Abstract
The Chinese hamster ovary (CHO) cell mutants ldlC and ldlB, which exhibit almost identical phenotypes, define two genes required for multiple steps in the normal medial and trans Golgi-associated processing of glycoconjugates. The LDLC gene encodes ldlCp, an ≈80-kDa protein, which in wild-type, but not ldlB, cells associates reversibly with the cytoplasmic surface of the Golgi apparatus. Here, we have used a retrovirus-based expression cloning system to clone a murine cDNA, LDLB, that corrects the pleiotropic mutant phenotypes of ldlB cells. The corresponding mRNA was not detected in ldlB mutants. LDLB encodes an ≈110-kDa protein, ldlBp, which lacks homology to known proteins and contains no common structural motifs. Database searches identified short segments of homology to sequences from Drosophila melanogaster, Arabidopsis thaliana, and Caenorhabditis elegans, and the essentially full-length homologous human sequence (82% identity); however, as was the case for ldlCp, no homologue was identified in Saccharomyces cerevisiae. We have found that in wild-type cell cytosols, ldlCp is a component of an ≈950-kDa “ldlCp complex,” which is smaller, ≈700 kDa, in ldlB cytosols. Normal assembly of this complex is ldlBp-dependent and may be required for Golgi association of ldlCp and for the normal activities of multiple luminal Golgi processes.
In eukaryotes, glycoproteins, proteoglycans, and glycolipids are processed during their transport through the Golgi complex (1–4). Two Chinese hamster ovary (CHO) cell mutants, ldlB and ldlC, initially selected because of their low-density lipoprotein receptor (LDLR) deficiency (5, 6), exhibit virtually identical, pleiotropic defects in medial and trans Golgi-associated glycoconjugate processing. In these mutants, virtually all N- and O-linked oligosaccharides on glycoproteins and most glycolipids are not processed properly. For instance, N-linked chains are converted to partial rather than complete endoglycosidase H-resistant forms. The global nature of the cell-surface glycosylation defects in these cells is highlighted by their altered sensitivity to a variety of toxic lectins; in particular, these mutants are hypersensitive to ricin relative to wild-type CHO cells (6).
The abnormal Golgi-associated glycoconjugate processing in these mutants is not caused by a substantial block in intracellular membrane trafficking. For example, the abnormal glycosylation of LDLR and decay accelerating factor results in their proteolytic cleavage and consequent secretion of the bulk of their extracellular domains (ref. 7; P. Reddy and M.K., unpublished data). In addition, overexpression of LDLRs in ldlC cells increases the otherwise very low steady-state level of unstable, abnormally glycosylated LDLRs on the cell surface, restoring LDL binding and endocytosis to essentially normal levels (7). Therefore, secretory and endocytic traffic do not appear to be significantly disrupted in the mutant cells. Analysis of glycoconjugate structures in ldlB and ldlC cells indicated that it was unlikely that the Golgi-associated defects were the result of deficiencies in single glycosidase, glycosyltransferase, or nucleotide sugar transporter activities (6). Furthermore, even though the subcellular localizations of the Golgi markers β-COP and mannosidase II in ldlB and ldlC cells appear normal at the resolution of light microscopy, the amount of these proteins in the Golgi is reduced relative to those in wild-type cells as determined by immunofluorescence microscopy (8), suggesting that the ldlB and ldlC defects affect other peripheral and integral membrane Golgi-associated proteins.
We previously reported the cloning of the human LDLC gene and its Caenorhabditis elegans homologue (8). The LDLC cDNA encodes a novel ≈80-kDa protein, ldlCp, which is expressed in wild-type CHO and ldlB cells but not in ldlC mutants. Sequence analysis revealed that ldlCp is likely to be a soluble, cytosolic protein. In wild-type cells, ldlCp exhibits a brefeldin A (BFA)-sensitive association with the Golgi apparatus similar to that reported for β-COP, a peripheral Golgi protein of the COPI complex involved in intracellular vesicular transport (9, 10). Although present in essentially normal amounts, ldlCp is not associated with the Golgi apparatus in ldlB mutants. These results suggest that ldlCp associates reversibly with the cytoplasmic surface of the Golgi and that this association depends on the LDLB gene and is required for normal Golgi function (8).
Here we used a highly efficient retrovirus-based expression cloning system to isolate a murine cDNA, LDLB, that corrects the pleiotropic defects in ldlB cells. The predicted product, ldlBp, is an ≈103- to 109-kDa protein which, like ldlCp (8), lacks homology to known proteins and contains no common structural motifs (e.g., signal sequences for endoplasmic reticulum translocation, membrane-spanning domains), suggesting that ldlBp is a soluble cytosolic protein. Its mRNA was undetectable in ldlB cells. In addition, we have identified a large (≈950-kDa) complex containing ldlCp in the cytosol of wild-type CHO cells and rat liver, and have found that ldlBp is required for the normal assembly or stability of this complex.
MATERIALS AND METHODS
Materials.
