Abstract
Erythrocyte invasion by Plasmodium vivax is completely dependent on binding to the Duffy blood group antigen by the parasite Duffy binding protein (DBP). The receptor-binding domain of this protein lies within a cysteine-rich region referred to as region II (DBPII). To examine whether antibody responses to DBP correlate with age-acquired immunity to P. vivax, antibodies to recombinant DBP (rDBP) were measured in 551 individuals residing in a village endemic for P. vivax in Papua New Guinea, and linear epitopes mapped in the critical binding region of DBPII. Antibody levels to rDBPII increased with age. Four dominant linear epitopes were identified, and the number of linear epitopes recognized by semiimmune individuals increased with age, suggesting greater recognition with repeated infection. Some individuals had antibodies to rDBPII but not to the linear epitopes, indicating the presence of conformational epitopes. This occurred in younger individuals or subjects acutely infected for the first time with P. vivax, indicating that repeated infection is required for recognition of linear epitopes. All four dominant B-cell epitopes contained polymorphic residues, three of which showed variant-specific serologic responses in over 10% of subjects examined. In conclusion, these results demonstrate age-dependent and variant-specific antibody responses to DBPII and implicate this molecule in partial acquired immunity to P. vivax in populations in endemic areas.
Plasmodium vivax malaria is widely distributed throughout the world and accounts for over half of all malaria infections outside of Africa and about 10% of malaria infections in Africa (15). The intraerythrocytic stage of the parasite in humans requires specific receptor-ligand interactions for successful invasion of its host (6). P. vivax invasion of erythrocytes is absolutely dependent on binding to the Duffy blood group. As a result, Duffy-negative individuals are completely resistant to P. vivax malaria (19). One member of a large family of erythrocyte binding proteins, referred to as the Duffy binding protein (DBP), mediates binding to the Duffy blood group or Duffy receptor for chemokines (DARC), since this Duffy blood group antigen has been identified to be a chemokine receptor (14). The critical binding motif of P. vivax DBP lies within a cysteine-rich domain referred to as region II between amino acids (aa) 291 and 460 (21). Region II of DBP (DBPII) may be a critical target for host protective immunity, based on several observations. First, certain regions of DBPII are highly polymorphic (24, 25) and appear to be maintained by immune selection (11). Second, antibodies to DBPII from populations in areas endemic for P. vivax inhibit binding of COS-7 cells that express DBPII ligand on their surface to DARC-positive erythrocytes (16, 23). Third, antibodies raised to region II of the Plasmodium knowlesi α protein, a molecule that is 70% homologous to P. vivax DBP and also mediates DARC-dependent infection of human erythrocytes, can inhibit P. knowlesi invasion of human erythrocytes (22).
Residents of areas endemic for P. vivax develop progressively stronger humoral and cellular immune responses with increasing age (13, 16, 18, 26), and dominant T-cell epitopes within the critical binding motif of DBPII have been identified (26). The relationship of these antibody responses in the context of concurrent P. vivax infection has not been evaluated in detail, and which regions of DBPII are preferentially recognized by antibodies has not been assessed. The present study aims to relate antibodies to DBP with age and infection in a population in an area of Papua New Guinea (PNG) endemic for P. vivax to identify linear B-cell epitopes within the critical binding motif of DBPII, to determine whether they correspond to polymorphic regions in the molecule, and to determine if variants are differentially recognized by sera from partially immune subjects.
MATERIALS AND METHODS
Study site and population.
Study subjects resided in three adjacent villages collectively referred to as Liksul, located 50 km north of Madang, PNG, directly across from Kar Kar Island. Residents belong to the Bargam ethnic and language group (http://www.sil.org/ethnologue/countries/Papu.html). All four human malaria species are transmitted in the area, and there is little seasonal variation in parasitemia rates (5). Residents are estimated to receive one infective bite approximately every other day, with the highest transmission during the wet season from October to May (4). Serum and whole blood samples were obtained from a cross-sectional survey of the entire village (n = 1,025), corresponding to >93% of the population in February 2000, and was immediately stored at −70°C. The Institutional Review Boards at Case Western Reserve University and the Papua New Guinea Institute of Medical Research approved the study.
DNA preparation and PCR amplification of genes encoding P. vivax DBPII.
