Abstract
We previously reported that infection by Mycobacterium tuberculosis, the causative agent of tuberculosis, leads to secretion of alpha/beta interferon (IFN-α/β). While IFN-α/β ordinarily stimulates formation of signal transducer and stimulator of transcription-1 (STAT-1) homodimers and IFN-stimulated gene factor-3 (ISGF-3), only ISGF-3 is found in infected human monocytes and macrophages. We have now investigated the basis for this unusual profile of transcription factor activation and its consequences for regulation of transcription, as well as the impact of infection on response to IFN-α. After infection, IFN-α stimulation of STAT-1 homodimers is inhibited in monocytes and macrophages, while stimulation of ISGF-3 increases in monocytes but tends to decline in macrophages. Effects of infection on the abundance of ISGF-3 subunits, STAT-1, STAT-2, and interferon regulatory factor 9, and on tyrosine phosphorylation of STAT-1 and STAT-2 explain the observed changes in DNA-binding activity, which correlate with increased or inhibited transcription of genes regulated by ISGF-3 and STAT-1. Infection by Mycobacterium bovis BCG does not inhibit IFN-α-stimulated tyrosine phosphorylation of STAT-1, formation of homodimers, or transcription of genes regulated by STAT-1 homodimers, suggesting that inhibition of the response to IFN-α/β by M. tuberculosis is an aspect of pathogenicity. Thus, this well-known feature of infection by pathogenic viruses may also be a strategy employed by pathogenic bacteria.
Significant advances in understanding the host defense against Mycobacterium tuberculosis have been made during the past decade. Mechanisms of innate and adaptive immunity that influence the initial course of infection and mediate control of disease have been identified (see references 16 and 49 for reviews). Both types of immunity involve alpha/beta interferons (IFN-α/β) (2, 4, 14, 18, 30, 47). The IFN-α/β system helps host defense against M. tuberculosis in mice (7), and M. tuberculosis infection induces production of IFN-α/β by human macrophages and dendritic cells (18, 60). Moreover, aerosolized IFN-α combined with conventional chemotherapy for tuberculosis improves patient outcome (19). However, few details are known about the interaction between the IFN-α/β system and infection by M. tuberculosis.
IFN-α/β influences cellular function largely by regulating gene expression as the culmination of a signal transduction cascade (3, 9, 12). High-affinity binding of IFN to the IFN-α/β cell surface receptor, mediated by dimerization of IFNAR1 and IFNAR2 subunits, is followed by tyrosine phosphorylation of associated protein tyrosine kinases, Janus kinase 1 (JAK1) and TYK2, another kinase in the JAK family. This activates the kinases, which in turn activate the latent cytoplasmic transcription factors signal transducer and activator of transcription-1 (STAT-1) and STAT-2 by catalyzing tyrosine phosphorylation of each at a single site. Phosphorylated STAT-1 can form a homodimer, and the homodimer can act alone or together with IFN regulatory factor 9 (IRF-9). Activated STAT-1 and activated STAT-2 can form a heterodimer, which associates with IRF-9 to form IFN-stimulated gene factor-3 (ISGF-3). After translocation to the nucleus, these transcription factors bind to different DNA sequences. STAT-1 homodimers bind the IFN-γ activation site (GAS), while ISGF-3 and the (STAT-1)2/IRF-9 complex bind the IFN-α/β-stimulated response element (ISRE). Many genes have one regulatory element or the other, although some have both. Thus, changes in STAT-1 homodimers and/or ISGF-3 can have distinct effects on gene expression.
To understand the effect of M. tuberculosis on the IFN-α/β system, it is important to examine both monocytes and macrophages, since both are exposed to infection by M. tuberculosis in vivo (50, 51). Moreover, differentiation affects the IFN-α/β system in ways that impinge on the cellular response to infection by M. tuberculosis and alter the interaction of human immunodeficiency virus and M. tuberculosis (60). Infection of the two cell types can be modeled in vitro by using primary cells and/or the THP-1 cell line, which has long been used to study phorbol ester-induced monocyte-to-macrophage differentiation (56). For convenience, and to distinguish them from primary cells, we refer to undifferentiated (untreated) and differentiated (phorbol ester-treated) THP-1 cells as THP-1 monocytes and THP-1 macrophages. THP-1 cells are also commonly used for studies of host response to infection by M. tuberculosis (for example, see references 6, 25, and 54).
We previously showed that ISGF-3, but not STAT-1 homodimers, can be detected in THP-1 macrophages and primary alveolar macrophages 1 day after infection by M. tuberculosis (60). In the present study, we examined the effect of infection by M. tuberculosis on the response to IFN-α/β. We found that IFN-α-stimulated signal transduction, and gene expression mediated by STAT-1 was inhibited. Mycobacterium bovis BCG did not inhibit the response to IFN-α, which suggests that the effect of M. tuberculosis is related to pathogenicity. IFN-α/β impinges on both innate immunity and the transition to Th1-cell-mediated immunity, and the role of IFN-α/β in both is thus likely to be affected by M. tuberculosis.
MATERIALS AND METHODS
Bacterial strains, cells, and cytokines.
All manipulations with viable M. tuberculosis cells were performed under biosafety level 3 containment. M. tuberculosis strains TN913, a clinical isolate, and H37Rv and M. bovis BCG (Pasteur) were obtained from the Public Health Research Institute Tuberculosis Center and were grown as previously described (63). H37Rv (ATCC 27294) was used for infection of peripheral blood monocytes (PBM) and PBM-derived macrophages as previously described (18).
THP-1 cells (57) obtained from the American Type Culture Collection were grown and infected as previously described (60) and harvested 3 days later. Infection was performed at a multiplicity of infection (MOI) of 1, unless otherwise stated. The MOI was confirmed by determining the CFU of each inoculum. By acid-fast staining of a small portion of infected cells at the time of harvest and by determining intracellular CFU from the extracts, similar uptake and growth of M. tuberculosis and M. bovis BCG in all experiments were confirmed. Infected cultures of THP-1 monocytes and macrophages were, respectively, 70 to 90% and 50 to 80% viable at the time of harvest. Uninfected cells were cultured and harvested in parallel, at which time they were 97 to 100% viable. Viability was determined by exclusion of trypan blue. Recovery of protein in cell extracts and recovery of radiolabeled RNA from the nuclear run-on assay are proportional to viability. Thus, use of equal amounts of protein for electrophoretic mobility shift assays (EMSAs) and immunoblotting and equal amounts of radiolabeled RNA for hybridization (see below) controls for differences in viability.
Primary cells were used in accordance with all applicable laws and regulations. Alveolar macrophages were obtained from a healthy volunteer and were cultured and infected with M. tuberculosis TN913 as previously described (60) and harvested 3 days later. Uninfected cells were cultured in parallel. PBM were obtained from a healthy volunteer as previously described (18). Infection with a single-cell suspension of M. tuberculosis H37Rv at an MOI of approximately 1 was begun immediately or after 4 days of adherence-induced differentiation and continued for 3 days. Alternatively, PBM were recovered from peripheral blood mononuclear cells by allowing them to adhere to culture flasks and then washing with phosphate-buffered saline (PBS) the next day to remove nonadherent cells. Cells were then infected or were cultured for 5 days with 30 ng of granulocyte-macrophage colony-stimulating factor (Biosource) per ml to obtain PBM-derived macrophages prior to infection. Cells were harvested 3 days after infection by M. tuberculosis H37Rv or M. bovis BCG. Uninfected cells obtained by either of these methods were cultured in parallel.
IFN-α was provided by Hoffman-LaRoche. As indicated, cells were stimulated with 500 U/ml for the final 30 min prior to harvest.
Extract preparation and EMSA.
All steps were performed at 0 to 4°C. THP-1 macrophages, PBM, and PBM-derived macrophages were scraped into their culture media. Cells that had been adherent or in suspension, including undifferentiated, uninfected THP-1 cells, were then collected by centrifugation and suspended in PBS. The centrifugation and suspension were repeated once more. Culture media containing nonadherent cells was removed from uninfected and infected cultures of cells from bronchoalveolar lavage fluid, and the adherent cells were washed gently with PBS. The remaining alveolar macrophages were 90 to 95% pure based on microscopic examination of morphology. The alveolar macrophages were then scraped into additional PBS. Extracts were prepared as described elsewhere (37). Briefly, washed cells were collected by centrifugation and lysed with nonionic detergent. Nuclei were recovered by centrifugation, and postnuclear supernatants were taken and adjusted to 0.3 M NaCl to produce the cytoplasmic extracts. The nuclei were extracted and then were removed by centrifugation at 13,000 × g for 10 min. The supernatants were recovered as the nuclear extracts. Extracts prepared from infected cells were sterilely filtered before removal from the biosafety level 3 laboratory. Extracts were frozen rapidly on crushed dry ice and stored at −80°C.