Reagents (and sources) were: Lipofectamine, G418, and Elongase (Life Technologies, Grand Island, NY); hygromycin (Boehringer Mannheim); cell culture media and supplements (Life Technologies, Grand Island, NY and JRH Biosciences, Lenexa, KS). Newborn calf lipoprotein-deficient serum (NCLPDS), LDL, and DiI-LDL (LDL labeled with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine iodide) were prepared as described (11, 12). Other reagents were obtained from Sigma or other standard chemical suppliers. Compactin was a gift from A. Endo (Tokyo Nodo University, Japan). Affinity-purified polyclonal anti-ldlCp antibody (anti-Cpep) was prepared as described (8). The anti-β-COP mAb M3A5 was obtained from R. Klausner and J. Donaldson (National Institutes of Health) (13). Rat liver cytosol, a gift from A. Fisher and T. Kirchhausen (Harvard Medical School), was prepared as described (14).
Cell Culture.
All incubations were performed at 37°C in a 5% CO2 incubator unless otherwise noted, and cells were harvested with trypsin/EDTA. Wild-type CHO, ldlB-11 and ldlB-WGAr (two independently derived clones of ldlB mutants), and ldlC cells (5, 6) were maintained in medium A [Ham’s F-12 containing penicillin (50 units/ml), streptomycin (50 μg/ml), and glutamine (2 mM)] with 5% (vol/vol) fetal bovine serum (FBS) (medium B). Phoenix ecotropic packaging cells (ATCC SD 3444) were a gift from G. Nolan (Stanford University Medical Center), and were maintained in medium C [Dulbecco’s Modified Eagle Medium containing penicillin, streptomycin, and glutamine as above and 10% (vol/vol) FBS]. ldlB[Eco] cells, expressing the ecotropic retrovirus receptor (15), were prepared by transfecting ldlB-11 cells with pCB7-Eco by using Lipofectamine (according to the manufacturer’s instructions) and were selected and maintained in medium B with 50 μg/ml hygromycin (medium D). ldlB[LDLB] transfectants (see below) were maintained in medium B supplemented with 1.3 ng/ml ricin (medium E). In preparation for DiI-LDL uptake assays, cells were grown for ≥48 h in medium A supplemented with 3% (vol/vol) NCLPDS (medium F) to induce LDLR expression.
Production of Retroviral Supernatants.
A size-fractionated (>2 kb) murine NIH 3T3 cell cDNA library containing >106 independent clones was prepared by using the SuperScript Plasmid System for cDNA Synthesis (Life Technologies, Grand Island, NY). BstXI/EcoRI adapters (Invitrogen) were ligated to the cDNA for nondirectional ligation into the BstXI sites of pMX-IRES-GFP (16, 17). The average insert size was ≈2 kb, and >95% of clones had inserts. Retroviral supernatants were prepared by transfecting Phoenix cells with the pMX-3T3-IRES-GFP library, pMX-GFP [control encoding the green fluorescent protein (GFP)] (17), or genomic DNA (virus mobilization assay) as described (18). For genomic DNA transfections, the cells were pretreated for 1 h with medium C with 24 μM chloroquine. Supernatants were collected 24 and 48 h posttransfection, frozen in liquid N2, and stored at −80°C.
Isolation of Ricin-Resistant, LDLR-Positive ldlB Retroviral Transfectants.
ldlB[Eco] cells were set on day 0 in medium D at 300,000 cells per 100-mm dish. On day 2, the cells were incubated overnight at 32°C with retroviral supernatants diluted 1:2 (pMX-3T3-IRES-GFP library) or 1:10 (pMX-GFP) with medium A containing 10% (vol/vol) FBS (medium G) and brought to 5 μg/ml polybrene. On day 3, the medium was replaced with fresh medium G (37°C), and on day 4, cells from each dish were reset into four 100-mm dishes in medium G. Samples from each infection dish were analyzed for GFP expression by flow cytometry, which showed similar infection rates (≈80%) for the library and control viral supernatants. On days 5 and 7, the medium was replaced with ricin selection medium (medium E) and on day 10 with medium F containing 2.5 ng/ml ricin (medium H). On day 15 we assessed LDLR activity by incubating the colonies for 1 h with medium F containing 1 μg protein/ml DiI-LDL, replacing the medium with medium H, and observing uptake of DiI-LDL in situ by using a Leitz inverted fluorescence microscope and a rhodamine filter package. Ricin-resistant, LDLR-positive colonies were picked and grown to mass culture in medium E.
Virus Mobilization Assay.
Low-titer retroviral supernatants were prepared as described above by transfecting Phoenix cells with 6 μg of genomic DNA isolated from individual ricin-resistant, LDLR-positive, retroviral transfectant clones produced in the primary infection of ldlB[Eco] cells with the cDNA library. On days 2 and 3, naive ldlB[Eco] cells (plated in six-well dishes at 80,000 cells per well in medium D on day 0) were treated with the undiluted Phoenix supernatants supplemented with 5 μg/ml polybrene and incubated overnight at 32°C. On day 4, the cells were reset into 100-mm dishes in medium B and selected with ricin in medium E followed by medium H as above. Surviving colonies from this second round of infection/selection were picked and grown to mass culture in LDLR selection medium [medium F supplemented with MeLoCo (250 μM mevalonate, 3 μg protein/ml LDL, and 40 μM compactin)] (19).
Recovery of Integrated Retroviral cDNAs.