DNA was extracted from 200 μl of whole blood samples individually by using spin blood kits (Qiagen Inc., Valencia, Calif.) according to the manufacturer's protocol. The final extract was eluted with 200 μl of deionized distilled water and stored at −20°C. Region II (aa 285 to 521) of the P. vivax DBP was amplified by nested PCR with primers complementary to conserved regions of this gene. Nest I forward and reverse primers were 5′- GATAAAACTGGGGAGGAAAAAGAT and 5′-CTTATCGGATTTGAATTGGTGGC, respectively. The nest I reaction (25-μl reaction volume) was carried out using 1.0 μl of template, 1.5 mM MgCl2, a 100 nM concentration of each deoxynucleotide triphosphate, 5 pmol of each primer, and 0.5 U of Taq polymerase (Life Technologies Inc., Rockville, Md.) in the supplied buffer. The nest I cycling conditions were as follows: initial denaturation of 2 min at 94°C; five cycles of 1 min at 94°C, 2 min at 59°C, and 2 min at 72°C; thirty cycles of 1 min at 94°C, 1 min at 54°C, and 2 min at 72°C; and a final extension of 10 min at 72°C. One microliter of the nest I reaction mixture was used in the nest II reaction mixture as template. Nest II forward and reverse primers were 5′-GATCGAAGATATCAATTATGTA and 5′-TATCATAAGGAGTTACGATAC, respectively. The reaction and cycling conditions for the nest II reaction (50-μl reaction volume) were similar to the nest I conditions, except that 53°C was used as the annealing temperature in the first 5 cycles and 48°C was used as the annealing temperature in the last 25 cycles. Three microliters of the nest II 712-bp amplicons were visualized by electrophoresis on a 1% agarose gel in 1× Tris-acetate-EDTA (TAE) buffer with 0.5 μg of ethidium bromide/μl.
DNA sequencing.
Ten microliters of the nest II 712-bp amplicons were visualized by electrophoresis on a 1% agarose gel with 0.5 μg of ethidium bromide/μl in 1× TAE buffer. The PCR products were purified from the agarose using a QiaQuick gel extraction kit (Qiagen Inc.) according to the manufacturer's protocol. The products were eluted with 30 μl of the supplied elution buffer and cloned directly into the pCR2.1-TOPO cloning vector by using a TOPO-TA cloning kit (Invitrogen, La Jolla, Calif.). Seven to 10 cloned species-specific inserts for each sample were sequenced from a Qiagen prepared template (Qiagen Inc.) by using vector-based extended M13 and forward and reverse primers. DNA sequencing was performed at Case Western Reserve University, Cleveland, Ohio, by fluorescence-based methodologies, using an Applied Biosystems 377 automated DNA sequencer.
Restriction fragment length polymorphism (RFLP) analysis.
A restriction site for AflII was identified within the amplified region of DBPII that cut depending on whether or not a mutation was present at codon 333. AflII recognized and cut the sequence in codon 333 (CTT [leucine]), producing 145- and 567-bp fragments; AflII did not cut when the single nucleotide polymorphism altered the codon encoding phenylalanine (TTT), since the restriction site was altered. Restriction digests were performed using 3 to 10 μl of dbpII amplicons generated from nested PCRs (as described above) and 0.5 U of restriction enzyme. The amplicons were digested for 2 h at 37°C and then visualized by gel electrophoresis on a 1-to-2.5% agarose gel with 0.5 μg of ethidium bromide/μl in 1× TAE buffer. If there was any doubt about the quality of a restriction digest, it was repeated or not included in the analysis.
Recombinant antigens and peptide.
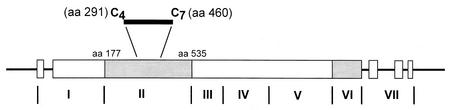
Expression and purification of recombinant DBP (rDBP) were described previously (13). Briefly, a portion of DBP (SalI; GenBank accession number M37514) from aa 177 to 815 that includes regions II to IV DBPII-IV was inserted in frame with glutathione S-transferase (GST) in the plasmid expression vector pGEX-2T (13). Figure 1 shows the structure of P. vivax DBP and the binding motif to DARC (aa 291 to 460), the region mapped for linear B-cell epitopes. To isolate DBPII-IV from the GST component, the expressed protein was cleaved with thrombin. The GST was then removed from the mixture with reduced glutathione agarose CL-4B beads (Fluka, Buchs, Switzerland). Polyclonal rabbit sera raised to rDBPII-IV bound to P. vivax schizonts could inhibit binding of COS-7 cells transfected with rDBP protein to DARC-positive erythrocytes (16). rDBPII was expressed in Escherichia coli, purified from inclusion bodies, refolded, and purified as described earlier. Refolded DBPII was shown to be conformationally correct based on its ability to bind Duffy-positive erythrocytes (23).