EMSA was performed with approximately 10 μg of extract protein (2 to 3 μl of extract), as previously described (60). The ISRE, GAS, and nonspecific oligonucleotides, as well as antisera used to identify proteins contained in protein-DNA complexes, have been described previously (37-39, 48, 58). The identity of all specific protein-DNA complexes has been confirmed by competition with excess specific oligonucleotides and lack of competition with excess unlabeled nonspecific oligonucleotides, as well as by reaction with specific antisera against STAT-1, STAT-2, and IRF-9, and lack of reaction with nonspecific antisera (reference 10 and data not shown). STAT-1 reacted only with antiserum against STAT-1. ISGF-3 reacted with antisera against STAT-1, STAT-2, and IRF-9, but not with nonspecific antiserum or antiserum against IRF-1. The nonspecific complex is defined as such by its failure to compete with any excess unlabeled oligonucleotides. The amount of this complex is proportional to total protein in the extracts from many types of cells, independent of any cytokine treatment or infection we have tested. Radioactivity in protein-DNA complexes was visualized and quantified with a PhosphorImager and ImageQuant software (Molecular Dynamics). The local average background correction was applied to each specific and nonspecific complex. Values for each specific complex were then normalized to the indicated nonspecific protein-DNA complex from the same extract, which served as an internal standard for preparation of extracts and performance of the assay. Student's t test was used to determine the statistical significance of differences in STAT-1 or ISGF-3 DNA-binding activity between pairs of conditions. Statistically significant differences between weak signals indicate reproducibility that is unlikely to arise coincidentally due to fluctuation of the signal-to-noise ratio, although statistically insignificant differences may reflect the limits of quantification of such signals.
Immunoblot analysis.
Protein concentrations in the samples were determined using the Bio-Rad Bradford reagent. Equal amounts of nuclear extracts (30 μg) or cytoplasmic extracts (50 μg [for IRF-9]) were resolved on sodium dodecyl sulfate (SDS)-8% polyacrylamide gel electrophoresis gels and electroblotted to nitrocellulose membranes (Bio-Rad). Coomassie blue staining of residual protein in the SDS-polyacrylamide gel was performed to confirm that equal amounts of protein were analyzed for each sample on a gel (data not shown). The membranes were blocked in PBS containing 2.5% nonfat milk and 0.2% Tween 20. Primary antibodies were diluted in the same solution. Rabbit antibodies against STAT-1 and STAT-2 (48), or IRF-9 (58), have been previously described. Mouse antibodies against pY-STAT-1 and pY-STAT-2 have been described elsewhere (34, 35). Membranes were washed with PBS and then incubated with horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse immunoglobulin G (Calbiochem) diluted in PBS. Membranes were washed with PBS and then incubated with LumiGLO chemiluminescent substrate (Kirkegaard & Perry Laboratories). Luminescence was detected with X-ray film (Midwest Scientific). The blots were stripped in buffer containing Tris (62.5 mM; pH 6.8), 2% SDS, and 100 mM β-mercaptoethanol for 1 h at 65°C and reused as above. For each independent experiment, STAT-1, pY-STAT-1, STAT-2, and pY-STAT-2 were all analyzed on one membrane and IRF-9 was analyzed on another.
Run-on transcription rate assays.
Cells were lysed, and nuclei were recovered as described above for preparation of extracts. Isolated nuclei were incubated for 10 min at 37°C with [α-32P]UTP to radiolabel nascent RNA. RNA was purified using TriReagent as recommended by the vendor (Molecular Research Center). Hybridization of a constant amount of radiolabeled RNA to excess plasmid probes fixed on nitrocellulose was performed as previously described (39). Images were obtained, and data were quantified with a PhosphorImager.
The probes were as follows. An ISG15 genomic fragment cloned in pGem1 has been previously described (40). An IP-10 cDNA (26) was subcloned into pGem1. Reverse transcription-PCR (RT-PCR) performed by standard methods (46) was used to obtain a 1.1-kb FcγR1 cDNA fragment from THP-1 macrophage RNA. RT was performed with oligo(dT) as a primer. PCR was performed with 5′ AACATGTGGTTCTAGACAACTCTG 3′ and 5′ GCTGTTCTTCTTTTGGATCCTGAC 3′ as the upstream and downstream primers, respectively. The amplification cycle (94°C, 1 min; 58°C, 1 min; 72°C, 1 min) was repeated 30 times. The PCR product was cloned into pTopo II as recommended by the vendor (Invitrogen) and subcloned into pGem1 for use as a probe. The FcγR1 sequence was confirmed. Specificity of hybridization was determined with pGem1 (Promega) as a probe. A human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe, clone 754537 (Research Genetics), was used as a positive control and as an internal standard. The results were quantified for each sample by subtracting the signal from pGem from all the other signals as a background correction and then normalizing the corrected signals to that of GAPDH.
RESULTS
Effects of M. tuberculosis infection on IFN-α-stimulated transcription factor DNA-binding activity.
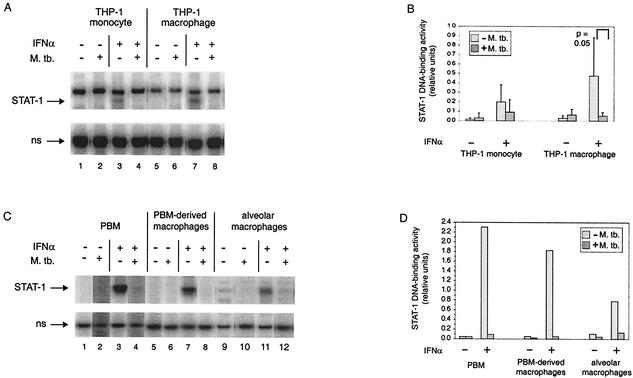
The previous observations (18, 60) that cells infected by M. tuberculosis secrete active IFN-α/β and contain ISGF-3 led us to ask whether the response to IFN-α was actually limited or inhibited by the infection, since the initial response to IFN-α/β can reduce the response to later exposure (24). We first assayed the DNA-binding activity of STAT-1 homodimers as a measure of homodimer formation in THP-1 cells (Fig. 1A). As expected, in the absence of IFN-α stimulation, activity was undetectable in THP-1 cells, regardless of whether they were uninfected or infected by M. tuberculosis and whether they were undifferentiated or differentiated (lanes 1, 2, 5, and 6). In the absence of infection, the cells were responsive to IFN-α stimulation (lanes 3 and 7). However, infection by M. tuberculosis inhibited the IFN-α-stimulated appearance of STAT-1 homodimer DNA-binding activity in the undifferentiated and differentiated cells (lanes 4 and 8). Quantification of the results from six experiments demonstrated that the extent of inhibition was reproducibly greater after differentiation (Fig. 1B). In THP-1 monocytes, the trend was the same in all of the experiments, although it was not statistically significant, and in THP-1 macrophages the difference was statistically significant. We confirmed that infection of PBM and PBM-derived macrophages from one donor and of alveolar macrophages from another resulted in similar or greater inhibition of IFN-α-stimulated STAT-1 homodimer DNA-binding activity (Fig. 1C and D). The same result was obtained with PBM-derived macrophages from two additional healthy volunteers (data not shown). Thus, the THP-1 cell line is a good model of what can happen in primary cells.
FIG. 1.
STAT-1 DNA-binding activity in monocytes and macrophages infected by M. tuberculosis and/or treated with IFN-α. (A) The complexes of the GAS probe with STAT-1 homodimers and with a nonspecific (ns) protein are shown for a representative experiment performed with THP-1 cells. THP-1 monocytes and THP-1 macrophages were infected and/or treated with IFN-α as indicated. (B) The STAT-1 DNA-binding activity from six experiments was quantified relative to the nonspecific DNA-binding activity. The average is shown for each condition, with the standard deviation shown by the error bar. Statistically significant differences between uninfected and infected cells are indicated next to the bracket over the respective pair. (C) The complexes of the GAS probe with STAT-1 homodimers and with a nonspecific (ns) protein are shown for extracts from PBM, PBM-derived macrophages, and alveolar macrophages that were infected and/or treated with IFN-α as indicated. (D) The STAT-1 DNA-binding activity shown in panel C was quantified relative to the nonspecific DNA-binding activity.
