Abstract
Control of anthrax toxin and capsule synthesis, the two major virulence factors of Bacillus anthracis, has been associated with two regulatory genes, atxA and acpA, located on virulence plasmids pXO1 and pXO2, respectively. We used transcriptional profiling to determine whether atxA and/or acpA control genes other than those already described and to investigate functional similarities of the regulators. Transcription was assessed in a pXO1+ pXO2+ parent strain and in isogenic mutants in which one or both regulatory genes were deleted. We determined that in addition to the toxin and capsule genes, atxA controls expression of numerous other genes on both plasmids and the chromosome. Generally, plasmid-encoded genes were more highly regulated than chromosomal genes, and both positive and negative effects were observed. Certain atxA-regulated genes were affected synergistically in an atxA acpA mutant. Yet overall, acpA appears to be a minor regulator with fewer targets than atxA. In contrast to previous reports of acpA function in attenuated strains, acpA had a minimal influence on capsule gene transcription and capsule synthesis in a genetically complete strain. Surprisingly, acpA expression was positively affected by atxA, although atxA-activated capsule gene transcription is not acpA dependent. The newly discovered atxA-regulated targets include genes predicted to encode secreted proteins and proteins with roles in transcriptional regulation and signaling. Regulation of chromosomal genes by atxA is particularly intriguing, given that many of the target genes have homologues in other Bacillus species that lack atxA homologues. Given the global effect of atxA on gene expression in B. anthracis, previous assumptions regarding reduced virulence of strains harboring single plasmids must be reassessed and the potential roles of newly identified atxA-regulated genes should be investigated.
The ability of Bacillus anthracis to cause anthrax is primarily attributed to its plasmid content. Plasmid pXO1 (182 kb) carries the anthrax toxin genes pagA, lef, and cya, while pXO2 (96 kb) harbors genes associated with synthesis of the antiphagocytic capsule, capB, capC, capA, and capD (16, 24, 28). The presence of pXO1 and pXO2 has been used for facile identification of the species, although the phylogenetic relationships between B. anthracis and the closely related Bacillus species B. cereus and B. thuringiensis are under continuing debate (12, 40).
For reasons of safety and facile manipulation in the laboratory, numerous investigators have employed attenuated strains carrying only one of the two plasmids in studies of toxin and capsule gene expression. The widely studied Sterne strain is toxigenic but noncapsulated, due to the presence of pXO1 and absence of pXO2 (16, 36). In addition to being used as the live animal vaccine in the United States, the Sterne strain serves as a source for toxin purification (21) and has proved useful for studies of anthrax toxin function. In a mouse model for anthrax, high doses of Sterne spores delivered subcutaneously result in a disease resembling systemic anthrax (33). Studies of toxin gene expression by the Sterne strain led to the discovery of atxA, a pXO1 gene required for transcription of the three toxin genes (7, 17, 42).
Investigations of capsule function and synthesis have been performed using a number of capsulated but nontoxigenic strains. Generally, pXO1− pXO2+ strains are avirulent in animal models (15, 41), and results of experiments employing cultured macrophages indicate an antiphagocytic role for the capsule (23, 24). Investigation of capsule gene expression in a pXO1− pXO2+ strain revealed that a pXO2-borne gene, acpA, was essential for cap operon transcription (44).
The mechanisms by which atxA and acpA control virulence gene expression are unknown. Some reports indicate limited functional similarity of the two regulators and a more expanded role for atxA in B. anthracis gene expression. The atxA gene cloned on a multicopy plasmid in a pXO1− pXO2+ strain positively regulates capB. In contrast, acpA cloned on a multicopy plasmid in a pXO1+ pXO2− strain does not affect toxin gene expression (43). Strains harboring both plasmids produce more capsule than pXO1− pXO2+ strains, and the enhanced capsule synthesis has been attributed to atxA (8, 11, 43). Results of proteomic studies and genetic screens for atxA-activated genes in a pXO1+ pXO2− strain indicate atxA-activated genes on pXO1 (13), but precise mapping and functional analysis of these additional target genes have not been performed.
Fully virulent strains of B. anthracis harbor pXO1 and pXO2, yet studies of expression and function of these important virulence gene regulators in a strain harboring both plasmids are lacking. To determine whether atxA and/or acpA controls genes other than the known virulence genes, and to further investigate the functional similarity of the regulators, we compared the transcriptional profiles of a genetically complete pXO1+ pXO2+ strain to those of isogenic single and double atxA and acpA mutants. Our results indicate a major role for atxA in B. anthracis gene expression and a synergistic effect of atxA and acpA on expression of certain genes.
MATERIALS AND METHODS
Strain construction.
We constructed a pXO1+ pXO2+ parent strain by transducing pXO2 from strain 6602 (Pasteur) (American Type Culture Collection) into strain 7702 (Sterne) (5), using CP51-mediated transduction as described previously (9). Cap+ transductants were selected using bacteriophage CP54, which lyses noncapsulated cells. The presence of pXO1 and pXO2 in a transductant, UT500, was confirmed by amplification of specific sequences using PCR. To confirm the structural integrity of pXO2 following transduction, numerous PCR products representing greater than 90% of the pXO2 DNA sequence were generated using multiple primer sets.
atxA- and acpA-null mutants in which coding sequences of the regulatory genes were replaced with DNA sequences conferring resistance to kanamycin (Ω-km) or spectinomycin (aad9) were constructed in UT500 (pXO1+ pXO2+) as follows. In the previously characterized 7702 mutant UT60 (7), internal sequences of atxA, from 14 nucleotides (nt) downstream of the first nucleotide to 86 nt upstream from the last nucleotide of the gene, were replaced with Ω-km. We used CP51-mediated transduction with selection for kanamycin resistance (7) to transfer the atxA-null mutation from UT60 to UT500, yielding UT501. The atxA-null mutation of UT501 was confirmed using PCR with primers corresponding to regions flanking the atxA gene and primers corresponding to sequences within the Ω-km element.