FIG. 1.
The molecular structure of the DBP shows that regions II and VI (dark shaded areas) are cysteine-rich regions of the molecule. There are 12 cysteines in region II. The region between cysteine 4 and cysteine 7 (aa 291 to 460) contains the critical binding motif to DARC (7, 8, 21).
A pin method of peptide synthesis (Chiron Mimotopes, Victoria Australia, [www.mimotopes.com]) generated 15-mer peptides displaced by 2 to 5 aa, for a total 19 peptides spanning the 170-aa critical binding region of DBPII. Peptide purity was >75% as determined by high-performance liquid chromatography (HPLC). The sequences were based on the most common alleles identified in the population (11). Additional peptides were synthesized that corresponded to the different variants for peptides 5 and 16 by Fmoc synthesis (GenoSys) (>85% purity by HPLC).
Measurement of antibody titers to rDBP.
Immulon 4 plates (Dynatech Laboratories, Sterling, Va.) were coated with 2 μg of rDBPII-IV or rDBPII per ml in phosphate-buffered saline (PBS; pH 7.4) overnight at 4°C and blocked with 1% bovine serum albumin (BSA) in PBS (blocking buffer). Antigen-coated wells were incubated overnight at 4°C with 100 μl of a 1:400 dilution of serum sample in blocking buffer/well. Plates were washed with PBS and 0.05% Tween 20 (wash buffer), followed by the addition of 100 μl of alkaline phosphatase-labeled goat-anti-human IgG (Jackson ImmunoResearch) at 0.5 μg/ml that was incubated for 1.5 h at room temperature and then washed three times prior to development with p-nitrophenyl phosphate as the substrate. The reaction was terminated by addition of 5% EDTA, and the optical density at 405 nm (OD405) in each well was recorded using an enzyme-linked immunosorbent assay (ELISA) microplate reader (Molecular Devices). For all test samples, both positive and negative control samples were added to each plate, and the reaction was stopped such that ODs varied less than 10% between test plates.
Antibody responses to peptides.
Peptides were adhered to modified polystyrene plates (Immulon-I microtiter plates; Dynatech Laboratories) based on a technique described by Ball et al. (1). Plates were first coated with poly-l-lysine (38 kDa; Sigma Scientific, St. Louis, Mo.) at 50 μg/ml in 0.05 M sodium bicarbonate buffer (pH 9.6). Plates were then washed with PBS after 1 h of incubation at room temperature, and 1% (vol/vol) glutaraldehyde (Sigma) was added for 15 min at room temperature. Plates were again washed in PBS, and test peptides were added in PBS at a concentration of 10 μg/ml and incubated overnight at room temperature. After washing twice with PBS, any remaining reactive aldehyde sites were blocked by addition of 1 M glycine for 1 h at room temperature and further blocked with 5% (wt/vol) dry milk and 1% (wt/vol) gelatin (Gibco, Detroit, Mich.) for 1 h. Following washing, test plasma was added at a 1:10 dilution, incubated overnight at 4°C, and again washed, and alkaline phosphatase-labeled goat-anti-human IgG (Jackson ImmunoResearch) was added at 0.5 μg/ml. The remaining steps of the ELISA were identical to those described for rDBP (see above). Positive and negative control samples were added to each plate.
Affinity purification of peptide-specific antibodies was undertaken to determine if they could bind to correctly folded rDBPII
Peptides were coupled to immobilized diaminodipropylamine (DADPA) on an agarose matrix using the linker 1-ethyl-3-(3-dimethylaminoprophyl) carbodiimide (EDC) by forming amide bonds between carboxyl groups on the linker and amines of the amino acid, according the manufacturer's instructions (EDC/DADPA immobilization kit; Pierce, Rockford, Ill.). One milliliter of pooled immune plasma (n = 3) containing high antibody titers to peptide 5 was added to the column and allowed to equilibrate for 1 h at room temperature after adding an additional 0.2 ml of PBS. The column was washed with 14 ml of PBS, and bound antibody was eluted with 0.1 M glycine at pH 3.0 in 1-ml fractions neutralized by the addition of 50 μl of 1 M Tris, pH 9.5. The absorbance of the fractions at 280 nm was determined to identify the protein peak. Fractions containing antibody were pooled and dialyzed against PBS prior to evaluating them for antibody levels.