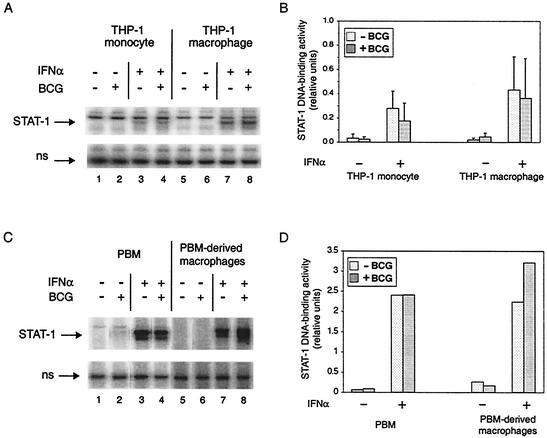
To determine if the inhibition of STAT-1 homodimer DNA-binding activity was related to pathogenicity, we used an EMSA to examine the effect of infection by a closely related, nonpathogenic mycobacterium, M. bovis BCG (Fig. 2A) and quantified the results from four replicate experiments (Fig. 2B). In contrast to M. tuberculosis, there was little or no effect on IFN-α-stimulated formation of STAT-1 homodimers in THP-1 monocytes (compare lanes 3 and 4) or in THP-1 macrophages (compare lanes 7 and 8). Even at a higher MOI (3 or 10), M. bovis BCG did not inhibit the response to IFN-α in either THP-1 monocytes or THP-1 macrophages, and when THP-1 macrophages were infected by one species or the other in the same experiment, the difference between M. tuberculosis and M. bovis BCG was again observed (data not shown). IFN-α stimulation of STAT-1 DNA-binding activity also was not inhibited in PBM and PBM-derived macrophages infected by M. bovis BCG (Fig. 2C and D). The difference between M. tuberculosis and M. bovis BCG suggests a pathogen-specific effect on IFN-α-stimulated STAT-1 homodimer DNA-binding activity.
FIG. 2.
STAT-1 DNA-binding activity in monocytes and macrophages infected by M. bovis BCG and/or treated with IFN-α. (A) The complexes of the GAS probe with STAT-1 homodimers and with a nonspecific (ns) protein are shown for a representative experiment performed with THP-1 cells. THP-1 monocytes and THP-1 macrophages were infected and/or treated with IFN-α as indicated. (B) The STAT-1 DNA-binding activity from four experiments was quantified relative to the nonspecific DNA-binding activity. The average is shown for each condition, with the standard deviation shown by the error bar. There were no statistically significant differences between respective pairs of uninfected and infected cells with or without IFN-α stimulation. (C) The complexes of the GAS probe with STAT-1 homodimers and with a nonspecific (ns) protein are shown for one experiment that was performed with PBM and PBM-derived macrophages to demonstrate that the difference between M. tuberculosis and M. bovis BCG was not only a feature of the THP-1 model. Cells were infected and/or treated with IFN-α as indicated. (D) The STAT-1 DNA-binding activity shown in panel C was quantified relative to the nonspecific DNA-binding activity.
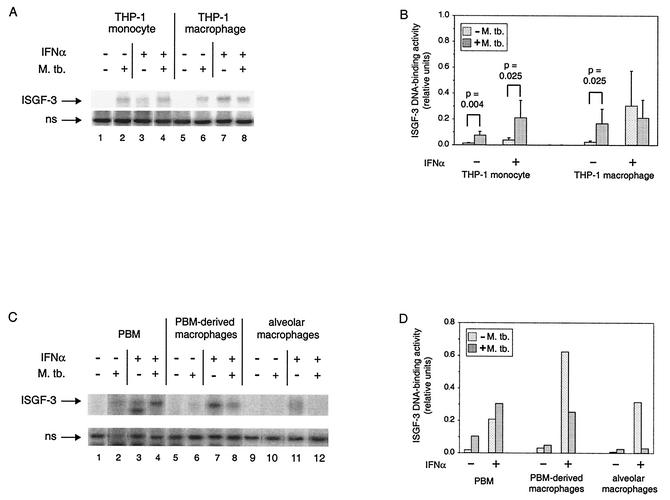
We next sought to determine if IFN-α stimulation of ISGF-3 DNA-binding activity would also be inhibited by M. tuberculosis infection (Fig. 3A), since it includes tyrosine-phosphorylated STAT-1. At 3 days postinfection, THP-1 monocytes and macrophages contained ISGF-3 (lanes 2 and 6), whereas it was only in macrophages 1 day postinfection (20). Stimulation with IFN-α induced ISGF-3 in THP-1 monocytes (lane 3) and in THP-1 macrophages (lane 7), although the induction in monocytes was minimal. Unexpectedly, the amount of ISGF-3 was greater in THP-1 monocytes that were stimulated with IFN-α after infection (lane 4) than in cells that were only stimulated. In contrast, the level of ISGF-3 in THP-1 macrophages that were stimulated after infection (lane 8) was lower than the level in cells that were only stimulated with IFN-α. Quantification of the results from replicate experiments is shown in Fig. 3B. Of the changes noted above, only the trend toward reduced IFN-α stimulation of ISGF-3 in THP-1 macrophages infected by M. tuberculosis was not statistically significant; the others had P values of ≤0.03.
FIG. 3.
ISGF-3 DNA-binding activity in monocytes and macrophages infected by M. tuberculosis and/or treated with IFN-α. (A) The complexes of the ISRE probe with ISGF-3 and with a nonspecific (ns) protein are shown for a representative experiment performed with THP-1 cells. THP-1 monocytes and THP-1 macrophages were infected and/or treated with IFN-α as indicated. (B) The ISGF-3 DNA-binding activity from five or six experiments was quantified relative to the nonspecific DNA-binding activity. Data from six experiments are included, except for THP-1 macrophages treated with IFN-α. The average is shown for each condition, with the standard deviation shown by the error bar. Statistically significant differences between uninfected and infected cells are indicated above brackets over the respective pairs. (C) The complexes of the ISRE probe with ISGF-3 and with a nonspecific (ns) protein are shown for extracts from PBM, PBM-derived macrophages, and alveolar macrophages that were infected and/or treated with IFN-α as indicated. (D) The ISGF-3 DNA-binding activity shown in panel C was quantified relative to the nonspecific DNA-binding activity.
We also determined whether IFN-α stimulation of ISGF-3 in primary cells was affected by M. tuberculosis, examining the same extracts as were tested for STAT-1 (Fig. 3C and D). At 3 days postinfection, ISGF-3 was detectable in PBM but not in PBM-derived macrophages and alveolar macrophages, while the opposite was true at 1 day postinfection (20). Moreover, infection of PBM increased IFN-α-stimulated formation of ISGF-3 by almost 50%, while infection of PBM-derived and alveolar macrophages inhibited it by more than 50 and 80%, respectively. IFN-α-stimulated formation of ISGF-3 in PBM-derived macrophages was inhibited as much or more by infection of cells from two additional donors (data not shown). Thus, what happens in THP-1 monocytes and THP-1 macrophages can happen in primary cells: with differentiation, there is a switch in the response of infected cells to exogenous IFN-α from augmented to inhibited formation of ISGF-3.