An acpA-null mutation, in which sequences from 34 nt downstream of the first nucleotide to 183 nt downstream of the translational stop codon of the gene were replaced with the spectinomycin resistance gene aad9, was generated using a method described previously (34). The nearest downstream open reading frame (ORF) is 1,695 nt from the 3′ end of acpA. The acpA-null mutation was first constructed in a strain harboring only pXO2. We used CP51-mediated transduction with selection for spectinomycin resistance (34) to transfer the mutation to UT500, yielding UT502. The acpA-null mutation in UT502 was confirmed by using PCR with primers corresponding to sequences flanking the acpA gene and primers corresponding to sequences within aad9.
To create a double mutant (atxA acpA), pXO2 harboring the acpA-null mutation was transferred to the Sterne atxA-null mutant UT60 by using CP51 transduction with selection for spectinomycin resistance. The presence of both mutations in UT503 was confirmed using PCR as described above.
RNA isolation.
RNA was extracted from cells grown in CA medium (39) containing 0.8% bicarbonate and 100 mM HEPES (pH 8.0) (CACO3) with shaking in a 5% CO2 atmosphere at 37°C, conditions known to promote toxin synthesis and used previously in investigations of toxin gene expression in B. anthracis (6, 7, 14). Briefly, cells were cultured overnight in Luria-Bertani broth containing 0.5% glycerol with antibiotics when appropriate (50 μg of kanamycin/ml or 100 μg of spectinomycin/ml). Cells were transferred to CACO3 without antibiotics such that the starting optical density at 600 nm (OD600) was 0.1. Following 3 h of incubation (OD600 ≈ 0.5), cells from mid-exponential-phase cultures were collected for RNA isolation. RNA was extracted with GramCracker reagents followed by RNAwiz according to protocols supplied by the manufacturer (Ambion, Austin, Tex.). RNA yields were quantitated by measuring absorbance at 260 nm. Typically, 15 to 30 μg of RNA was obtained from 1 ml of culture.
Western hybridizations.
Cells were grown in CACO3 as described for RNA isolation. At mid-exponential phase (OD600 ≈ 0.5) and late exponential phase (OD600 ≈ 0.8), culture supernates were filtered through cellulose acetate membranes (pore size, 0.2 μm) (Nalgene, Rochester, N.Y.) and frozen in a dry ice-ethanol bath. Western hybridizations were performed as described previously (34) with the following exceptions. Membranes were blocked overnight at 4°C in Tris-buffered saline with Tween (TBS-T) containing 5% milk. Primary antibody (rabbit anti-PA, anti-LF, or anti-EF) was added at a dilution of 1:500 for 1 h at room temperature. Membranes were washed in TBS-T and then treated with the secondary antibody (horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G; Bio-Rad) at a dilution of 1:5,000 in TBS-T-5% milk for 1 h at room temperature. After a final wash in TBS-T, cross-reactive material was visualized using an enhanced chemiluminescent detection kit (Amersham, Piscataway, N.J.).
Microarraying procedure.
A total of 3,290 chromosomal genes and 138 plasmid-borne genes were represented as PCR products (303 to 999 bp) on the “genomic” microarray. Thus, the DNA microarray contained DNA fragments corresponding to approximately 60% of the predicted genes of the 5.23-Mbp B. anthracis genome (http://www.tigr.org). Preparation of DNA microarrays, slide processing, cDNA probe synthesis, and hybridization were similar to those described previously and detailed as follows (32).
PCR products (200 ng in 50% dimethyl sulfoxide) were deposited onto 25-mm by 75-mm CMT-UltraGAPS amino silane-coated glass microscope slides (Corning, Acton, Mass.) using an Amersham Lucidea printer. Humidity was maintained at ∼55% during printing. After printing, slides were air dried for 30 min, and then the DNA was cross-linked to the surface by using a Stratagene Stratalinker to deliver 50 mJ of short-wavelength UV energy. Slides were stored in a desiccator. To wash unbound nucleic acids and salts from the slides, they were prehybridized for 2 h at 42°C in a 50-ml solution of 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), and 1.0% bovine serum albumin. Slides were then washed four times in MilliQ water and three times in isopropanol and dried.
Microarray probe preparation.
Six micrograms of Random Hexamer Primers (Invitrogen, Rockville, Md.) were annealed to 2 μg of total RNA in a total volume of 18.5 μl by heating the reaction mixture to 70°C for 10 min, followed by chilling on ice. To this reaction mixture, 6 μl of 5× First Strand reverse transcriptase (RT) buffer (250 mM Tris-HCl [pH 8.3], 375 mM KCl, 15 mM MgCl2), 3 μl of 0.1 M dithiothreitol (Invitrogen), 0.6 μl of a deoxynucleoside triphosphate mixture (25 mM concentrations of dATP, dGTP, and dCTP, and 8 mM dTTP [Invitrogen] with 17 mM aa-dUTP [Sigma]), and 400 U of Superscript II RT (Invitrogen) were added. The reactions were then placed at 42°C overnight. The RNA template was hydrolyzed by adding 10 μl of 1 M NaOH and 10 μl of 0.5 M EDTA and incubating at 65°C for 15 min. This reaction was then neutralized with 25 μl of 1 M Tris (pH 7.0). Probes were purified over QIAquick PCR purification columns by replacing the Tris-based buffers with phosphate wash (5 mM KPO4 [pH 8.0], 80% ethanol) and elution buffers (4 mM KPO4 [pH 8]). Reactions were dried in a Speed-Vac to completion. Dried aminoallyl-labeled cDNA was resuspended in 4.5 μl of 0.1 M sodium carbonate buffer, pH 9.3. Next, 4.5 μl of the appropriate resuspended ester Cy dye was added to the cDNA and allowed to couple for 2 h at room temperature in the dark. After coupling, 35 μl of 100 mM NaOAc (pH 5.2) was added and reactions were again purified with QIAquick PCR purification columns, but this time using supplied buffers. The appropriate samples, one labeled with Cy3 and the other with Cy5, were mixed and dried to completion.
Microarray hybridization.
Dried probes were resuspended in 30-μl volumes (50% formamide, 5× SSC, 0.1% SDS, and 100 g of salmon sperm DNA/ml). This mixture was heated for 10 min at 95°C and then added to a prehybridized slide under a coverslip. The slide was then placed at 42°C for 16 h in a sealed hybridization chamber (Corning 2551) which was humidified with 20 μl of 5× SSC. Arrays were then washed at 55°C, once in a solution of 2× SSC-0.1% SDS for 10 min, once in 0.1× SSC-0.1% SDS for 10 min, and thrice in 0.1× SSC for 2 min, followed by a final quick wash in MilliQ water.