Genomic DNA from a ricin-resistant, LDLR-positive ldlB retroviral transfectant isolated from the mobilization assay was amplified by PCR using Elongase with 2 mM Mg2+, 2% dimethyl sulfoxide, and primers located within pMX-IRES-GFP (5′ primer, CCACCGCCCTCAAAGTAGACG; 3′ primer, CCAACTTAATCGCCTTGCAGCA). The full-length PCR product was subcloned into EcoRV-digested pcDNA3.1 (Invitrogen) to generate the pLDLB-1 expression vector.
Sequencing and Analysis.
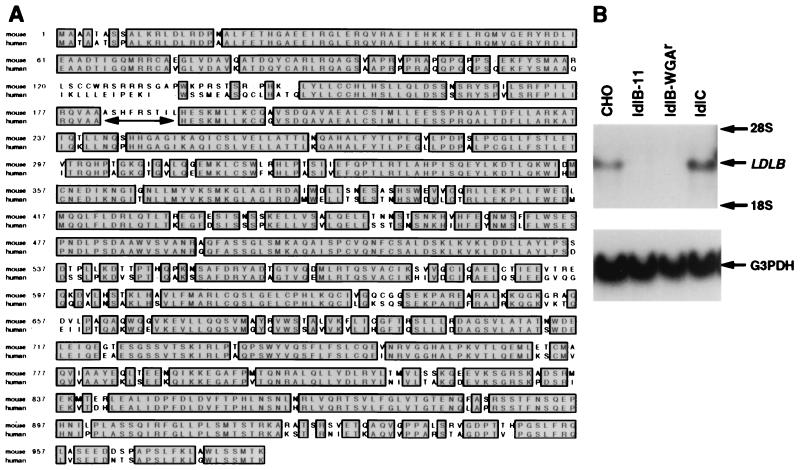
Both strands of pLDLB-1 were sequenced by using oligonucleotide primers and automated sequencers (Research Genetics, Huntsville, AL). Expressed sequence tags (ESTs) corresponding to the human homologue of ldlBp were identified in the National Center for Biotechnology (NCBI) database by using blast (20): AA442565, AA701034, AA436810, AI244664, AA96967, AA251405, AA826944, AA621133, AA287379, AA287478, N77768, AA192573, AA251404, N62862, AA319188, R01841, AA040337, R71213, AA32326, AA040338, AA853738, D20818, and N77918. Except for a 9-bp interval (see Fig. 1A), the entire human ldlBp sequence was assembled. Single bases were inserted into the human EST sequence at seven positions corresponding to murine amino acid positions 208, 410, 441, 449, 517, 577, and 588 to maintain the appropriate reading frame.
Figure 1.
Sequences of the murine and human LDLB cDNAs and LDLB expression in wild-type and mutant CHO cells. (A) Predicted amino acid sequence of murine ldlBp and alignment with a human ldlBp sequence assembled from the EST database (see Materials and Methods). The predicted amino acid sequence is numbered assuming that the first potential initiator codon is utilized. The human and mouse genes were aligned utilizing clustx. Identical amino acids are shaded and boxed. The double-headed, horizontal arrow indicates a gap in the human ldlBp sequence for which no information was available in the EST database. (B) Northern blot analysis (see Materials and Methods) of the LDLB mRNA in wild-type and mutant CHO cells by using LDLB (Upper) or G3PDH (Bottom) cDNA probes.
Transfection of ldlB Cells with pLDLB-1.
ldlB-11 cells were set at 1.6 × 105 cells per well in six-well dishes and transfected 2 days later with 340 ng of pLDLB-1 or a control vector (pcDNA3.1) by using Lipofectamine. Both pLDLB-1 and pcDNA3.1 contain a neoR selectable marker allowing for selection with G418. Two days later, the cells were replated (7.5 × 105 cells per 100-mm dish) and selected with ricin (medium E) or G418 [medium B supplemented with 75 μg/ml G418 (medium I)]. After approximately 2 weeks of selection, surviving colonies were assayed for uptake of DiI-LDL as described above. Several colonies of ricin-resistant, LDLR-positive ldlB cells stably transfected with pLDLB-1 were isolated, and one, designated ldlB[LDLB], was used in the experiments reported below. All results with this colony were confirmed by using independent colonies.
Gel Chromatographic Analysis of ldlCp.
Cytosol was prepared from 40 confluent 150-mm dishes each of wild-type CHO, ldlB, and ldlB[LDLB] cells by using a modification of the method of Balch et al. (ref. 21 and W. Balch, personal communication). Cells were harvested and sequentially washed, first with medium G then with 140 mM potassium acetate/10 mM triethylamine–acetic acid (pH 7.2) containing 20 μg/ml trypsin inhibitor type IIs, and then 125 mM potassium acetate/25 mM Hepes⋅KOH (pH 7.2) containing 20 μg/ml trypsin inhibitor type IIs. Cells were homogenized in 125 mM potassium acetate/25 mM Hepes⋅KOH (pH 7.2) containing 0.1 units/ml aprotinin, 5 μg/ml leupeptin, and 2 μg/ml trypsin inhibitor type IIs by using a ball bearing homogenizer (H&Y Enterprise, Redwood City, CA) (21). After centrifugation of the homogenate for 1 h at 200,000 × g, the supernatant (cytosol) was concentrated approximately 2.5-fold by centrifugation using Centricon-3 concentrators (Amicon). Cytosol also was isolated from wild-type CHO cells incubated for 5 min with medium B containing 5 μg/ml BFA, in which case all buffers used also contained 5 μg/ml BFA. Two hundred microliters of concentrated cytosols were size-fractionated by using gel chromatography at a flow rate of 0.25 ml/min on a 24-ml Superose 6 column (Amersham Pharmacia) equilibrated with PBS (22). Forty-eight 0.5-ml fractions were collected and 30-μl aliquots were analyzed by electrophoresis on 10% SDS/PAGE gels and immunoblotting with anti-ldlCp antibody as described (8), except that antibody binding was detected by using an enhanced chemiluminescence (ECL) Western blotting analysis system (Amersham Pharmacia).