Statistics.
Statistical significance between means was compared using Student's t test with log-transformed values. Simple linear regression analysis of log-transformed data examined the correlation between variables and was verified by the Wilcoxon rank sum test.
RESULTS
Infection status of the study subjects.
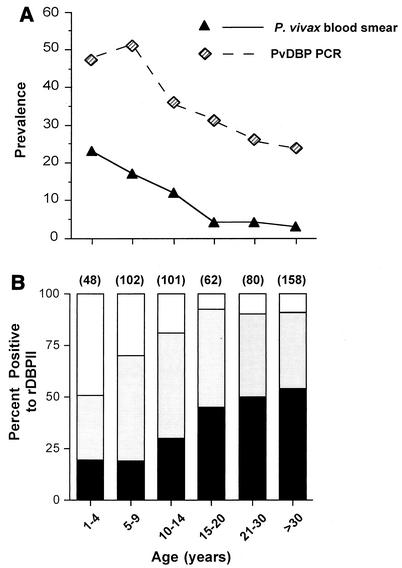
Overall, 91 of 1,025 (9%) residents in the study village were blood smear positive for P. vivax, and 358 (35%) were infected with P. vivax as measured by nested PCR for dbpII. Children ages 1 to 4 years had the highest prevalence of infection as determined by blood smear. The prevalence of infection peaked at ages 5 to 9 years when infection was determined by PCR (Fig. 2A). Two- to sevenfold more individuals, especially among adults, were infected with P. vivax as assessed by PCR compared to that determined by blood smear, indicating the presence of many infections below the level of detection by microscopy. Ninety-three percent of samples that were positive by blood smear for P. vivax were also PCR positive.
FIG. 2.
(A) The relationship of age and P. vivax prevalence as determined by PCR for P. vivax dbp or by blood smear for the whole study population (n = 1,025). (B) Antibody responses to rDBPII were measured in 551 of the subjects from the same samples used to measure parasite infection, and the levels are plotted with respect to age. Dark regions represent ODs of >0.51 (the highest tercile or third of antibody responses), shaded regions represent ODs of 0.178 to 0.5, and the open box shows the absence of a significant antibody responses (OD < 0.178, the mean + 3 SD of control plasma; n = 11). The number of individuals examined in each group is indicated in parentheses above each bar.
Age-specific antibody responses to rDBP are inversely related to P. vivax prevalence.
Antibody levels to rDBPII were examined in 551 subjects (54% of the population) selected to represent all age groups (Fig. 2B). The proportion of subjects that showed any significant antibody reactivity to rDBPII (greater than mean + 3 standard deviations [SD] of adult controls subjects never exposed to P. vivax) or high reactivity, defined as antibody levels corresponding to the highest third or tercile of antibody levels, increased with age until it reached a plateau at ages of 15 years and older. Antibody reactivity to rDBPII was inversely related to P. vivax prevalence in the population (r2 = −0.41; P < 0.0001; Wilcoxon rank sum test).
Since all previous reports of naturally occurring antibodies to DBP in populations in endemic areas used a different construct, rDBPII-IV (13, 16, 18), antibodies to this recombinant molecule were measured in the same samples. The levels as estimated by OD were significantly correlated (r2 = 0.23; P < 0.001; Wilcoxon rank sum test) for the two recombinant antigens. The age-specific levels of antibody reactivity to the two proteins were also similar, and the age-specific prevalence of high antibody levels to rDBPII was statistically equivalent to that of antibody levels directed to rDBPII-IV (P = 0.3; chi-square test).
There was no difference in rDBPII-IV or rDBPII titers between individuals with concurrent P. vivax or Plasmodium falciparum infection compared to uninfected individuals when adjusted for age (data not shown).
Determination of linear B-cell epitopes.