Statistical analysis.
To allow appropriate statistical analysis of the results, RNA preparations from at least three independent cultures were tested for each strain. For each hybridization, RNA was obtained from cultures of the UT500 parent strain and the mutant strains grown in parallel. Each RNA preparation was used in at least two separate hybridizations, and slides contained at least two spots for each gene in the array. A subarray containing PCR products representing 276 chromosomal and 30 plasmid genes, each spotted 15 times, was used to generate a larger number of replicate ratios for statistical purposes. The genes of the subarray included nonregulated (control) genes and putative regulated genes.
Analysis was similar for both genomic arrays and subarrays. Each mutant was analyzed separately. After quantitation and global normalization using average spot intensity, log ratios of mutant to wild type (strain UT500) were calculated for each spot. Replicate spots on the same slide were averaged to obtain a single value for each ORF for each slide, with spots that failed the quality criteria being omitted. Values from slide pairs from the same culture were averaged to generate a single value for each ORF for each independent culture (n = 3). ORFs were considered to be eligible for analysis if all three cultures were represented by at least one spot of acceptable quality. Spots with a reference fluorescence signal at least three times the local background were accepted for quantitation. Unacceptable spots were those with no signal or those associated with a slide problem such as dust or a scratch. The number of spots contributing to an analysis-eligible ORF ranged from 3 to 12. ORFs were considered significantly regulated if (i) the overall fold change was at least 2 (i.e., absolute value of average log ratio was greater than 0.693) and (ii) the P value from a one-sample t test, testing whether the grand mean log ratio was different from 0.0, was significant at the α = 0.05 level or better.
Sequence analysis.
We used the following resources to characterize the predicted products of genes of interest: BLAST (www.ncbi.nlm.nih.gov/BLAST), SMART (http://smart.embl-heidelberg.de/), TMpred (http://www.ch.embnet.org/software/TMPRED_form.html), and PredictProtein (http://www.embl-heidelberg.de/predictprotein/predictprotein.html).
RESULTS
Global effects of atxA and acpA.
Two attenuated strains used frequently to assess B. anthracis gene expression are the Sterne strain 7702 (pXO1+ pXO2−) (5) and the Pasteur strain 6602 (pXO1− pXO2+) (9, 44). To investigate the roles of the plasmid-encoded regulatory genes atxA and acpA in a genetically complete strain, we constructed a pXO1+ pXO2+ parent strain, UT500, by transducing pXO2 from 6602 into 7702. atxA- and acpA-null mutations were introduced into UT500 to yield the isogenic mutants UT501 (atxA), UT502 (acpA), and UT503 (atxA acpA). UT500 and the mutants exhibited similar growth rates in all media tested (data not shown). For microarray analysis, cells were grown under conditions used previously to assess toxin gene regulation, buffered CA medium in a 5% CO2 atmosphere at 37°C (6, 7, 14), and RNA was extracted when cultures reached mid-exponential phase.
The transcriptome of UT500 indicated that 30% of all ORFs represented on the array were expressed during growth under our culture conditions. Data showing the fold induction and P values for all regulated ORFs (≥2-fold induction; P < 0.05) from experiments comparing UT500 to the isogenic mutants can be viewed at the website http://mmg.uth.tmc.edu/mmg2/facpages/tkoehler/tkoehler.html. Comparison of the transcription profiles of UT500 and UT501 revealed that, in addition to the pXO1-encoded toxin genes and the capsule genes of pXO2, atxA regulated numerous other genes on pXO1, pXO2, and the chromosome. Remarkably, of the 38 plasmid genes that were expressed, 18 of these were controlled by atxA.
In contrast to the substantial effect of atxA on gene expression in the pXO1+ pXO2+ background, the transcription profiles of UT500 and the acpA mutant UT502 indicated relatively few acpA-regulated genes for cells grown under our culture conditions. Genes of the capsule biosynthesis operon, capBCAD, on pXO2 and amiA, a pXO2 gene encoding an amidase that hydrolyzes peptidoglycan (26), were the only genes whose transcription differed in UT500 and UT502. Expression of the amiA gene was 10- to 25-fold higher in UT500 than in the acpA mutant UT502, while expression of the cap operon genes was only 2-fold higher in UT500 than in UT502 (P < 0.05 on subarray). Notably, comparison of the transcription profiles of UT500 and UT501 indicated that the capsule genes and amiA were also regulated by atxA.
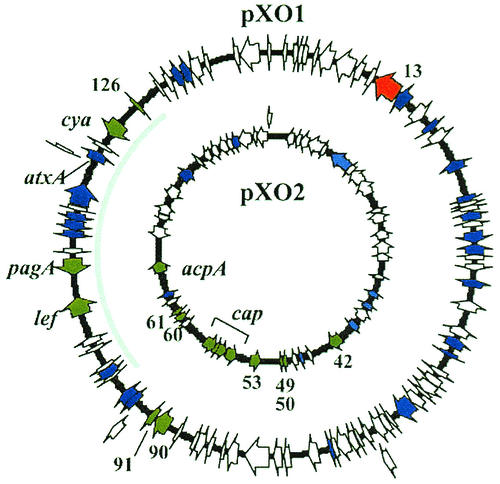
Generally, plasmid-borne genes were more highly regulated than genes on the chromosome. atxA regulated various plasmid genes from 3- to 60-fold, while different chromosomal genes were affected no more than 8-fold. With one exception, pXO1-13, atxA exerted a positive effect on the plasmid gene targets (Fig. 1 and Table 1). Negative and positive effects of atxA were apparent for regulated genes on the chromosome. As shown in Fig. 1, the spatial distribution of regulated genes on pXO1 and pXO2 mapped to distinct regions on the plasmids. Regulated genes of pXO1 clustered to a 54-kb region that included the pathogenicity island (28). The regulated genes of pXO2 localized to a 33-kb region surrounding the capsule biosynthetic operon. Unlike the plasmid genes, the loci of regulated chromosomal genes were generally scattered.
FIG. 1.