Other Methods.
Immunofluorescence microscopy was performed as described (8) except that affinity-purified polyclonal anti-ldlCp antibody was used at 10 μg/ml and the samples were mounted in VectaShield mounting medium (Vector Laboratories). In vitro transcription/translation was performed by using the TNT T7 Quick Coupled Transcription/Translation System (Promega). Other techniques were performed as described (22) except as noted. Genomic DNA was prepared by using a Blood and Cell Culture DNA Midi kit (Qiagen, Chatsworth, CA). Southern blots were prepared from NheI-digested genomic DNA by using Hybond N+ filters (Amersham Pharmacia) and hybridized with a 32P-labeled GFP cDNA probe [EcoRI/NotI fragment of pEGFP-1 (CLONTECH)] at 65°C in QuickHyb (Stratagene). Northern blots were prepared (30 μg of total RNA per sample) by using GeneScreen Plus filters (DuPont/NEN) and hybridized as described (8) by using 32P-labeled LDLB (3-kb NotI fragment of pLDLB-1) or G3PDH (CLONTECH) probes.
RESULTS
Expression Cloning of a Murine cDNA, LDLB, That Complements the Defects in ldlB Cells.
The ricin hypersensitivity of ldlB mutants relative to wild-type cells (6) was exploited to clone by functional expression the wild-type LDLB cDNA. The ldlB[Eco] cells were infected either with a high-titer retroviral supernatant from an NIH 3T3-cell cDNA library prepared in the pMX-IRES-GFP vector or with a control virus, pMX-GFP, encoding GFP alone (see Materials and Methods). The infected cells were subjected to ricin selection conditions that kill ldlB mutants but not wild-type cells. Whereas only 14 colonies were obtained from a single dish of cells infected with the control virus, 257 ricin-resistant colonies were obtained from four dishes of cells infected with the library supernatant. Of these, 158 were LDLR-positive, as measured by DiI-LDL uptake, whereas none of the colonies from the control dish were LDLR-positive. Genomic DNA was individually prepared from 24 of the 158 ricin-resistant, LDLR-positive retroviral transfectant colonies and subjected to Southern blot analysis by using a restriction enzyme expected to excise fragments that contain the inserted cDNA together with a 3.5-kb segment of the vector, including the GFP-encoding cDNA. A GFP cDNA probe hybridized to between 1 and 6 restriction fragments in each of the 24 samples, establishing that these cells had been infected with one or more retroviruses.
An ≈6.5-kb hybridizing restriction fragment was present in 21 of the 24 clones. To determine whether this fragment contained a cDNA that could correct the defects in ldlB cells, we performed a virus mobilization assay (see Materials and Methods). Ecotropic retrovirus-packaging cells were transfected with genomic DNA from each of these 24 individual clones to generate low-titer retroviral supernatants from proviruses contained within the genomic DNAs. As negative controls, retroviral supernatants also were generated using genomic DNAs from uninfected ldlB[Eco] cells and from ldlB[Eco] cells infected with the control virus (pMX-GFP). Naive ldlB[Eco] cells then were infected with these retroviral supernatants and subjected to ricin selection. We observed 40–200 ricin-resistant colonies per mobilization assay dish for supernatants produced from each of the 24 experimental samples and only 2–8 colonies per dish for the controls. Sixteen colonies from 4 of the 24 mobilization assay dishes were isolated and all were able to grow in MeLoCo selection medium, which permits proliferation of LDLR-positive, but not LDLR-negative, cells (19). Southern blot analysis with a GFP probe of genomic DNAs from these 16 clones revealed a single, ≈6.5-kb restriction fragment in all 16 samples. This mobilization analysis indicated that the ricin-resistant, LDLR-positive colonies arose as a consequence of infection by retroviruses encoding a single type of cDNA insert that complements the defects in ldlB mutants. After accounting for vector sequence contained within the ≈6.5-kb restriction fragment, the size of the cDNA insert was estimated to be ≈3 kb.