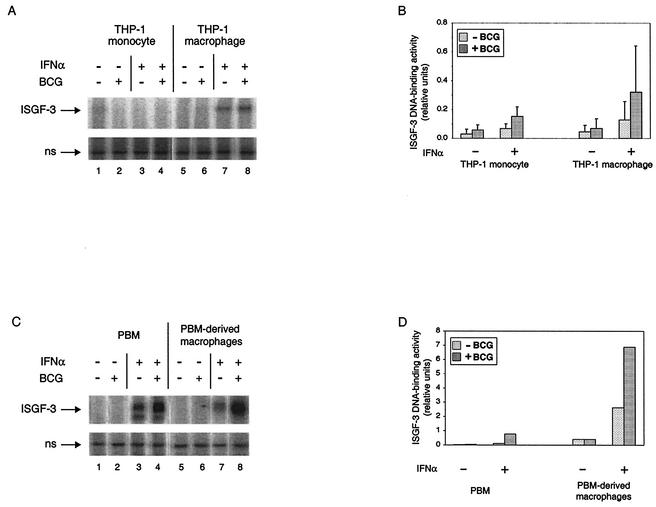
To determine whether the effects of M. tuberculosis on ISGF-3 might be related to pathogenicity, we again examined infection of THP-1 cells by M. bovis BCG. ISGF-3 DNA-binding activity was essentially undetectable by EMSA (Fig. 4A) following infection of either THP-1 monocytes (compare lanes 1 and 2) or THP-1 macrophages (compare lanes 5 and 6) with M. bovis BCG. Also in contrast to the effects of infection by M. tuberculosis on response to IFN-α, there was a trend toward augmented formation of ISGF-3 in both THP-1 monocytes and THP-1 macrophages after infection by M. bovis BCG (lanes 3, 4, 7, and 8). However, as demonstrated by quantification of the results from four replicate experiments, there were no statistically significant differences arising from infection by M. bovis BCG in cells that were not stimulated with IFN-α or in the response of cells to IFN-α (Fig. 4B). There was also no detectable ISGF-3 in PBM and PBM-derived macrophages infected by M. bovis BCG, and there was an increase in IFN-α stimulation of ISGF-3 in both groups (Fig. 4C and D). These data demonstrate that formation of ISGF-3 is a specific response to a pathogenic mycobacterium. Moreover, as seen with STAT-1, there are differences between pathogenic and nonpathogenic mycobacteria in their effects on the ability of cells to form ISGF-3, as revealed by stimulation with exogenous IFN-α.
FIG. 4.
ISGF-3 DNA-binding activity in monocytes and macrophages infected by M. bovis BCG and/or treated with IFN-α. (A) The complexes of the ISRE probe with ISGF-3 and with a nonspecific (ns) protein are shown for a representative experiment performed with THP-1 cells. THP-1 monocytes and THP-1 macrophages were infected and/or treated with IFN-α as indicated. (B) The ISGF-3 DNA-binding activity from four experiments was quantified relative to the nonspecific DNA-binding activity. The average is shown for each condition, with the standard deviation shown by the error bar. There were no statistically significant differences between respective pairs of uninfected and infected cells with or without IFN-α stimulation. (C) The complexes of the ISRE probe with ISGF-3 and with a nonspecific (ns) protein are shown for one experiment that was performed with PBM and PBM-derived macrophages to demonstrate that the difference between M. tuberculosis and M. bovis BCG was not only a feature of the THP-1 model. Cells were infected and/or treated with IFN-α as indicated. (D) The ISGF-3 DNA-binding activity shown in panel C was quantified relative to the nonspecific DNA-binding activity.
Effect of M. tuberculosis infection on the abundance and tyrosine phosphorylation of ISGF-3 subunits.
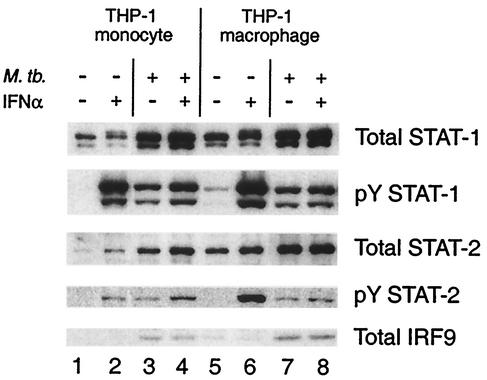
We examined the abundance of ISGF-3 subunits, STAT-1, STAT-2, and IRF-9, and tyrosine phosphorylation of the STAT proteins to determine whether the observed changes in DNA-binding activity after infection by M. tuberculosis might be due to altered protein levels, altered signaling, or both (Fig. 5). Infection led to an increase in the abundance of STAT-1, STAT-2, and IRF-9 in both THP-1 monocytes (lane 3) and THP-1 macrophages (lane 7). The increases due to infection of THP-1 macrophages were beyond the increases that occurred due to differentiation alone (lane 5). There was more tyrosine phosphorylation of STAT-1 in response to IFN-α stimulation (lanes 2 and 6) than in response to infection, yet infection followed by IFN-α stimulation (lanes 4 and 8) resulted in less STAT-1 tyrosine phosphorylation than after stimulation alone. In THP-1 macrophages, STAT-2 tyrosine phosphorylation was also greater in response to IFN-α stimulation (lane 6) than to infection (lane 7) and less with IFN-α stimulation after infection (lane 8) than with stimulation alone. The exception to this pattern was tyrosine phosphorylation of STAT-2 in THP-1 monocytes, which was higher in cells that were stimulated with IFN-α after infection (lane 4) than in cells that were only stimulated (lane 2) and which was similar after IFN-α stimulation or infection alone (lane 3). These changes in subunit abundance and tyrosine phosphorylation can account for the observed effects on STAT-1 homodimer and ISGF-3 DNA-binding activity.
FIG. 5.
Abundance and tyrosine phosphorylation of STAT-1 and STAT-2 and abundance of IRF-9 in THP-1 cells infected by M. tuberculosis and/or treated with IFN-α. THP-1 monocytes and THP-1 macrophages were infected with M. tuberculosis and/or treated with IFN-α as indicated. Antibodies against STAT-1, tyrosine phosphorylated STAT-1 (pY STAT-1), STAT-2, and tyrosine phosphorylated STAT-2 (pY STAT-2) were used to detect the respective targets in nuclear extracts, and antibody against IRF-9 was used to detect it in cytoplasmic extracts. The nuclear extracts were the same as those assayed for DNA-binding activity. The same filters were stripped and reprobed for each target protein. Equal amounts of protein were analyzed for each sample (see Materials and Methods). Representative results are shown. Similar results were obtained in four experiments.
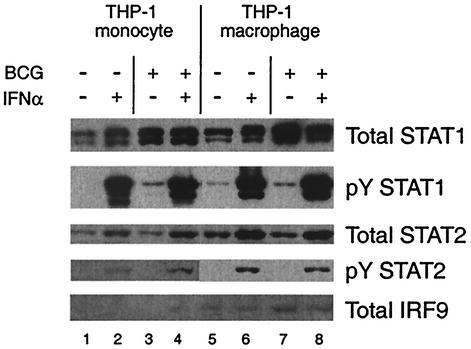
We determined whether the effects of infection by M. tuberculosis on ISGF-3 subunit abundance and tyrosine phosphorylation were specific, again by comparison to infection by M. bovis BCG (Fig. 6). M. bovis BCG infection of THP-1 monocytes and macrophages (lanes 3 and 7) led to increased STAT-1 and IRF-9 protein abundance and slight STAT-1 tyrosine phosphorylation, although the increase in IRF-9 was minimal in THP-1 monocytes. However, there was little if any change in STAT-2 abundance and no detectable STAT-2 tyrosine phosphorylation. Moreover, infection by M. bovis BCG had no effect on IFN-α stimulation of STAT-1 and STAT-2 tyrosine phosphorylation (compare lanes 2 and 4, or 6 and 8). Thus, M. tuberculosis has specific effects on the subunits of ISGF-3, and the effects of M. bovis BCG on the subunits also are consistent with its effects on ISGF-3 and STAT-1 DNA-binding activity.
FIG. 6.
Abundance and tyrosine phosphorylation of STAT-1 and STAT-2 and abundance of IRF-9 in THP-1 cells infected by M. bovis BCG and/or treated with IFN-α. THP-1 monocytes and THP-1 macrophages were infected with M. bovis BCG and/or treated with IFN-α as indicated. Immunoblot analysis of equal amounts of protein from each sample was performed as described in the legend to Fig. 5. Representative results are shown. Similar results were obtained in four experiments.
Effect of M. tuberculosis infection on IFN-α-stimulated gene expression.