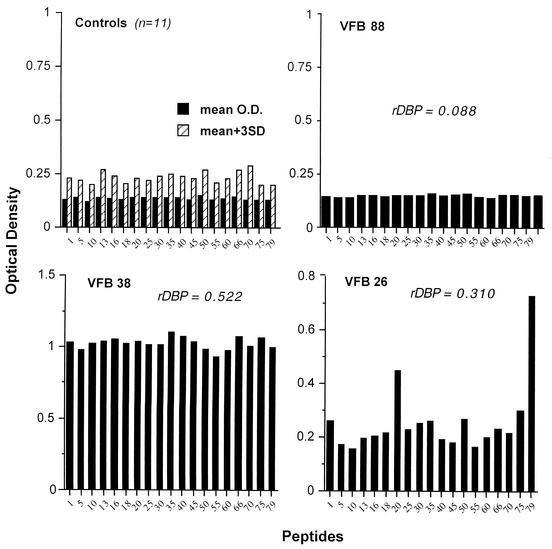
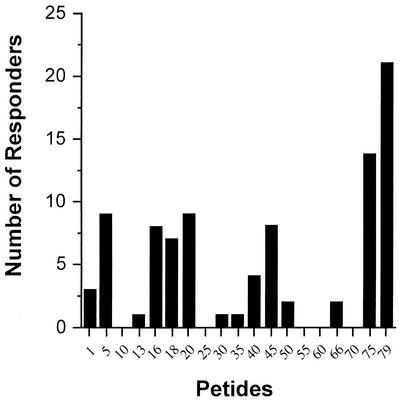
In order to evaluate the presence of linear epitopes spanning the 170-aa binding motif of DBPII (aa 291 to 460), 15-mer synthetic peptides, overlapping by 5 aa, were adhered to poly-l-lysine-coated plates to assure that all peptides used were bound in similar quantities. Some peptides examined were offset by only 2 or 3 aa (e.g., peptides 13, 16, 18, and 20), since these peptides had been previously identified to contain T-cell epitopes (26). Peptide-specific antibody responses were examined in plasma from 11 North Americans never exposed to malaria and a subset of 69 subjects from the endemic villages. This subset of individuals was a stratified random sample of the whole population, to represent individuals in all age groups. Figure 3 shows antibody reactivity for all North Americans (upper left panel) and three representative individuals to demonstrate the different patterns of peptide-specific responses. Overall, seven P. vivax-exposed subjects had no peptide-specific antibody responses, e.g., levels for all peptides were equivalent to those of North American controls (e.g., VFB 88). Twenty-six individuals had OD readings that exceeded controls, but the antibody responses were equivalent among the peptides (e.g., VFB 38), and the remaining subjects (n = 36) showed preferential binding to one or more peptides (e.g., VFB 26). Peptides 5, 16, 18, 20, 45, and 75 and 79 (Fig. 4) were preferentially recognized by plasma in the latter group of 36 individuals, indicating the presence of dominant linear B-cell epitopes.
FIG. 3.
Plasma from individuals residing in areas endemic for P. vivax recognizes linear epitopes. Nineteen 15-mer peptides overlapping by 2 to 5 aa spanning the 170-aa critical binding motif of region II of DBP were used to examine antibody reactivity from individual plasma, as shown on the x axis. The upper left panel shows the mean values (dark bars) + 3 SD (hatched bars) for control plasma (n = 11) for each peptide. The remaining panels show representative antibody responses to all 19 peptides in three representative individuals. The OD of the same serum sample directed to rDBPII-IV is shown for the three study subjects.
FIG. 4.
The cumulative number of individuals showing selective antibody reactivity to 15-mer peptides spanning the 170-aa binding motif of DBPII. Some serum samples recognized two or more epitopes. An individual was considered to preferentially recognize one peptide over others when the OD was greater than the mean + 3 SD of eight peptides with the lowest OD. This differed from criteria used to define a positive response to any peptide shown in Fig. 3.
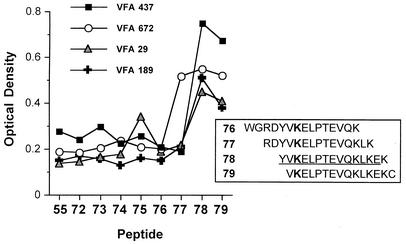
Finer mapping of the B-cell epitope located within peptides 75 and/or 79 is shown in Fig. 5. Plasma from individuals containing antibodies that preferentially bound peptide 79 demonstrated that the probable linear epitope in this region of the molecule is fully contained in peptide 78.