An ≈3.2-kb fragment of the provirus integrated into the genomic DNA of 1 of the 16 clones obtained in the mobilization assay was amplified by PCR, ligated into pcDNA3.1 to generate the expression vector pLDLB-1, and sequenced. This fragment comprised a 3,034-bp cDNA insert as well as ≈200 bp derived from the pMX-IRES-GFP vector and the adapters used in the cDNA library construction. The cDNA contains a 15-bp 5′-untranslated region, a 2,940-bp ORF, a 56-bp 3′-untranslated region, and a 23-bp poly(A) tail. The predicted amino acid sequence of the corresponding 980-residue protein (calculated molecular mass of 109 kDa), designated ldlBp, is shown in Fig. 1A. An alternative translation start site at position 51 would generate a 103-kDa protein. In vitro transcription/translation using pLDLB-1 as the template (see Materials and Methods) generated a protein of ≈110 kDa (not shown). ldlBp’s predicted amino acid sequence contains no common sequence motifs such as candidate transmembrane domains or signal sequences for endoplasmic reticulum translocation, suggesting that, like ldlCp (8), ldlBp is a soluble, cytoplasmic protein. Database analyses revealed no similarities to known genes or proteins; however, multiple human ESTs have permitted us to construct an essentially full-length protein sequence for human ldlBp (Fig. 1A) that is 82% identical to the murine sequence. In addition, we identified short EST or genomic DNA sequences from Drosophila melanogaster (AI109997, AI134952, AI260021, and AI258322), Arabidopsis thaliana (AB005242), and C. elegans (Y54E10.Contig116) which are highly homologous to portions of ldlBp, raising the possibility that there may be ldlBp homologues in these organisms. In contrast to these multicellular organisms, the yeast Saccharomyces cerevisiae has no LDLB homologue (data not shown).
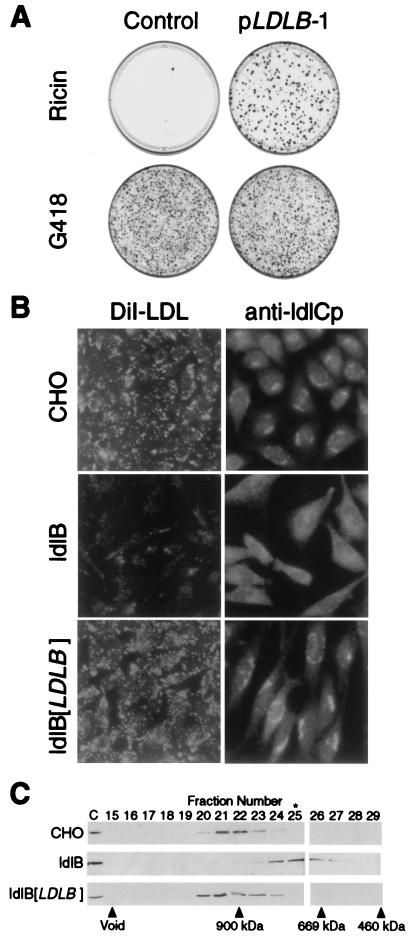
To determine whether expression of the cloned LDLB gene was defective in ldlB cells, we performed Northern blot analysis on RNA from wild-type CHO, ldlB-11, ldlB-WGAr, and ldlC cells by using LDLB and G3PDH (loading control) cDNAs as probes (Fig. 1B). There was a strong LDLB signal in the wild-type CHO and ldlC samples but no detectable signal from the two independently isolated ldlB mutants. Thus, loss of LDLB expression in ldlB mutants is likely to be the source of the mutant phenotypes in these cells. To confirm this, we transfected ldlB cells with the pLDLB-1 expression vector or the control plasmid, pcDNA3.1. Even though their transfection efficiencies into ldlB-11 cells were similar, as measured by selection with G418, pLDLB-1, but not the control plasmid, conferred resistance to ricin (1.3 ng/ml) on ldlB cells (Fig. 2A). Similar results were obtained when an independently isolated ldlB mutant clone, ldlB-WGAr, was analyzed. However, pLDLB-1 did not confer resistance to ricin on ldlC cells (data not shown). Furthermore, a clone of ldlB-11 cells stably transfected with pLDLB-1 (ldlB[LDLB]) exhibited LDLR activity, as measured by DiI-LDL uptake, that was similar to that of wild-type CHO cells and significantly greater than that of the LDLR-deficient, untransfected ldlB mutants (Fig. 2B, Left). Previous immunofluorescence studies established that ldlCp associates with the cytoplasmic surface of the Golgi apparatus in wild-type CHO cells, but not in ldlB cells where it apparently is distributed throughout the cytoplasm (ref. 8 and Fig. 2B, Right). Golgi association of ldlCp was restored in the ldlB[LDLB] cells (Fig. 2B, Right). Thus, the three hallmark features of ldlB mutants, (i) abnormal glycoconjugate synthesis and cell surface expression as measured by selection with ricin, (ii) reduced LDLR activity as measured by DiI-LDL uptake, and (iii) loss of Golgi association of ldlCp (6, 8), were corrected by transfection with pLDLB-1.
Figure 2.