Plasmid gene expression profile. Only genes printed on the DNA array are represented. Cells were grown to mid-exponential phase in CACO3 with shaking at 37°C in a 5% CO2 atmosphere. Genes positively regulated by atxA are shown in green. The negatively regulated gene is shown in red. Nonregulated genes are shown in blue. Genes shown in white were not expressed. The pathogenicity island of pXO1 is indicated by a grey line. pXO1 is 181.6 kb (accession no. AF065404), and pXO2 is 96.2 kb (accession no. AF188935).
TABLE 1.
atxA-regulated plasmid genes.
| Plasmid and ORF | Fold regulation | P value | Known or putative function |
|---|---|---|---|
| pXO1 | |||
| 13 | −3 | 0.01 | TM and DNA binding domains |
| 90 | 61 | <0.01 | SLH domain |
| 91 | 25 | <0.01 | Acid phosphatase |
| 107 | 11 | 0.02 | Lethal factor |
| 110 | 52 | <0.01 | Protective antigen |
| 122 | 3 | 0.05 | Edema factor |
| 126 | 7 | 0.02 | Unknown |
| pXO2 | |||
| 42 | 5 | 0.03 | Amidase |
| 49-50 | 9 | <0.01 | Transposase |
| 53 | 9 | 0.02 | Similar to AcpA |
| 55 | 8 | 0.05 | Capsule depolymerase |
| 56 | 8 | 0.03 | Capsule biosynthesis protein CapA |
| 57 | 8 | 0.02 | Capsule biosynthesis protein CapC |
| 58 | 9 | 0.02 | Capsule biosynthesis protein CapB |
| 60 | 41 | <0.01 | Unknown |
| 61 | 54 | <0.01 | Unknown |
| 64 | 12 | 0.01 | AcpA |
atxA acpA synergy.
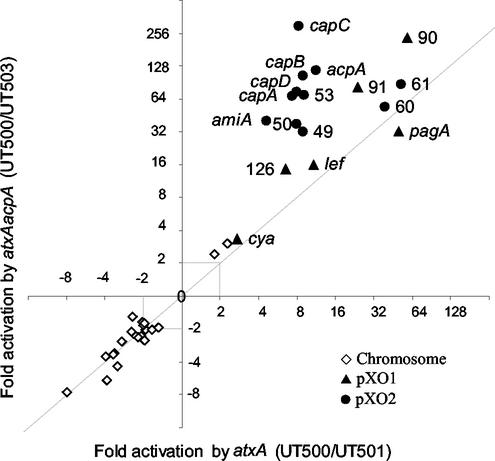
Expression of certain atxA-controlled genes was more greatly affected in the double mutant UT503 (atxA acpA) than in the single mutant UT501 (atxA). As depicted in Fig. 2, we compared differences in gene expression detected in profiles of UT500 and UT501 to those detected in profiles of UT500 and UT503. As presented in the figure, data points representing genes regulated by atxA and not affected by acpA fall on the diagonal line of the graph. A shift from the line indicates a positive or negative effect of acpA on gene expression in the atxA mutant background. Interestingly, data points representing a number of plasmid-encoded genes fall above the diagonal line, indicating that in an atxA mutant background the acpA gene had a positive effect on gene expression.
FIG. 2.
atxA and acpA synergy. Data points shown represent genes regulated (P < 0.05) in all UT500-UT501 and UT500-UT503 experiments. Numbers shown represent the plasmid ORF names. Both axes represent fold regulation (log scale). ◊ = chromosomal genes; ▴ = pXO1 genes; • = pXO2 genes.
For some atxA-regulated genes, comparison of the transcription profiles of UT500 and UT502 (acpA) revealed no apparent regulation by acpA. However, expression of these genes in the double mutant UT503 (atxA acpA) differed significantly from that observed in the single mutant UT501 (atxA). For example, pXO1-90 was regulated 60-fold when the profiles of UT500 and UT501 (atxA) were compared but 238-fold when profiles of UT500 and UT503 (atxA acpA) were compared. pXO2-53 was regulated 9-fold when the profiles of UT500 and UT501 (atxA) were compared but 74-fold when profiles of UT500 and UT503 (atxA acpA) were compared.
The apparent synergy of atxA and acpA for control of the capsule operon genes capBCAD and amiA may be due in part to control of acpA expression by atxA. We observed a 7- to 19-fold positive effect of atxA on acpA expression when profiles of UT500 and UT501 (atxA) were compared (Table 1). Although previous investigations have suggested that AcpA is limiting for capsule gene expression (44), atxA has been reported to elevate cap gene expression in the absence of acpA (43). Therefore, it is unlikely that the atxA-mediated increase in capsule gene expression, and possibly amiA expression, can be attributed solely to increased expression of the acpA gene.
Capsule and toxin synthesis.
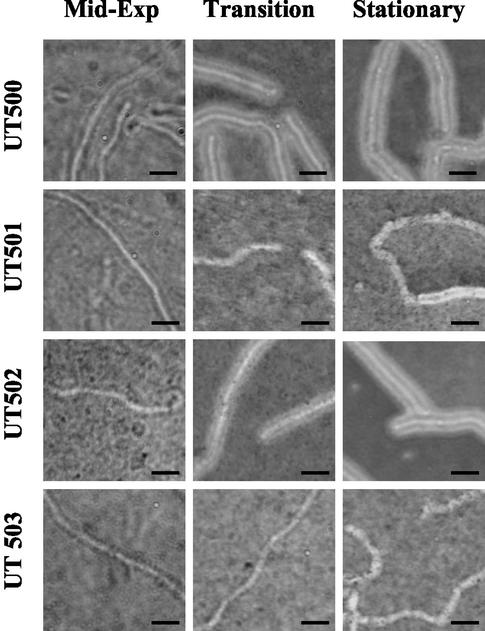
Previous reports of capsule gene expression have concluded that although atxA positively affects capsule synthesis, acpA is the major regulator of the capsule genes. Our transcriptional profiling data indicate that in a strain harboring both plasmids, atxA has a greater effect on cap gene expression than acpA (7- to 9-fold compared to 2- to 3-fold). To determine if our transcriptional analysis was in agreement with capsule synthesis by the genetically complete strain, we evaluated capsule production in UT500 and the isogenic mutants. India ink preparations of cells cultured under the same conditions used for transcriptional profiling experiments are shown in Fig. 3. At mid-exponential phase, UT500 and the mutant strains did not appear capsulated. However, at the transition to stationary phase, significant differences in UT500 and the mutants were observed. UT500 cells were fully capsulated, and the capsule material completely excluded the ink particles. At the same time point, no capsule was visible on cells of UT501 and UT503. UT502 (acpA) exhibited capsule formation at the transition into stationary phase, but ink particles were not fully excluded and the capsule material appeared thinner than that observed on UT500 cells. At stationary phase, the differences between UT500 and UT502 were no longer apparent. Cells of UT503 remained noncapsulated at stationary phase, while some UT501 cells appeared to produce a thin layer of capsule material.