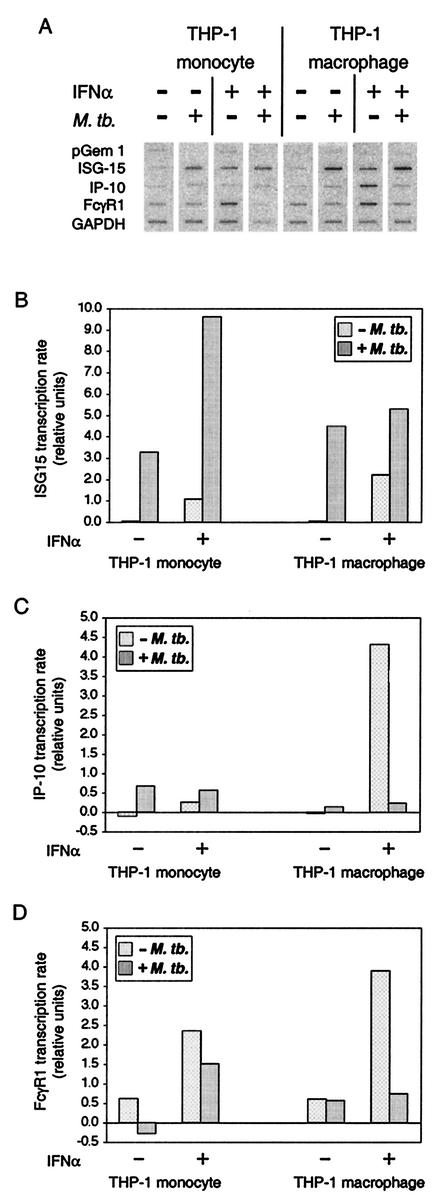
To determine the functional consequences of the changes in abundance and phosphorylation of ISGF-3 subunits that result from infection by M. tuberculosis and its effect on response to IFN-α, beyond the corresponding changes in ISGF-3 and STAT-1 DNA-binding activity, we measured the transcription rate of three genes, ISG15, IP-10, and FcγR1 (Fig. 7A). ISG15 is a well-characterized gene regulated by ISGF-3 in response to IFN-α (41, 42). IP-10 and FcγR1 have been characterized as IFN-γ-inducible genes (26, 36), but both are also induced by IFN-α (17, 33). IP-10 is regulated by STAT-1 and IRF-9 acting at an ISRE (27), and STAT-1 homodimers formed in response to IFN-α can bind to a GAS element in the FcγR1 promoter (1).
FIG. 7.
Transcription of genes regulated by ISGF-3 and by STAT-1 homodimers in THP-1 cells infected by M. tuberculosis and/or stimulated with IFN-α. THP-1 monocytes and THP-1 macrophages were infected and/or treated with IFN-α as indicated. Nascent RNA was radiolabeled in isolated nuclei, extracted, and hybridized to membranes on which the indicated probes were fixed. The data were quantified as described in Materials and Methods. The values shown are relative to the housekeeping GAPDH gene for each sample. Comparable results were obtained in two replicate experiments.
Quantification of the results for ISG15 (Fig. 7B), IP-10 (Fig. 7C), and FcγR1 (Fig. 7D) highlights the differences among these genes in the effect of infection alone on transcription and in the effect of infection on the transcriptional response to IFN-α. Infection by M. tuberculosis activated transcription of ISG15 and IP-10, but not FcγR1, in both THP-1 monocytes and THP-1 macrophages. Transcription of ISG15 was consistent in both cell types with the formation of ISGF-3 in cells infected by M. tuberculosis. However, ISGF-3 and another transcription factor, IRF-3, together may account for the observation that transcription of ISG15 was greater in cells that were only infected than in cells that were only stimulated with IFN-α. IRF-3 is also activated by M. tuberculosis infection (43), and it can directly activate the ISG15 gene in response to viral infection (8, 59). Transcription of IP-10 may be attributable to activation of NF-κB in cells infected by M. tuberculosis (29, 60), since IP-10 transcription is regulated by NF-κB in response to lipopolysaccharide stimulation (31), it is not regulated by ISGF-3 (33), and the increase in IRF-9 due to infection is not accompanied by formation of STAT-1 homodimers. The lack of FcγR1 transcription was consistent with the absence of STAT-1 homodimers in cells infected by M. tuberculosis. Transcription of all three genes was activated by IFN-α stimulation in monocytes and macrophages, but the effect of infection on response to IFN-α distinguished ISG15 from IP-10 and FcγR1, the genes regulated by STAT-1 homodimers.
IFN-α stimulation of infected THP-1 monocytes resulted in a transcription rate for ISG15 that was approximately twice the sum of the rates in cells that were only infected or stimulated (Fig. 7B). In contrast, in THP-1 macrophages that were infected and then stimulated with IFN-α, transcription was less than the sum of the rates in cells that were either infected or stimulated and only slightly greater than the rate due to infection alone. The fold increase in ISG15 transcription due to stimulation alone was greater in macrophages than in monocytes, although the induction in macrophages was less than might have been expected from the amount of ISGF-3 detected (Fig. 3). Importantly, in THP-1 monocytes and THP-1 macrophages, transcription of ISG15 reflects the respective enhancing or limiting effect of infection on IFN-α stimulation of ISGF-3 DNA-binding activity shown in Fig. 3.
IP-10 transcription in THP-1 monocytes infected by M. tuberculosis was slightly greater than in cells that were infected and stimulated with IFN-α, which in turn was more induced than in cells that were only stimulated with IFN-α (Fig. 7C), while FcγR1 transcription was greater in THP-1 monocytes that were only stimulated with IFN-α than in cells that were infected by M. tuberculosis and then stimulated with IFN-α (Fig. 7D). In THP-1 macrophages, both genes were very responsive to IFN-α stimulation in uninfected cells, and infection essentially blocked IFN-α stimulation of their transcription. Thus, in THP-1 monocytes and THP-1 macrophages, infection by M. tuberculosis reduces or eliminates IFN-α stimulation of transcription for both genes, which correlates with the effect on STAT-1 DNA-binding activity.
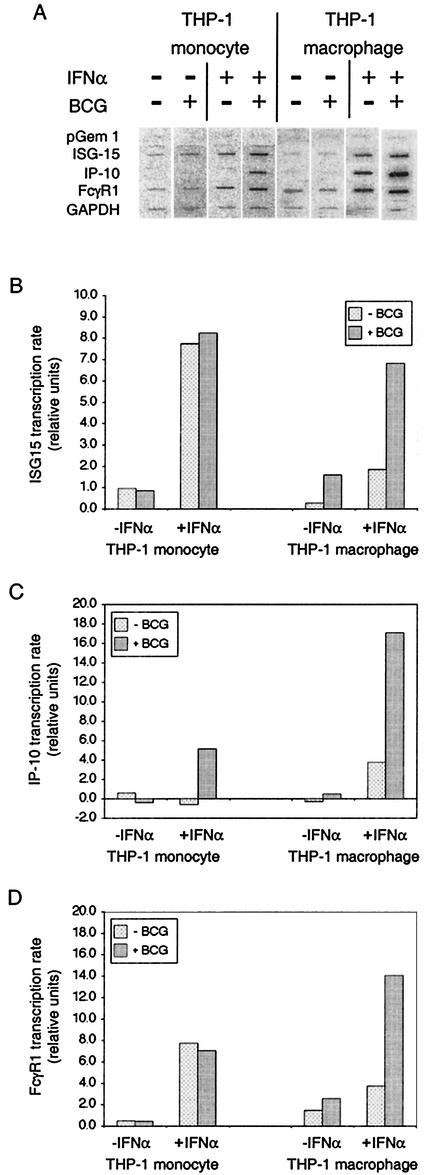
We also examined the effects of infection by M. bovis BCG on transcription of ISG15, IP-10, and FcγR1 (Fig. 8). Infection had no effect on unstimulated or IFN-α-stimulated ISG15 transcription in THP-1 monocytes, but both were increased by infection of THP-1 macrophages. IP-10 and FcγR1 transcription in THP-1 monocytes and THP-1 macrophages was essentially unaffected by M. bovis BCG infection. However, in both, IFN-α-stimulated transcription of IP-10 was increased by infection. Moreover, while IFN-α-stimulated transcription of FcγR1 was unaffected in THP-1 monocytes, it was also increased in THP-1 macrophages by M. bovis BCG infection. Thus, the transcriptional response to M. bovis BCG was quite different from the response to M. tuberculosis. In particular, there was no inhibition of IFN-α-stimulated transcription.
FIG. 8.
Transcription of genes regulated by ISGF-3 and by STAT-1 homodimers in THP-1 cells infected by M. bovis BCG and/or stimulated with IFN-α. THP-1 monocytes and THP-1 macrophages were infected at an MOI of approximately 3 and/or treated with IFN-α as indicated. Nascent RNA was radiolabeled in isolated nuclei, extracted, and hybridized to membranes on which the indicated probes were fixed. The data were quantified as described in Materials and Methods. The values shown are relative to the housekeeping GAPDH gene for each sample. Comparable results were obtained in two replicate experiments.