LDLB cDNA corrects the mutant phenotypes of ldlB cells. (A) Ricin sensitivity. ldlB cells were transfected with either pLDLB-1 or pcDNA3.1 (control). Two days later, the cells were replated and selected with ricin (medium E) or G418 (medium I, transfection efficiency control). Twelve days later, the dishes were fixed and stained with crystal violet. (B) LDLR activity and Golgi localization of ldlCp. Left, wild-type CHO, ldlB, and ldlB[LDLB] cells were grown on glass cover slips in medium F, and LDLR activity, as measured by uptake of fluorescent DiI-LDL (1 μg protein per ml, 1 hr), was observed by using a ×40 objective lens as described (see Materials and Methods). Right, Wild-type CHO, ldlB, and ldlB[LDLB] cells were grown on glass cover slips in medium B and immunostained with a polyclonal anti-ldlCp antibody followed by FITC-labeled goat anti-rabbit IgG and observed by using a ×63 objective lens as described (see Materials and Methods). Golgi localization of ldlCp in CHO and ldlB[LDLB] cells was confirmed by double staining with anti-β-COP antibody (not shown). (C) Gel chromatographic analysis of ldlCp. Cytosol was prepared from wild-type CHO, ldlB, and ldlB[LDLB] cells and analyzed by Superose 6 chromatography as described (see Materials and Methods). Samples were subjected to SDS/PAGE and immunoblotting with affinity-purified polyclonal anti-ldlCp antibody. Fractions containing the ≈80-kDa ldlCp band, as well as unfractionated, diluted (1:10) cytosol (C) are shown. The column was calibrated with Mr standards ranging from 65 to 900 kDa, including apoferritin (460 kDa), thyroglobulin (669 kDa), and IgM (900 kDa) (vertical arrows). The peak fraction of β-COP-containing coatomer, determined by immunoblotting with a monoclonal antibody against β-COP (not shown), is indicated by ∗.
ldlCp Is Part of a Large Macromolecular Complex.
To further investigate the role of ldlBp in ldlCp’s attachment to the Golgi, we examined the oligomerization state of ldlCp in homogenates of wild-type CHO, ldlB, and ldlB[LDLB] cells. Preliminary cell-fractionation studies (S. Podos and M.K., unpublished data) suggested that under the conditions we used to disrupt the cells (see Materials and Methods), most of the ldlCp (apparent monomer mass of ≈80 kDa determined by SDS/PAGE, ref. 8 and see below) is found in the cytosolic fraction and not the membrane fraction, presumably because the Golgi-bound ldlCp dissociates during the procedure. Samples of cytosol from each cell type as well as rat liver cytosol were size-fractionated by using a Superose 6 column, and the fractions were subjected to SDS/PAGE and immunoblotting analysis by using anti-ldlCp and anti-β-COP antibodies. In cytosol from wild-type CHO cells (Fig. 2C, Top) and rat liver (data not shown), ldlCp eluted with the surprisingly large apparent mass of ≈950 kDa, a size substantially greater than that of the ≈700-kDa, β-COP-containing coatomer complex (data not shown) (23). Thus, ldlCp appears to exist exclusively as a component of an ≈950-kDa homo- or heterooligomeric protein complex in cytosol from wild-type CHO cells and rat liver. Before homogenization, we treated some of the cultures of wild-type CHO cells with BFA, a drug that induces several peripheral Golgi proteins (e.g., ldlCp, β-COP-containing coatomer) to dissociate from the Golgi (8, 9). BFA treatment did not affect the Superose 6 elution profile of cytosolic ldlCp (data not shown), and thus presumably does not induce the dissociation of ldlCp from the Golgi by dissociating the ldlCp complex. Similar analysis of ldlCp in cytosol from ldlB cells established that ldlCp eluted in a smaller complex, ≈700 kDa (Fig. 2C, Middle). The ldlCp complex was restored to its normal size in ldlB[LDLB] cells (Fig. 2C, Bottom). Thus, ldlBp is required for the assembly, processing, or stability of intact ldlCp complexes, and this presumably accounts for both the inability of ldlCp to associate properly with the Golgi apparatus in ldlB cells and the pleiotropic functional Golgi defects in these cells.
DISCUSSION
We have used a retrovirus-based expression cloning strategy to clone a murine cDNA, LDLB, which corrects the abnormalities of ldlB mutants. Expression of the endogenous hamster LDLB message was virtually undetectable in two independently isolated ldlB mutant clones, suggesting that mutations in LDLB itself, or a gene required for the expression of LDLB, are responsible for the pleiotropic Golgi defects in ldlB mutants. The retroviral expression library cloning system used here has several benefits over traditional genomic DNA or cDNA library transfection techniques used to clone genes from mammalian cell mutants (8, 24–26). The very high efficiency of retrovirus-mediated gene transfer permits the screening of the entire complexity (≈106 unique cDNAs) of a large cDNA library about 16 times by using only four 100-mm dishes of cells (≈4 × 106 cells infected with an average of four viruses per cell). In addition, the mobilization assay described here permits the efficient identification and PCR-based rescue from multiply infected target cells of the retrovirally encoded cDNA responsible for the phenotypic correction. This mobilization approach is analogous to the secondary and tertiary transfections used in genomic DNA transfection methods (8).
The pleiotropic medial- and trans-Golgi-associated glycoconjugate processing defects in ldlB cells result from the absence of the mRNA encoding ldlBp, the protein product of the LDLB gene. In ldlBp’s absence, ldlCp does not associate with the Golgi apparatus. As shown here, ldlBp is an ≈103- to 109-kDa protein that, like ldlCp (8), lacks homology to any other known proteins and contains no common structural motifs, such as transmembrane domains or an endoplasmic reticulum translocation signal sequence. As was the case for ldlCp (8), there is no readily apparent homologue for ldlBp in yeast. The presence in the EST and genomic databases of potential D. melanogaster and C. elegans ldlBp homologues with significant similarity to murine and human ldlBp suggests that ldlBp mediates a well-conserved cellular function in multicellular eukaryotes.