FIG. 5.
Fine B-cell epitope mapping of the C-terminal region of the DBPII erythrocyte adhesion motif, using plasma from four subjects containing antibodies that recognized peptides 75 and/or 79. 15-mer peptides (offset by 1 or 2 aa) were bound to polyvinyl plates, which are described in detail in Materials and Methods.
Antibody responses to peptides correlate with that observed for rDBP.
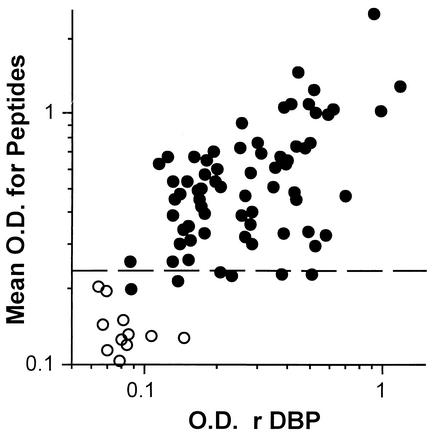
The mean peptide-specific OD (for all peptides) correlated with plasma antibody levels to rDBPII (r2 = 0.47; P < 0.001) (Fig. 6) and rDBPII-IV (r2 = 0.51; P < 0.001) (data not shown). Plasma from some individuals had antibodies that recognized rDBP but had little reactivity to peptides. Antibodies from these individuals may recognize conformational and not linear epitopes. To examine this possibility, a pool of human serum obtained from expatriates that had become acutely infected with P. vivax (kindly provided by W.E. Collins, Centers for Disease Control and Prevention, Atlanta, Ga.) was analyzed against rDBP and the 19 test peptides. The pooled serum recognized rDBPII-IV (OD = 0.507) and none of the peptides (OD values of <0.159). Similarly, acute-phase serum collected from a North American resident infected with P. vivax for the first time with a 2-week history of symptoms and peripheral blood smear of 0.07% parasitemia reacted to rDBPII-IV (OD = 0.612) and failed to react to any peptides. When rDBPII-IV was run on denaturing gel containing 5 mM dithiothreitol (to break disulfide bonds) and probed with the same antibody pool from acutely infected subjects (Western blot analysis), no reactivity to the antigen was observed (data not shown). Overall, these observations demonstrate that acute, primary P. vivax infection recognizes conformational epitopes of DBPII.
FIG. 6.
Relationship between mean peptide-specific antibody levels with that to rDBPII in residents of a village endemic for P. vivax in PNG. Peptide-specific antibody levels were estimated by taking the mean OD to all 19 peptides. Antibody levels to rDBPII were determined as described in Materials and Methods at 1:400. Each point represents plasma from a single individual (n = 69; closed circles) and controls (n = 11; open circles). The dashed line indicates a positive peptide-specific antibody response (e.g., >0.242, which is the mean + 3 SD for all peptides in controls, as shown in Fig. 3). Plasma from some individuals had antibodies that recognized rDBP but little reactivity to peptides (spots at or below dashed line).
The number of linear epitopes recognized and the peptide-specific antibody levels increase with age.
The observation that acute-phase serum from initial P. vivax infection recognized only conformational epitopes suggests that as individuals become repeatedly infected, more antibodies recognize linear epitopes. To examine this possibility, the number of linear epitopes recognized by semiimmune sera was examined with respect to age. Individuals that recognized >4 linear epitopes were significantly older (median, 31 years; 95% confidence interval [CI], 24 to 38 years) than individuals that recognized 1 to 4 linear epitopes (median, 12 years; CI, 9 to 16 years) or no linear epitopes (median, 4 years; CI, 0 to 8 years). Of note, five of seven subjects that failed to recognize any peptide had significant antibody responses to rDBPII-IV. Four out of the five subjects that recognized rDBPII-IV but no peptides were aged 5 years or less.
Mean peptide-specific antibody increased with age (Table 1); however, this age-related increase in antibody levels was delayed compared to that observed with rDBPII (Fig. 2), consistent with a delay in developing antibody responses to linear epitopes. This age-related change in antibody responses was observed for three of the four dominant linear B-cell epitopes (peptides 5, 16, and 45). By contrast, peptides that did not show any preferential recognition, such as peptides 25 and 55 (Fig. 4), had overall lower optical density levels and did not show significant changes with age. Antibody reactivity to peptide 78 was higher than that observed to the other peptides and was also equivalent among the different age groups.