DISCUSSION
The work presented above describes effects of M. tuberculosis on the human IFN-α/β system. In order to determine whether infection would affect the IFN-α/β signal transduction pathway, we compared IFN-α stimulation of infected cells with infection or stimulation. We found that M. tuberculosis specifically inhibits the IFN-α/β signal transduction pathway, since infection of primary cells and THP-1 cells by M. tuberculosis inhibits activation of STAT-1 by IFN-α but not by IFN-γ (55; Y. Qiao, S. Prabhakar, and R. Pine, unpublished observations). We examined monocytes or macrophages that were uninfected, infected by M. tuberculosis, or infected by M. bovis BCG, without or with further stimulation by IFN-α, to determine a profile for several aspects of the IFN-α/β system. The abundance of STAT-1, STAT-2, and IRF-9, the tyrosine phosphorylation of STAT-1 and STAT-2, the DNA-binding activity of STAT-1 homodimers and ISGF-3, and the transcription of genes regulated by these transcription factors constituted a distinct profile for each of these 12 experimental conditions. The correlations among the individual aspects within each experimental condition strongly suggest that each of the observed effects of infection on the IFN-α/β system is responsible for the next one downstream.
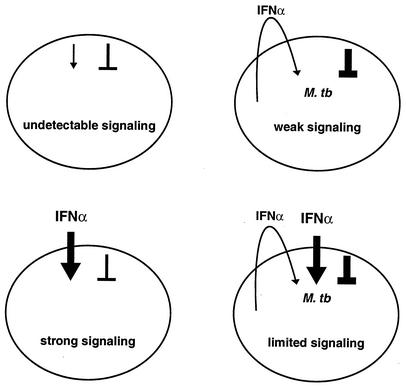
Overall, the results of our prior study (60) and this report suggest a simple schematic model (Fig. 9). In uninfected, unstimulated cells, whatever signaling occurs stochastically or even due to extremely low levels of autocrine IFN-α/β is counterbalanced by stochastic negative regulation, and the net effect is an undetectable level of signaling. If cells are infected, secreted IFN-α/β leads to some increase in signaling, which is low but detectable despite increased negative regulation. Stimulation alone leads to strong signaling. However, the increased negative regulation caused by infection becomes apparent as a limit in signal transduction when it is tested by further stimulation.
FIG. 9.
Schematic representation of interaction between M. tuberculosis and the IFN-α/β system. IFN signaling is balanced by positive and negative regulation. Changes in either may affect the extent of response and the dose response to IFN. Infection by M. tuberculosis leads to signaling through secretion of IFN-α/β and to induction of negative regulation. The negative regulation would likely weaken the response to secreted IFN, as suggested by the inhibited response that was observed when it was tested with additional IFN after infection.
The specific negative regulation of IFN-α/β signaling might occur through down-regulation of the cell surface IFN-α/β receptor, but this is not the case (S. Prabhakar, D. Tse, and R. Pine, unpublished observations). It may involve protein tyrosine phosphatases, suppressor of cytokine signaling (SOCS) proteins, or both, through specific interaction with the IFN-α/β receptor and/or TYK-2. The src-homology domain 2 containing protein tyrosine phosphatases PTP-1C and PTP-1D has been implicated in negative regulation of IFN-α/β signaling (10, 11, 61), although there is contradictory data about the role of PTP-1D (62). More recently, the membrane tyrosine phosphatase CD45 has been implicated in negative regulation of macrophage response to IFN-α/β (22). Of these, a role for PTP-1C is perhaps most likely, since mycobacterial lipoarabinomannan can activate this phosphatase (23). It has also been demonstrated that SOCS-1, SOCS-2, and SOCS-3 can inhibit the response to IFN-α/β, although the observation of inhibition by any particular SOCS protein may depend on the context in which it is examined (44, 45, 52, 53). Interleukin-10 (IL-10) and IFN-α/β, among other cytokines secreted during infection, can induce SOCS expression (5, 52). The inhibition of signal transduction by infection that we report here is partly a response to the IFN-α/β secreted during infection (S. Prabhakar and R. Pine, unpublished observations), but we have not ruled out a role for IL-10. Whether the immunosuppressive effect of IL-10 during infection (for example, see references 15 and 21) involves induction of SOCS proteins and/or suppression of IFN-α/β signaling and whether the negative feedback regulation of IFN-α/β signaling is immunosuppressive independent of any role for IL-10 remain to be determined.
The effects of infection by M. tuberculosis on STAT-1 homodimer and ISGF-3 formation are distinct from those caused by other bacteria. Infection of human PBM-derived macrophages by pathogenic Streptococcus pyogenes leads to formation of not only ISGF-3 but also STAT-1 homodimers, while infection by nonpathogenic Lactobacillus rhamnosus GG results in formation of STAT-1 homodimers but not ISGF-3 (30). Most notably, M. bovis BCG has very different effects than M. tuberculosis on the IFN-α/β system. The qualitative and quantitative differences we have observed between infection by M. tuberculosis and by M. bovis BCG suggest that a paradigm for pathogenic viruses extends to a major human bacterial pathogen: inhibition of host response to IFN-α/β is an aspect of pathogenicity (20). Comparison of M. tuberculosis with M. bovis BCG is especially helpful in supporting this concept, since M. bovis BCG is clearly nonpathogenic for people who have immune systems that function in the normal range. It would be of interest to also compare the virulent and avirulent pair of M. tuberculosis strains H37Rv and H37Ra to determine whether this difference between species extends to these different strains of M. tuberculosis. If so, the bacterial feature(s) that accounts for the effect of M. tuberculosis might be more readily identified by comparing strains than by comparing mycobacterial species or by comparing M. tuberculosis with even-less-related bacteria.
The influence of M. tuberculosis on signal transduction and on gene expression in response to IFN-α/β could impinge on both the innate immune response and the transition from innate immunity to Th1-cell-mediated immunity. Formation of STAT-1 homodimers in response to IFN-α/β plays a role in the innate immune response to bacterial infections and may play a role in the transition from innate to Th1-cell-mediated immunity (30, 32). In humans, activation of STAT-2 also plays a role in development of Th1-cell-mediated immunity through the function of a STAT-2 domain that is absent from the mouse homologue (14). Normal formation of STAT-1 homodimers is essential for human resistance to normally avirulent mycobacteria, as shown by the increased susceptibility of humans heterozygous for a STAT-1 mutation that reduces formation of STAT-1 homodimers by 75% in cells stimulated by IFN-α or IFN-γ (13). Notably, this mutation reduces IFN-α-stimulated formation of ISGF-3 by approximately 25%. Thus, inhibition by M. tuberculosis of STAT-1 homodimer formation and STAT-2 tyrosine phosphorylation in response to IFN-α/β might adversely affect host defense against this pathogen.
Reports suggesting that, in mice, induction of IFN-α/β by M. tuberculosis infection may be pathogenic (28) and that the ability to respond to endogenous IFN-α/β is required for normal host defense (7) may be reconciled by our observations on the response of infected cells to IFN-α. It was found that more-pathogenic strains of M. tuberculosis induce more IFN-α/β mRNA than less-pathogenic strains and that intranasal administration of IFN-α/β to infected mice increases bacterial burden and decreases median survival time compared to infection alone (28). The extent to which any of the strains examined inhibit response to IFN-α/β is not known. As judged by the effects of infection by M. tuberculosis and M. bovis BCG on activation of ISGF-3 in THP-1 cells, the data presented here also suggest that virulent mycobacteria cause more production of IFN-α/β than avirulent mycobacteria. Cells infected by M. tuberculosis at an MOI of 1 contained ISGF-3, as did cells infected by M. bovis BCG at an MOI of 10 (data not shown) but not at an MOI of 1. In another study, a statistically significant increase in bacterial burden was observed during the first 40 days postinfection in the lungs of mice having a null mutation in the IFN-α/β receptor (7), suggesting that IFN-α/β produced during infection can play a defensive role. A protective response to endogenous IFN-α/β induced by infection might occur even though infection inhibits and thereby is likely to limit the response. Overall, differences between more- and less-pathogenic mycobacteria in their effect on response to IFN-α/β may be at least as significant as differences in their induction of IFN-α/β.
Acknowledgments
We thank Michael Brunda for a gift of Roferon-α and Christian Schindler and David E. Levy for gifts of antibodies. We also thank Karl Drlica for critically reading the manuscript.