Immunofluorescence microscopy demonstrates that ldlCp is a peripheral Golgi protein in wild-type CHO cells (8), however, preliminary cell-fractionation experiments (S. Podos and M.K., unpublished data) revealed that most of the ldlCp is present in the cytosolic fraction as opposed to the membrane fraction. This suggests that either the conditions used to homogenize the cells cause dissociation of ldlCp from the Golgi or that there are two pools of ldlCp in wild-type cells, a Golgi-associated pool and a cytosolic pool. The ldlBp-dependent nature of ldlCp Golgi-association suggests that ldlBp could serve as a Golgi anchor for ldlCp, participate with ldlCp in a complex of cytosolic proteins that bind to the Golgi surface, or process ldlCp into a form competent to bind to the Golgi. We have demonstrated that ldlCp is part of a large, ≈950-kDa complex in cytosol prepared from wild-type CHO cells. Furthermore, in cytosol prepared from ldlB cells, which lack LDLB mRNA, the ldlCp complex is significantly smaller (≈700 kDa). Transfection of ldlB cells with pLDLB-1 restores the ldlCp complex to the size observed in wild-type cells. These results demonstrate that ldlBp is required for the normal assembly, processing, or stability of the ldlCp complex, and suggest that an intact ldlCp complex is required for Golgi association of ldlCp. Parsimony suggests that ldlBp is likely to be an essential component of the ldlCp complex, although it may influence the size of the ldlCp complex without being directly integrated in the complex by participating directly or indirectly in the assembly, processing, or stabilization of the complex.
The diversity of the defects in ldlB and ldlC mutants suggests that the mutations may affect the regulation, compartmentalization, or activity of several different Golgi enzymes or enzyme substrates (6). The primary biochemical defect in these cells may cause Golgi disruptions by either (i) blocking the synthesis of small and/or macromolecular substrates or their access to Golgi enzymes, (ii) blocking Golgi enzyme transport to or retention at the appropriate site, (iii) preventing posttranslational modifications of Golgi enzymes required for their function, (iv) disrupting the basic structure of the Golgi or its luminal environment (pH, ion concentrations) so that multiple, distinct enzymes cannot function in situ, or (v) some combination of these. Alternatively, the ldlB and ldlC mutations could interfere subtly with intraGolgi membrane transport, such that the glycoconjugate substrates themselves lack access to certain compartments within the Golgi (8). The identification of the LDLC and LDLB genes should help define the mechanisms underlying the phenotypes of these mutants.
The ldlCp complex joins a growing list of soluble multiprotein complexes associated with the cytoplasmic surfaces of vesicles and organelles in the secretory pathway. These include the well studied coatomer complexes COPI and COPII (10, 23, 27) and the recently identified Exocyst (28, 29), TRAPP (transport protein particle) (30), and GTC (Golgi transport) (31) complexes. GTC is a five-component (110-, 109-, 90-, 82-, and 71-kDa subunits), ≈800-kDa Golgi-associated heterooligomer isolated from mammalian tissue (31). The cDNA for its 90-kDa subunit, GTC-90, has been cloned. GTC was identified based on its ability to stimulate cis- to medial-Golgi transport activity in an in vitro assay. Processing of vesicular stomatitis virus (VSV) G protein in this assay requires both the fusion of VSV-G containing, N-acetylglucosaminyltransferase I (NAGT I)-defective donor Golgi membranes with wild-type acceptor Golgi membranes, and the subsequent transfer of [3H]N-acetylglucosamine to VSV-G by NAGT I in the acceptor Golgi membranes. Therefore, GTC could have influenced the assay by affecting transport per se, glycosylation only, or both (31). Several common features raise the possibility that GTC and ldlCp complex may be identical: (i) They are both large, Golgi-associated oligomers of apparently similar size that can be detected in the cytosol of disrupted specimens. (ii) Two of the subunits of GTC (≈109 and 82 kDa) are similar in mass to ldlBp and ldlCp. (iii) ldlB and ldlC cells, which both lack a functional ldlCp complex, fail to process VSV-G to its normal endoglycosidase H-resistant form (6), and GTC stimulates the same process in an in vitro assay (31). Finally, (iv) whereas there apparently are C. elegans and D. melanogaster homologues of ldlBp, ldlCp, and GTC-90, there are no obvious homologues for these proteins in the S. cerevisiae genome (refs. 8 and 31, and see above). Determination of the relationships between the ldlCp complex and GTC and additional characterization of their effects on the secretory pathway should provide additional insights into the structure and function of the Golgi apparatus.
Acknowledgments
We thank M. Penman and S. Xu for excellent technical assistance; W. Balch for advice on preparing cytosol; G. Nolan for his gift of Phoenix packaging cells; R. Klausner and J. Donaldson for providing the anti-β-COP antibody (M3A5, originally from T. Kreis); T. Kirchhausen and A. Fisher for providing rat liver cytosol; R. Horvitz and members of his laboratory (C. Ceol, G. Stanfield, and E. Hartwieg) for use of the fluorescence microscope, advice, and assistance; and S. Sanders and C. Kaiser for critically reading this manuscript. We are especially grateful to X. Liu (Whitehead Institute, Cambridge, MA) for providing the pMX-IRES-GFP vector and advice about PCR methods. This work was supported by National Institutes of Health Grant HL 41484. J.C. was supported by National Institutes of Health National Research Service Award GM 17591. D.H. was supported by National Institutes of Health National Research Service Award CA 09541.