FIG. 3.
Capsule production by the B. anthracis parent strain (UT500) and isogenic mutants. Cells were grown in CACO3 with shaking at 37°C in a 5% CO2 atmosphere. At the growth phases indicated, cells were removed and capsule was visualized with India ink. UT500, parent; UT501, atxA; UT502, acpA; UT503, atxA acpA. Bar = 5 μm.
Consistent with the microscopic observations, UT500 and UT502 (acpA) produced mucoid colonies when grown for 12 h on solid CACO3 medium. In contrast, colonies of UT501 and UT503 were nonmucoid. Prolonged incubation of UT501, but not UT503, resulted in some capsule material, but the amount was significantly reduced compared to the parent strain (data not shown.) These phenotypes are in agreement with the transcription profile data, indicating a small effect of acpA on cap expression in the genetically complete strain. Capsule synthesis by the pXO1+ pXO2+ strain is dependent upon the presence of one of the two regulators, and atxA has a greater positive effect on capsule synthesis than acpA.
In contrast to the new findings regarding capsule synthesis, toxin protein synthesis by the pXO1+ pXO2+ UT500 strain and the mutants was consistent with previous reports in which toxin gene expression in the pXO1+ pXO2− Sterne strain was measured using other methods (17). We detected protective antigen, lethal factor, and edema factor in supernates of UT500 and UT502 cultures, but not in supernates of UT501 and UT503 cultures (Fig. 4). Thus, toxin protein synthesis by UT500 and the mutants reflected the results of the microarray analysis; atxA is essential for toxin gene expression. It is noteworthy that although the cya gene, encoding edema factor, was regulated only threefold (P < 0.046) by atxA in the microarray analysis, a clear difference in protein synthesis in the parent and mutant strain was apparent.
FIG. 4.
Toxin synthesis by the B. anthracis parent strain (UT500) and isogenic mutants. Cells were grown as described in the legend for Fig. 1. Filtered culture supernates from the growth phases indicated were subjected to Western hybridization using antitoxin antiserum as indicated. PA, protective antigen; LF, lethal factor; EF, edema factor. UT500, parent; UT501, atxA; UT502, acpA; UT503, atxA acpA.
Bioinformatic analysis of newly discovered atxA-regulated ORFs.
The predicted products of the newly discovered atxA-regulated ORFs were evaluated using a variety of computational tools. We tested for similarity to genes of other organisms, the presence of signature motifs, and sequences indicative of cellular localization. A list of regulated genes and their predicted functions or homologues can be viewed at the website http://mmg.uth.tmc.edu/mmg2/facpages/tkoehler/tkoehler.html. Among the atxA-regulated plasmid-borne ORFs were four ORFs, pXO1-13, pXO1-90, pXO1-91, and pXO2-42 (the amiA gene), predicted to encode surface-exposed proteins. One ORF may encode a regulatory protein; pXO2-53 is predicted to encode a protein that is similar to AcpA (62%) and AtxA (48%). Two atxA-regulated plasmid ORFs are similar to the well-characterized insertion element IS231. The sequences and organization of pXO2-49 and pXO2-50 indicate a single gene of the IS231 V or W subgroup (22). A translational shift would result in a single protein encoded by these ORFs, with pXO2-49 encoding the amino-terminal portion of the protein and pXO2-50 encoding the carboxy-terminal part of the protein. Other atxA-regulated plasmid-borne ORFs, pXO2-60, pXO2-61, and pXO1-126, are predicted to encode soluble proteins, but our analysis did not yield clues regarding function.
BLAST searches of the atxA-regulated chromosomal genes revealed homologues in Bacillus subtilis (20) and other species. Many of these genes are predicted to have roles in amino acid biosynthesis. For example, ORFs showing similarity to genes encoding biosynthetic enzymes for branched-chain amino acids, ilvC (ORF 97) and ilvB (ORF 99), and for aromatic amino acids, aroA, aroF, hisC, tyrA, and aroE (ORFs 4962 to 4967) were negatively regulated by atxA. Other atxA-regulated chromosomal ORFs (ORFs 982 to 985, 364, 4183, and 6016) are predicted to encode components of oligopeptide transport systems found in B. subtilis and other gram-positive species (18, 19). Another set of regulated ORFs (ORFs 2700 to 2702) are similar to acetoin utilization genes of B. subtilis. Interestingly, in B. subtilis the app and opp oligopeptide transport genes, certain amino acid biosynthesis genes, and acetoin utilization genes are part of the scoC regulon. ScoC is a pleiotropic regulator that affects changes in gene expression throughout growth (4). The B. anthracis genome contains a scoC homologue which was expressed under our growth conditions but was not regulated by atxA.
DISCUSSION
Our investigation focused on the roles of two regulatory genes, atxA and acpA, in B. anthracis gene expression. These genes were identified and initially characterized in attenuated B. anthracis strains harboring single virulence plasmids (6, 7, 17, 42, 44). Plasmid pXO1 carries the structural genes for the anthrax toxin proteins and atxA, the anthrax toxin activator. Plasmid pXO2 harbors genes required for capsule biosynthesis and acpA, the anthrax capsule activator. Our analysis of the transcriptional profiles of a genetically complete (pXO1+ pXO2+) toxigenic capsulated B. anthracis strain and isogenic mutants deleted for the regulatory genes indicates that these regulators affect expression of numerous B. anthracis genes on the plasmids and on the chromosome.