This work was supported by a grant from the National Institutes of Health (AI37877) to R.P. S.P. was supported by a fellowship from the Heiser Program for Research in Leprosy and Tuberculosis. E.C. and E.G. were supported by grants from the Istituto Superiore di Sanitá.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Beadling, C., D. Guschin, B. A. Witthuhn, A. Ziemiecki, J. N. Ihle, I. M. Kerr, and D. A. Cantrell. 1994. Activation of JAK kinases and STAT proteins by interleukin-2 and interferon alpha, but not the T cell antigen receptor, in human T lymphocytes. EMBO J. 13:5605-5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belardelli, F., and I. Gresser. 1996. The neglected role of type I interferon in the T-cell response: implications for its clinical use. Immunol. Today 17:369-372. [DOI] [PubMed] [Google Scholar]
- 3.Bluyssen, H. A. R., J. E. Durbin, and D. E. Levy. 1996. ISGF3γ p48, a specificity switch for interferon activated transcription factors. Cytokine Growth Factor Rev. 7:11-17. [DOI] [PubMed] [Google Scholar]
- 4.Bogdan, C. 2000. The function of type I interferons in antimicrobial immunity. Curr. Opin. Immunol. 12:419-424. [DOI] [PubMed] [Google Scholar]
- 5.Brender, C., M. Nielsen, C. Ropke, M. H. Nissen, A. Svejgaard, N. Billestrup, C. Geisler, and N. Odum. 2001. Interferon-alpha induces transient suppressors of cytokine signalling expression in human T cells. Exp. Clin. Immunogenet. 18:80-85. [DOI] [PubMed] [Google Scholar]
- 6.Chan, J., X. Fan, S. W. Hunter, P. J. Brennan, and B. R. Bloom. 1991. Lipoarabinomannan, a possible virulence factor involved in persistence of Mycobacterium tuberculosis within macrophages. Infect. Immun. 59:1755-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper, A. M., J. E. Pearl, J. V. Brooks, S. Ehlers, and I. M. Orme. 2000. Expression of the nitric oxide synthase 2 gene is not essential for early control of Mycobacterium tuberculosis in the murine lung. Infect. Immun. 68:6879-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daly, C., and N. C. Reich. 1995. Characterization of specific DNA-binding factors activated by double-stranded RNA as positive regulators of interferon alpha/beta-stimulated genes. J. Biol. Chem. 270:23739-23746. [DOI] [PubMed] [Google Scholar]
- 9.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 10.David, M., H. E. Chen, S. Goelz, A. C. Larner, and B. G. Neel. 1995. Differential regulation of the alpha/beta interferon-stimulated Jak/Stat pathway by the SH2 domain-containing tyrosine phosphatase SHPTP1. Mol. Cell. Biol. 15:7050-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David, M., G. Zhou, R. Pine, J. E. Dixon, and A. C. Larner. 1996. The SH2 domain-containing tyrosine phosphatase PTP1D is required for interferon α/β-induced gene expression. J. Biol. Chem. 271:15862-15865. [DOI] [PubMed] [Google Scholar]
- 12.Domanski, P., and O. R. Colamonici. 1996. The type-I interferon receptor. The long and short of it. Cytokine Growth Factor Rev. 7:143-151. [DOI] [PubMed] [Google Scholar]
- 13.Dupuis, S., C. Dargemont, C. Fieschi, N. Thomassin, S. Rosenzweig, J. Harris, S. M. Holland, R. D. Schreiber, and J. L. Casanova. 2001. Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science 293:300-303. [DOI] [PubMed] [Google Scholar]
- 14.Farrar, J. D., J. D. Smith, T. L. Murphy, S. Leung, G. R. Stark, and K. M. Murphy. 2000. Selective loss of type I interferon-induced STAT4 activation caused by a minisatellite insertion in mouse Stat2. Nat. Immunol. 1:65-69. [DOI] [PubMed] [Google Scholar]
- 15.Flesch, I. E., and S. H. Kaufmann. 1993. Role of cytokines in tuberculosis. Immunobiology 189:316-339. [DOI] [PubMed] [Google Scholar]
- 16.Flynn, J. L., and J. Chan. 2001. Immunology of tuberculosis. Annu. Rev. Immunol. 19:93-129. [DOI] [PubMed] [Google Scholar]
- 17.Fultz, M. J., and S. N. Vogel. 1992. Interferon alpha-induced changes in Fc gamma R-specific mRNA expression and isotype-specific, Fc gamma R-mediated phagocytosis in C3H/OuJ (Lpsn) and C3H/HeJ (Lpsd) macrophages. J. Leukoc. Biol. 51:300-304. [DOI] [PubMed] [Google Scholar]
- 18.Giacomini, E., E. Iona, L. Ferroni, M. Miettinen, L. Fattorini, G. Orefici, I. Julkunen, and E. M. Coccia. 2001. Infection of human macrophages and dendritic cells with Mycobacterium tuberculosis induces a differential cytokine gene expression that modulates T cell response. J. Immunol. 166:7033-7041. [DOI] [PubMed] [Google Scholar]
- 19.Giosue, S., M. Casarini, L. Alemanno, G. Galluccio, P. Mattia, G. Pedicelli, L. Rebek, A. Bisetti, and F. Ameglio. 1998. Effects of aerosolized interferon-alpha in patients with pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 158:1156-1162. [DOI] [PubMed] [Google Scholar]
- 20.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 21.Hickman, S. P., J. Chan, and P. Salgame. 2002. Mycobacterium tuberculosis induces differential cytokine production from dendritic cells and macrophages with divergent effects on naive T cell polarization. J. Immunol. 168:4636-4642. [DOI] [PubMed] [Google Scholar]
- 22.Irie-Sasaki, J., T. Sasaki, W. Matsumoto, A. Opavsky, M. Cheng, G. Welstead, E. Griffiths, C. Krawczyk, C. D. Richardson, K. Aitken, N. Iscove, G. Koretzky, P. Johnson, P. Liu, D. M. Rothstein, and J. M. Penninger. 2001. CD45 is a JAK phosphatase and negatively regulates cytokine receptor signalling. Nature 409:349-354. [DOI] [PubMed] [Google Scholar]
- 23.Knutson, K. L., Z. Hmama, P. Herrera-Velit, R. Rochford, and N. E. Reiner. 1998. Lipoarabinomannan of Mycobacterium tuberculosis promotes protein tyrosine dephosphorylation and inhibition of mitogen-activated protein kinase in human mononuclear phagocytes. Role of the Src homology 2 containing tyrosine phosphatase 1. J. Biol. Chem. 273:645-652. [DOI] [PubMed] [Google Scholar]
- 24.Larner, A. C., A. Chaudhuri, and J. E. Darnell, Jr. 1986. Transcriptional induction by interferon. New protein(s) determine the extent and length of the induction. J. Biol. Chem. 261:453-459. [PubMed] [Google Scholar]
- 25.Lopez-Ramirez, G. M., W. N. Rom, C. Ciotoli, A. Talbot, F. Martiniuk, B. Cronstein, and J. Reibman. 1994. Mycobacterium tuberculosis alters expression of adhesion molecules on monocytic cells. Infect. Immun. 62:2515-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luster, A. D., J. C. Unkeless, and J. V. Ravetch. 1985. Gamma-interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature 315:672-676. [DOI] [PubMed] [Google Scholar]
- 27.Majumder, S., L. Z. Zhou, P. Chaturvedi, G. Babcock, S. Aras, and R. M. Ransohoff. 1998. p48/STAT-1alpha-containing complexes play a predominant role in induction of IFN-gamma-inducible protein, 10 kDa (IP-10) by IFN-gamma alone or in synergy with TNF-alpha. J. Immunol. 161:4736-4744. [PubMed] [Google Scholar]
- 28.Manca, C., L. Tsenova, A. Bergtold, S. Freeman, M. Tovey, J. M. Musser, C. E. Barry III, V. H. Freedman, and G. Kaplan. 2001. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proc. Natl. Acad. Sci. USA 98:5752-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Means, T. K., S. Wang, E. Lien, A. Yoshimura, D. T. Golenbock, and M. J. Fenton. 1999. Human toll-like receptors mediate cellular activation by Mycobacterium tuberculosis. J. Immunol. 163:3920-3927. [PubMed] [Google Scholar]
- 30.Miettinen, M., A. Lehtonen, I. Julkunen, and S. Matikainen. 2000. Lactobacilli and streptococci activate NF-κB and STAT signaling pathways in human macrophages. J. Immunol. 165:3733-3740. [DOI] [PubMed] [Google Scholar]
