johnson. 2001. Development of Streptococcus thermophilus lacZ as a reporter gene for Candida albicans. Microbiology 147:1189-1195. [DOI] [PubMed] [Google Scholar]
- 101.Umeyama, T., Y. Nagai, M. Niimi, and Y. Uehara. 2002. Construction of FLAG tagging vectors for Candida albicans. Yeast 19:611-618. [DOI] [PubMed] [Google Scholar]
- 102.Varma, A., and K. J. Kwon-Chung. 1998. Construction of stable episomes in Cryptococcus neoformans. Curr. Genet. 34:60-66. [DOI] [PubMed] [Google Scholar]
- 103.Viaene, J., P. Tiels, M. Logghe, S. Dewaele, W. Martinet, and R. Contreras. 2000. MET15 as a visual selection marker for Candida albicans. Yeast 16:1205-1215. [DOI] [PubMed] [Google Scholar]
- 104.Weber, Y., U. J. Santore, J. F. Ernst, and R. K. Swoboda. 2001. Divergence of eukaryotic secretory components: the Candida albicans homolog of the Saccharomyces cerevisiae Sec20 protein is N terminally truncated, and its levels determine antifungal drug resistance and growth. J. Bacteriol. 183:46-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Weidner, G., C. d'Enfert, A. Koch, P. C. Mol, and A. A. Brakhage. 1998. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr. Genet. 33:378-385. [DOI] [PubMed] [Google Scholar]
- 106.Whelan, W. L., R. M. Partridge, and P. T. Magee. 1980. Heterozygosity and segregation in Candida albicans. Mol. Gen. Genet. 180:107-113. [DOI] [PubMed] [Google Scholar]
- 107.Whiteway, M., D. Dignard, and D. Y. Thomas. 1992. Dominant negative selection of heterologous genes: isolation of Candida albicans genes that interfere with Saccharomyces cerevisiae mating factor-induced cell cycle arrest. Proc. Natl. Acad. Sci. USA 89:9410-9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wickes, B. L., and J. C. Edman. 1995. The Cryptococcus neoformans GAL7 gene and its use as an inducible promoter. Mol. Microbiol. 16:1099-1109. [DOI] [PubMed] [Google Scholar]
- 109.Wilson, R. B., D. Davis, B. M. Enloe, and A. P. Mitchell. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65-70. [DOI] [PubMed] [Google Scholar]
- 110.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Woods, J. P., and W. E. Goldman. 1993. Autonomous replication of foreign DNA in Histoplasma capsulatum: role of native telomeric sequences. J. Bacteriol. 175:636-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Woods, J. P., E. L. Heinecke, and W. E. Goldman. 1998. Electrotransformation and expression of bacterial genes encoding hygromycin phosphotransferase and beta-galactosidase in the pathogenic fungus Histoplasma capsulatum. Infect. Immun. 66:1697-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Woods, J. P., D. M. Retallack, E. L. Heinecke, and W. E. Goldman. 1998. Rare homologous gene targeting in Histoplasma capsulatum: disruption of the URA5Hc gene by allelic replacement. J. Bacteriol. 180:5135-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Worsham, P. L., and W. E. Goldman. 1990. Development of a genetic transformation system for Histoplasma capsulatum: complementation of uracil auxotrophy. Mol. Gen. Genet. 221:358-362. [DOI] [PubMed] [Google Scholar]
- 115.Ye, X., B. Feng, and P. J. Szaniszlo. 1999. A color-selectable and site-specific integrative transformation system for gene expression studies in the dematiaceous fungus Wangiella (Exophiala) dermatitidis. Curr. Genet. 36:241-247. [DOI] [PubMed] [Google Scholar]
- 116.Yesland, K., and W. A. Fonzi. 2000. Allele-specific gene targeting in Candida albicans results from heterology between alleles. Microbiology 146:2097-2104. [DOI] [PubMed] [Google Scholar]
- 117.Zhou, P., M. S. Szczypka, R. Young, and D. J. Thiele. 1994. A system for gene cloning and manipulation in the yeast Candida glabrata. Gene 142:135-140. [DOI] [PubMed] [Google Scholar]