The newly discovered atxA-regulated genes may have direct or indirect roles in B. anthracis pathogenesis. Some of the highly regulated target genes on the plasmids are predicted to encode secreted proteins or proteins associated with the bacterial cell membrane. pXO1-13 is predicted to encode a membrane-associated protein, and sequences near the carboxy terminus contain a putative DNA-binding domain (3). The amino terminus of the pXO1-90 protein has three highly conserved SLH (S-layer homology) motifs that could potentially serve to anchor the protein to the bacterial cell surface (25), possibly rendering the protein immunogenic. pXO1-91 is predicted to encode a membrane-associated phosphatase with a PAP-2 family signature (38). Such enzymes are phosphoesterases that function at acidic pH and may be relevant if expressed when B. anthracis resides in the phagolysosome (10). The pXO1-90 and pXO1-91 genes are adjacent and in the same orientation. Northern hybridization analysis confirmed that the genes are atxA regulated but revealed distinct transcripts, indicating that the genes are not cotranscribed (data not shown). The amiA gene (pXO2-42) was recently shown to be an enzyme that hydrolyzes peptidoglycan and may be a B. anthracis autolysin (26). The AmiA protein also has an SLH motif at the amino terminus. Such surface-exposed proteins may be relevant for virulence.
Previous reports of sequence and functional similarity between atxA and acpA brought the physiological significance of acpA into question. The amino acid sequences of AtxA (56 kDa) and AcpA (55 kDa) are 25% identical and 47% similar. atxA was reported to positively control expression of the toxin and capsule genes (11, 43), while acpA-mediated control was limited to capsule gene expression (43). Thus, the existence of acpA indicated an apparent functional redundancy. Our data reveal two significant findings regarding the relationship between these regulators. First, atxA positively regulates acpA expression while acpA does not affect atxA expression. Second, the two regulators have a synergistic effect on expression of certain target genes.
Our transcriptional profiling results and microscopic studies also revealed a significant role for atxA in capsule gene expression. Prior to our investigation, published studies established that (i) acpA is essential for capsule gene expression in a pXO1− pXO2+ strain (44), (ii) an acpA-null mutant in a pXO1− pXO2+ strain can be complemented by atxA cloned in trans on a multicopy plasmid (43), and (iii) pXO1+ pXO2+ strains produce more capsule than pXO1− pXO2+ strains (8, 9, 11, 43). The overall conclusion of these reports was that, while atxA contributes to capsule gene expression, apcA is the major regulator of the cap operon. Nonetheless, the relative contributions of atxA and acpA to capsule gene expression in a fully virulent strain had not been determined, and there were no reports indicating the capsule phenotype of a pXO1+ pXO2+ strain harboring an acpA-null mutation. Our transcriptional profiling data and microscopic observations indicate a major role for atxA in capsule synthesis. Under our growth conditions, capsule production by the pXO1+ pXO2+ acpA-null mutant was slightly reduced compared to that of the parent strain, while the atxA-null mutant exhibited delayed and severely diminished capsule production relative to the parent.
The absence of any information regarding the mechanism by which these two regulators enhance gene expression limits our ability to propose a model for gene control. There are no reports of specific protein-DNA interactions for either protein, and similarities in promoter regions of regulated genes have not been found. If AtxA and AcpA directly control expression of some target genes by binding to promoter sequences, it is possible the two proteins are functionally similar but have different affinities for promoter DNA sequences. A simple system in which atxA controls acpA-regulated genes solely by exerting a positive effect on acpA expression is unlikely because, in our experiments and in results reported previously, atxA functions in the absence of acpA (7, 17, 42).
Our data greatly expand the list of atxA- and acpA-regulated genes in B. anthracis. Considering that all previously known targets of atxA were positively regulated, it is somewhat surprising that atxA also negatively regulates a number of genes. AtxA may function as both a repressor and an activator, as has been reported for some transcriptional regulators (31). Moreover, some or all of the target genes may be controlled by an unidentified downstream regulator. Certain atxA-regulated genes identified in our experiments are predicted to encode proteins with similarity to known regulators. For example, the atxA-regulated gene pXO2-53 is predicted to encode a protein with sequence similarity to AtxA and AcpA. Synthesis of this protein may add another layer of complexity to gene regulation in B. anthracis. Another candidate for a downstream regulator is pagR. This 300-nt gene was not represented on the microarray due to technical difficulties. Previous work in our lab has shown that pagA and pagR are cotranscribed (14). pagR is responsible for autogenous control of the atxA-regulated pag operon and may have other targets as well.
Perhaps most intriguing is the discovery of atxA-regulated genes on the B. anthracis chromosome. Many of these genes are predicted to have roles in amino acid biosynthesis and transport of amino acids and small peptides into the bacterial cell. atxA control of genes encoding transporters may be of significance for interactions of B. anthracis with its environment, be it the mammalian host or the soil. Compared to the B. subtilis genome, B. anthracis harbors a large number of genes predicted to be important for amino acid and peptide utilization (33a). It has been hypothesized that these genes are useful for growth in protein-rich environments, such as decaying animal matter (33a). Genes predicted to encode components of Opp- and App-like ABC transporters of B. subtilis and other gram-positive species (18) were negatively regulated by atxA. These include four ORFs at one locus that are predicted to encode all components of an App-like transporter and other ORFs at distinct loci that are predicted to encode homologues of individual App transporter proteins. Transporters of this type function as oligopeptide permease systems (29, 30). There have been no reports concerning these systems in B. anthracis. However, an oligopeptide permease system has been implicated in virulence gene expression in B. thuringiensis. The B. thuringiensis oligopeptide permease Opp imports a small peptide that, following processing, interacts with the pleiotropic virulence gene regulator PlcR, allowing it to bind its target DNA (35). Although B. anthracis does not produce a functional PlcR protein, it does carry an ORF predicted to encode a homologue of the B. thuringiensis peptide precursor.
It is noteworthy that the cross talk between pXO1 and the B. anthracis chromosome is bidirectional. In other work, Saile and Koehler have shown that the toxin genes of pXO1 are controlled by the chromosomal genes abrB and spo0A (34). Like scoC, abrB and spo0A function in a complex network, well studied in B. subtilis, that is involved in environmental signaling and adaptive responses (37). The transcriptional profiling data presented here do not indicate epistatic relationships between the plasmid-borne and chromosome-borne regulators.