- 31.Ohmori, Y., and T. A. Hamilton. 1993. Cooperative interaction between interferon (IFN) stimulus response element and kappa B sequence motifs controls IFN gamma- and lipopolysaccharide-stimulated transcription from the murine IP-10 promoter. J. Biol. Chem. 268:6677-6688. [PubMed] [Google Scholar]
- 32.Ohmori, Y., and T. A. Hamilton. 2001. Requirement for STAT1 in LPS-induced gene expression in macrophages. J. Leukoc. Biol. 69:598-604. [PubMed] [Google Scholar]
- 33.Park, C., S. Li, E. Cha, and C. Schindler. 2000. Immune response in Stat2 knockout mice. Immunity 13:795-804. [DOI] [PubMed] [Google Scholar]
- 34.Paulson, M., S. Pisharody, L. Pan, S. Guadagno, A. L. Mui, and D. E. Levy. 1999. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J. Biol. Chem. 274:25343-25349. [DOI] [PubMed] [Google Scholar]
- 35.Paulson, M., C. Press, E. Smith, N. Tanese, and D. E. Levy. 2002. IFN-stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nat. Cell Biol. 4:140-147. [DOI] [PubMed] [Google Scholar]
- 36.Pearse, R. N., R. Feinman, and J. V. Ravetch. 1991. Characterization of the promoter of the human gene encoding the high-affinity IgG receptor: transcriptional induction by gamma-interferon is mediated through common DNA response elements. Proc. Natl. Acad. Sci. USA 88:11305-11309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pine, R. 1997. Convergence of TNFα and IFNγ signalling pathways through synergistic induction of IRF-1/ISGF-2 is mediated by a composite GAS/κB promoter element. Nucleic Acids Res. 21:4346-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pine, R., A. Canova, and C. Schindler. 1994. Tyrosine phosphorylated p91 binds to a single element in the ISGF2/IRF-1 promoter to mediate induction by IFN α and IFN γ, and is likely to autoregulate the p91 gene. EMBO J. 13:158-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pine, R., T. Decker, D. S. Kessler, D. E. Levy, and J. E. Darnell, Jr. 1990. Purification and cloning of interferon-stimulated gene factor 2 (ISGF2): ISGF2 (IRF-1) can bind to the promoters of both beta interferon- and interferon-stimulated genes but is not a primary transcriptional activator of either. Mol. Cell. Biol. 10:2448-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pine, R., D. E. Levy, N. Reich, and J. E. Darnell, Jr. 1988. Transcriptional stimulation by CaPO4-DNA precipitates. Nucleic Acids Res. 16:1371-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reich, N., B. Evans, D. Levy, D. Fahey, E. Knight, Jr., and J. E. Darnell, Jr. 1987. Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc. Natl. Acad. Sci. USA 84:6394-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reich, N. C., and J. E. Darnell, Jr. 1989. Differential binding of interferon-induced factors to an oligonucleotide that mediates transcriptional activation. Nucleic Acids Res. 17:3415-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Remoli, M. E., E. Giacomini, G. Lutfalla, E. Dondi, G. Orefici, A. Battistini, G. Uze, S. Pellegrini, and E. M. Coccia. 2002. Selective expression of type I IFN genes in human dendritic cells infected with Mycobacterium tuberculosis. J. Immunol. 169:366-374. [DOI] [PubMed] [Google Scholar]
- 44.Sakai, I., K. Takeuchi, H. Yamauchi, H. Narumi, and S. Fujita. 2002. Constitutive expression of SOCS3 confers resistance to IFN-alpha in chronic myelogenous leukemia cells. Blood 100:2926-2931. [DOI] [PubMed] [Google Scholar]
- 45.Sakamoto, H., H. Yasukawa, M. Masuhara, S. Tanimura, A. Sasaki, K. Yuge, M. Ohtsubo, A. Ohtsuka, T. Fujita, T. Ohta, Y. Furukawa, S. Iwase, H. Yamada, and A. Yoshimura. 1998. A Janus kinase inhibitor, JAB, is an interferon-gamma-inducible gene and confers resistance to interferons. Blood 92:1668-1676. [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Sareneva, T., I. Julkunen, and S. Matikainen. 2000. IFN-alpha and IL-12 induce IL-18 receptor gene expression in human NK and T cells. J. Immunol. 165:1933-1938. [DOI] [PubMed] [Google Scholar]
- 48.Schindler, C., K. Shuai, V. R. Prezioso, and J. E. Darnell, Jr. 1992. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science 257:809-813. [DOI] [PubMed] [Google Scholar]
- 49.Schluger, N. W., and W. N. Rom. 1998. The host immune response to tuberculosis. Am. J. Respir. Crit. Care Med. 157:679-691. [DOI] [PubMed] [Google Scholar]
- 50.Schmitt, E., G. Meuret, and L. Stix. 1977. Monocyte recruitment in tuberculosis and sarcoidosis. Br. J. Haematol. 35:11-17. [DOI] [PubMed] [Google Scholar]
- 51.Schwander, S. K., E. Sada, M. Torres, D. Escobedo, J. G. Sierra, S. Alt, and E. A. Rich. 1996. T lymphocytic and immature macrophage alveolitis in active pulmonary tuberculosis. J. Infect. Dis. 173:1267-1272. [DOI] [PubMed] [Google Scholar]
- 52.Shen, X., F. Hong, V. A. Nguyen, and B. Gao. 2000. IL-10 attenuates IFN-alpha-activated STAT1 in the liver: involvement of SOCS2 and SOCS3. FEBS Lett. 480:132-136. [DOI] [PubMed] [Google Scholar]
- 53.Song, M. M., and K. Shuai. 1998. The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins inhibit interferon-mediated antiviral and antiproliferative activities. J. Biol. Chem. 273:35056-35062. [DOI] [PubMed] [Google Scholar]
- 54.Tchou-Wong, K. M., O. Tanabe, C. Chi, T. A. Yie, and W. N. Rom. 1999. Activation of NF-κB in Mycobacterium tuberculosis-induced interleukin-2 receptor expression in mononuclear phagocytes. Am. J. Respir. Crit. Care Med. 159:1323-1329. [DOI] [PubMed] [Google Scholar]
- 55.Ting, L. M., A. C. Kim, A. Cattamanchi, and J. D. Ernst. 1999. Mycobacterium tuberculosis inhibits IFN-gamma transcriptional responses without inhibiting activation of STAT1. J. Immunol. 163:3898-3906. [PubMed] [Google Scholar]
- 56.Tsuchiya, S., Y. Kobayashi, Y. Goto, H. Okumura, S. Nakae, T. Konno, and K. Tada. 1982. Induction of maturation in cultured human monocytic leukemia cells by a phorbol diester. Cancer Res. 42:1530-1536. [PubMed] [Google Scholar]
- 57.Tsuchiya, S., M. Yamabe, Y. Yamaguchi, Y. Kobayashi, T. Konno, and K. Tada. 1980. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 26:171-176. [DOI] [PubMed] [Google Scholar]
- 58.Veals, S., C. Schindler, D. Leonard, X.-Y. Fu, R. Aebersold, J. E. Darnell, Jr., and D. E. Levy. 1992. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol. Cell. Biol. 12:3315-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiden, M., N. Tanaka, Y. Qiao, B. Y. Zhao, Y. Honda, K. Nakata, A. Canova, D. E. Levy, W. N. Rom, and R. Pine. 2000. Differentiation of monocytes to macrophages switches the Mycobacterium tuberculosis effect on HIV-1 replication from stimulation to inhibition: modulation of interferon response and CCAAT/enhancer binding protein β expression. J. Immunol. 165:2028-2039. [DOI] [PubMed] [Google Scholar]
- 61.Yetter, A., S. Uddin, J. J. Krolewski, H. Jiao, T. Yi, and L. C. Platanias. 1995. Association of the interferon-dependent tyrosine kinase Tyk-2 with the hematopoietic cell phosphatase. J. Biol. Chem. 270:18179-18182. [DOI] [PubMed] [Google Scholar]
- 62.You, M., D. H. Yu, and G. S. Feng. 1999. Shp-2 tyrosine phosphatase functions as a negative regulator of the interferon-stimulated Jak/STAT pathway. Mol. Cell. Biol. 19:2416-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao, B. Y., R. Pine, J. Domagala, and K. Drlica. 1999. Fluoroquinolone action against clinical isolates of Mycobacterium tuberculosis: effects of a C-8 methoxyl group on survival in liquid media and in human macrophages. Antimicrob. Agents Chemother. 43:661-666. [DOI] [PMC free article] [PubMed] [Google Scholar]