ABBREVIATIONS
- LDL
low density lipoprotein
- LDLR
low density lipoprotein receptor
- NCLPDS
newborn calf lipoprotein deficient serum
- DiI
1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine iodide
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- EST
expressed sequence tag
- CHO
Chinese hamster ovary
- BFA
brefeldin A
- GTC
Golgi transfer complex
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF109377).
References
- 1.Pryer N K, Wuestehube L J, Schekman R. Annu Rev Biochem. 1992;61:471–516. doi: 10.1146/annurev.bi.61.070192.002351. [DOI] [PubMed] [Google Scholar]
- 2.Rothman J E. Nature (London) 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 3.Kornfeld R, Kornfeld S. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 4.Hirschberg C B, Snider M D. Annu Rev Biochem. 1987;56:63–87. doi: 10.1146/annurev.bi.56.070187.000431. [DOI] [PubMed] [Google Scholar]
- 5.Kingsley D M, Krieger M. Proc Natl Acad Sci USA. 1984;81:5454–5458. doi: 10.1073/pnas.81.17.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kingsley D M, Kozarsky K F, Segal M, Krieger M. J Cell Biol. 1986;102:1576–1585. doi: 10.1083/jcb.102.5.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddy P, Krieger M. Mol Cell Biol. 1989;9:4799–4806. doi: 10.1128/mcb.9.11.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Podos S D, Reddy P, Ashkenas J, Krieger M. J Cell Biol. 1994;127:679–691. doi: 10.1083/jcb.127.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donaldson J G, Lippincott-Schwartz J, Bloom G S, Kreis T E, Klausner R D. J Cell Biol. 1990;111:2295–2306. doi: 10.1083/jcb.111.6.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serafini T, Stenbeck G, Brecht A, Lottspeich F, Orci L, Rothman J E, Wieland F T. Nature (London) 1991;349:215–220. doi: 10.1038/349215a0. [DOI] [PubMed] [Google Scholar]
- 11.Krieger M. Cell. 1983;33:413–422. doi: 10.1016/0092-8674(83)90423-3. [DOI] [PubMed] [Google Scholar]
- 12.Pitas R E, Innerarity T L, Weinstein J N, Mahley R W. Arteriosclerosis. 1981;1:177–185. doi: 10.1161/01.atv.1.3.177. [DOI] [PubMed] [Google Scholar]
- 13.Allan V J, Kreis T E. J Cell Biol. 1986;103:2229–2239. doi: 10.1083/jcb.103.6.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Traub L M, Ostrom J A, Kornfeld S. J Cell Biol. 1993;123:561–573. doi: 10.1083/jcb.123.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker B W, Boettiger D, Spooncer E, Norton J D. Nucleic Acids Res. 1992;20:5234. doi: 10.1093/nar/20.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Onishi M, Kinoshita S, Morikawa Y, Shibuya A, Phillips J, Lanier L L, Gorman D M, Nolan G P, Miyajima A, Kitamura T. Exp Hematol. 1996;24:324–329. [PubMed] [Google Scholar]
- 17.Liu X, Sun Y, Constantinescu S N, Karam E, Weinberg R A, Lodish H F. Proc Natl Acad Sci USA. 1997;94:10669–10674. doi: 10.1073/pnas.94.20.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein J L, Helgeson A S, Brown M S. J Biol Chem. 1979;254:5403–5409. [PubMed] [Google Scholar]
- 20.Altschul S F, Gish W, Miller W, Myers E W, Lipman D. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Balch W E, Dunphy W G, Braell W A, Rothman J E. Cell. 1984;39:405–416. doi: 10.1016/0092-8674(84)90019-9. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 23.Waters M G, Serafini T, Rothman J E. Nature (London) 1991;349:248–251. doi: 10.1038/349248a0. [DOI] [PubMed] [Google Scholar]
- 24.Guo Q, Vasile E, Krieger M. J Cell Biol. 1994;125:1213–1224. doi: 10.1083/jcb.125.6.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rawson R B, Zelenski N G, Nijhawan D, Ye J, Sakai J, Hasan M T, Chang T Y, Brown M S, Goldstein J L. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 26.Velazquez L, Fellous M, Stark G R, Pellegrini S. Cell. 1992;70:313–322. [PubMed] [Google Scholar]
- 27.Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach M F, Ravazzola M, Amherdt M, Schekman R. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 28.TerBush D R, Maurice T, Roth D, Novick P. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu S, Ting A E, Hazuka C D, Davanger S, Kenny J W, Kee Y, Scheller R H. Neuron. 1996;17:1209–1219. doi: 10.1016/s0896-6273(00)80251-2. [DOI] [PubMed] [Google Scholar]
- 30.Sacher M, Jiang Y, Barrowman J, Scarpa A, Burston J, Zhang L, Schieltz D, Yates J R, III, Abeliovich H, Ferro-Novick S. EMBO J. 1998;17:2494–2503. doi: 10.1093/emboj/17.9.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walter D M, Paul K S, Waters M G. J Biol Chem. 1998;273:29565–29576. doi: 10.1074/jbc.273.45.29565. [DOI] [PubMed] [Google Scholar]