B. anthracis is a member of the group 1 bacilli, which also includes the less harmful species B. cereus and B. thuringiensis (2). Historically, the three species were grouped phylogenetically on the basis of their similar physiology. Recent studies employing multienzyme electrophoresis and amplified fragment length polymorphism analysis indicate significant genetic diversity within B. cereus and B. thuringiensis isolates, while B. anthracis is relatively monomorphic (12, 40). The phylogeny of these species continues to be deliberated. Nevertheless, in terms of virulence, the plasmid content of the species has served for facile distinction of the species. Major virulence factors, such as certain insecticidal toxins of B. thuringiensis and the anthrax toxin proteins and capsule of B. anthracis, are plasmid encoded.
While more-detailed analysis of the genomes of the group 1 species will likely reveal numerous differences in gene content, increasing evidence suggests that the distinct phenotypes of the species with regard to pathogenesis may be due in some part to differential gene expression related to the presence or absence of functional regulatory proteins. The chromosomal regulator plcR of B. thuringiensis and B. cereus is a transcriptional activator that controls expression of a number of known and potential virulence genes, including those encoding lecithinase, proteases, and hemolysins (1). B. anthracis harbors homologues of many plcR-regulated genes, some of which are preceded by consensus sites for PlcR binding (27, 33a). However, these genes do not appear to be highly expressed in B. anthracis, because B. anthracis lacks a functional PlcR protein (1). In an analogous manner, B. cereus and B. thuringiensis do not appear to harbor atxA or acpA homologues. However, these species contain homologues of the atxA target genes on the chromosome. Although the roles of the newly identified atxA-regulated genes described here are not known, we view the species-specific regulators as important players in the transcriptomes of the species.
Our results represent a snapshot of the B. anthracis transcriptome for one time point under one specific growth condition. Certainly the transcription profile of B. anthracis will vary during growth under other conditions, including infection. It is noteworthy that in a mouse model for anthrax, a pXO1+ pXO2− atxA-null mutant is avirulent and the antibody response to all three toxin proteins is decreased significantly (7). These data indicate that atxA function is important in vivo. No studies of acpA function during infection have been reported. In light of results presented here, studies of gene expression in B. anthracis and other plasmid-containing species should be interpreted with caution when cured strains are employed. It will be of interest to test the pXO1+ pXO2+ parent and regulatory gene mutants in animal models to determine relative virulence and to compare toxin and capsule synthesis in vivo. The significant cross talk between the B. anthracis plasmids and chromosome is likely to be important for virulence and other aspects of B. anthracis physiology.
Acknowledgments
We thank Robin Cline and Erik Snesrud for assistance with the microarray and Elke Saile, Yahua Chen, and Maria Hadjifrangiskou for insightful comments during the course of the work and critical reading of the manuscript. Antisera were kindly provided by R. John Collier.
This work was supported by National Institutes of Health grant AI 33537 (to T.K.). Grants supporting array analysis were U24 DK58862-01A1 and DAMD 17-01-1-0132 from the NIDDK and the Department of Defense (to S.G.H.). Development of the DNA microarray was funded by the Office of Naval Research grant N000140010663 (to S.P.).
Editor: V. J. DiRita
REFERENCES
- 1.Agaisse, H., M. Gominet, O. A. Okstad, A. B. Kolsto, and D. Lereclus. 1999. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 32:1043-1053. [DOI] [PubMed] [Google Scholar]
- 2.Ash, C., J. A. Farrow, M. Dorsch, E. Stackebrandt, and M. D. Collins. 1991. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 41:343-346. [DOI] [PubMed] [Google Scholar]
- 3.Brennan, R. G., and B. W. Matthews. 1989. The helix-turn-helix DNA binding motif. J. Biol. Chem. 264:1903-1906. [PubMed] [Google Scholar]
- 4.Caldwell, R., R. Sapolsky, W. Weyler, R. R. Maile, S. C. Causey, and E. Ferrari. 2001. Correlation between Bacillus subtilis scoC phenotype and gene expression determined using microarrays for transcriptome analysis. J. Bacteriol. 183:7329-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cataldi, A., E. Labruyere, and M. Mock. 1990. Construction and characterization of a protective antigen-deficient Bacillus anthracis strain. Mol. Microbiol. 4:1111-1117. [DOI] [PubMed] [Google Scholar]
- 6.Dai, Z., and T. M. Koehler. 1997. Regulation of anthrax toxin activator gene (atxA) expression in Bacillus anthracis: temperature, not CO2/bicarbonate, affects AtxA synthesis. Infect. Immun. 65:2576-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai, Z., J.-C. Sirard, M. Mock, and T. M. Koehler. 1995. The atxA gene product activates transcription of the anthrax toxin genes and is essential for virulence. Mol. Microbiol. 16:1171-1181. [DOI] [PubMed] [Google Scholar]
- 8.Fouet, A., and M. Mock. 1996. Differential influence of the two Bacillus anthracis plasmids on regulation of virulence gene expression. Infect. Immun. 64:4928-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green, B. D., L. Battisti, T. M. Koehler, and C. B. Thorne. 1985. Demonstration of a capsule plasmid in Bacillus anthracis. Infect. Immun. 49:291-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guidi-Rontani, C., M. Weber-Levy, E. Labruyere, and M. Mock. 1999. Germination of Bacillus anthracis spores within alveolar macrophages. Mol. Microbiol. 31:9-17. [DOI] [PubMed] [Google Scholar]
- 11.Guignot, J., M. Mock, and A. Fouet. 1997. AtxA activates the transcription of genes harbored by both Bacillus anthracis virulence plasmids. FEMS Microbiol. Lett. 147:203-207. [DOI] [PubMed] [Google Scholar]
- 12.Helgason, E., O. A. Okstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A.-B. Kolsto. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmaster, A. R., and T. M. Koehler. 1997. The anthrax toxin activator gene atxA is associated with CO2-enhanced non-toxin gene expression in Bacillus anthracis. Infect. Immun. 65:3091-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmaster, A. R., and T. M. Koehler. 1999. Autogenous regulation of the Bacillus anthracis pag operon. J. Bacteriol. 181:4485-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivins, B. E., J. W. Ezzell, Jr., J. Jemski, K. W. Hedlund, J. D. Ristroph, and S. H. Leppla. 1986. Immunization studies with attenuated strains of Bacillus anthracis. Infect. Immun. 52:454-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koehler, T. M. 2002. Bacillus anthracis genetics and virulence gene regulation, p. 143-164. In T. M. Koehler (ed.), Anthrax, vol. 271. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 17.Koehler, T. M., Z. Dai, and M. Kaufman-Yarbray. 1994. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one of two promoters. J. Bacteriol. 176:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koide, A., and J. A. Hoch. 1994. Identification of a second oligopeptide transport system in Bacillus subtilis and determination of its role in sporulation. Mol. Microbiol. 13:417-426. [DOI] [PubMed] [Google Scholar]
- 19.Koide, A., M. Perego, and J. A. Hoch. 1999. ScoC regulates peptide transport and sporulation initiation in Bacillus subtilis. J. Bacteriol. 181:4114-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 21.Leppla, S. H. 1995. Anthrax toxins, p. 543-572. In J. Moss, B. Iglewski, M. Vaughan, and A. T. Tu (ed.), Bacterial toxins and virulence factors in disease. Marcel Dekker, New York, N.Y.
- 22.Mahillon, J., R. Rezsohazy, B. Hallet, and J. Delcour. 1994. IS231 and other Bacillus thuringiensis transposable elements: a review. Genetica 93:13-26. [DOI] [PubMed] [Google Scholar]
- 23.Makino, S., M. Watarai, H. I. Cheun, T. Shirahata, and I. Uchida. 2002. Effect of the lower molecular capsule released from the cell surface of Bacillus anthracis on the pathogenesis of anthrax. J. Infect. Dis. 186:227-233. [DOI] [PubMed] [Google Scholar]
- 24.Makino, S.-I., I. Uchida, N. Terakado, C. Sasakawa, and M. Yoshikawa. 1989. Molecular characterization and protein analysis of the cap region, which is essential for encapsulation in Bacillus anthracis. J. Bacteriol. 171:722-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mesnage, S., T. Fontaine, T. Mignot, M. Delepierre, M. Mock, and A. Fouet. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19:4473-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mesnage, S., and A. Fouet. 2002. Plasmid-encoded autolysin in Bacillus anthracis: modular structure and catalytic properties. J. Bacteriol. 184:331-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mignot, T., M. Mock, D. Robichon, A. Landier, D. Lereclus, and A. Fouet. 2001. The incompatibility between the PlcR- and AtxA-controlled regulons may have selected a nonsense mutation in Bacillus anthracis. Mol. Microbiol. 42:1189-1198. [DOI] [PubMed] [Google Scholar]
- 28.Okinaka, R. T., K. Cloud, O. Hampton, A. Hoffmaster, K. K. Hill, P. Keim, T. M. Koehler, G. Lamke, S. Kumano, J. Mahillon, D. Manter, Y. Martinez, D. Ricke, R. Svensson, and P. J. Jackson. 1999. The sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol. 181:6509-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peltoniemi, K., E. Vesanto, and A. Palva. 2002. Genetic characterization of an oligopeptide transport system from Lactobacillus delbrueckii subsp. bulgaricus. Arch. Microbiol. 177:457-467. [DOI] [PubMed] [Google Scholar]
- 30.Perego, M. 1997. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. Proc. Natl. Acad. Sci. USA 94:8612-8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez-Martin, J., F. Rojo, and V. de Lorenzo. 1994. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol. Rev. 58:268-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson, S., R. T. Cline, H. Tettelin, V. Sharov, and D. A. Morrison. 2000. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J. Bacteriol. 182:6192-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pezard, C., P. Berche, and M. Mock. 1991. Contribution of individual toxin components to virulence of Bacillus anthracis. Infect. Immun. 59:3472-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Økstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolstø, and C. M. Fraser. The genome sequence of Bacillus anthracis Ames and comparison to closely-related bacteria. Nature, in press. [DOI] [PubMed]
- 34.Saile, E., and T. M. Koehler. 2002. Control of anthrax toxin gene expression by the transition state regulator abrB. J. Bacteriol. 184:370-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slamti, L., and D. Lereclus. 2002. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 21:4550-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sterne, M. 1939. The use of anthrax vaccines prepared from avirulent (uncapsulated) variants of Bacillus anthracis. Onderstepoort J. Vet. Sci. Anim. Ind. 13:307-312. [Google Scholar]
- 37.Strauch, M. A. 1993. Abr, a transition state regulator, p. 757-764. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. ASM Press, Washington, D.C.
- 38.Stukey, J., and G. M. Carman. 1997. Identification of a novel phosphatase sequence motif. Protein Sci. 6:469-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorne, C. B., and F. C. Belton. 1957. An agar-diffusion method for titrating Bacillus anthracis immunizing antigen and its application to a study of antigen production. J. Gen. Microbiol. 17:505-516. [DOI] [PubMed] [Google Scholar]
- 40.Ticknor, L. O., A. B. Kolsto, K. K. Hill, P. Keim, M. T. Laker, M. Tonks, and P. J. Jackson. 2001. Fluorescent amplified fragment length polymorphism analysis of Norwegian Bacillus cereus and Bacillus thuringiensis soil isolates. Appl. Environ. Microbiol. 67:4863-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uchida, I., K. Hashimoto, and N. Terakado. 1986. Virulence and immunogenicity in experimental animals of Bacillus anthracis strains harbouring or lacking 110 MDa and 60 MDa plasmids. J. Gen. Microbiol. 132(Pt. 2):557-559. [DOI] [PubMed] [Google Scholar]
- 42.Uchida, I., J. M. Hornung, C. B. Thorne, K. R. Klimpel, and S. H. Leppla. 1993. Cloning and characterization of a gene whose product is a trans-activator of anthracis toxin synthesis. J. Bacteriol. 175:5329-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uchida, I., S. Makino, T. Sekizaki, and N. Terakado. 1997. Cross-talk to the genes for Bacillus anthracis capsule synthesis by atxA, the gene encoding the trans-activator of anthrax toxin synthesis. Mol. Microbiol. 23:1229-1240. [DOI] [PubMed] [Google Scholar]
- 44.Vietri, N. J., R. Marrero, T. A. Hoover, and S. L. Welkos. 1995. Identification and characterization of a trans-activator involved in the regulation of encapsulation by Bacillus anthracis. Gene 152:1-9. [DOI] [PubMed] [Google Scholar]