TABLE 1.
Age-related changes in peptide-specific antibody responses
| Age range (years) | n | Response (OD) to peptidea
|
||||||
|---|---|---|---|---|---|---|---|---|
| 5 | 16 | 25 | 45 | 55 | 78 | Mean (all peptides) | ||
| 2-9 | 23 | 0.58 ± 0.11 | 0.51 ± 0.10 | 0.38 ± 0.08 | 0.49 ± 0.09 | 0.39 ± 0.13 | 0.75 ± 0.13 | 0.55 ± 0.10 |
| 10-19 | 18 | 0.49 ± 0.05 | 0.46 ± 0.05 | 0.35 ± 0.03 | 0.47 ± 0.06 | 0.32 ± 0.03 | 0.69 ± 0.9 | 0.48 ± 0.05 |
| ≥20 | 28 | 0.74 ± 0.07 | 0.71 ± 0.06 | 0.47 ± 0.09 | 0.67 ± 0.07 | 0.51 ± 0.11 | 0.81 ± 0.07 | 0.93 ± 0.26 |
| Pb | 0.02 | 0.02 | NS | 0.03 | NS | NS | 0.04 | |
Mean ± SEM.
Significance levels, using Student's t test to compare individuals 2 to 19 years old with those ≥20 years of age. NS, not significant.
Peptide-specific antibodies recognize rDBPII..
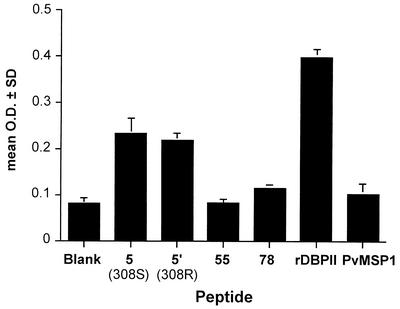
In order to determine whether linear B-cell epitopes to DBPII are accessible to the host immune response, we examined the abilities of peptide-specific antibodies to bind rDBPII. A sample of pooled immune sera (n = 4) containing high titers of antibodies that preferentially recognized peptide 5 was affinity purified on beads linked to peptide 5. Preabsorbed sera also recognized recombinant P. vivax MSP119 (OD = 0.614). After repeated absorption, peptide-specific sera preferentially recognized peptide 5 as well as rDBPII (Fig. 7). The loss of antibody reactivity to P. vivax MSP119 from the column elute demonstrates that nonspecific antibody retention on the column was unlikely. A similar analysis was performed for peptides 16 and 78. Affinity-purified antibody directed to peptide 16 but not 78 bound to rDBPII (data not shown).
FIG. 7.
Affinity-purified antibody to peptide 5 binds correctly folded rDBPII. Individual plasma samples with elevated levels of antibody directed to peptide 5 (S variant, n = 4) were affinity purified as described in Materials and Methods. Eluted antibody did not bind other peptides (55 and 78 are shown as examples) or P. vivax MSP119. Bars represent the mean ± SEM of triplicate wells.
In parallel experiments, individual peptides (5, 16, and 78) were evaluated as to whether they could inhibit antibody binding to the same high-titer sera prior to affinity purification. This would evaluate the relative contribution of the specific peptide to recognition of rDBPII. At an optimal concentration of 100 μg/ml, peptide 5 inhibited antibody binding by 26% ± 5% (mean ± standard error of the mean [SEM]; P = 0.02; n = 4) and peptide 16 by 17% ± 6% (P = 0.05; n = 4). No significant inhibition was observed with peptide 78.
Polymorphisms in linear epitopes alter antibody reactivity.
Polymorphisms in DBPII may have arisen from immune selection by altering the host immune response to these variants. To examine this hypothesis, we first examined whether polymorphisms occurred in the dbpII gene, encoding identified major linear B-cell epitopes based on sequence and RFLP analysis of isolates, from the same population (11). All five major linear B-cell epitopes contained polymorphic residues (Table 2). Peptides 5 and 78 contained one polymorphic residue corresponding to two alleles identified in the study population. Peptides 16 and 20 contained polymorphisms at codon 333, a leucine (L) to phenylalanine (F) at positions 13 (peptide 16) and position 5 (peptide 20). Peptide 45 had three polymorphic residues corresponding to codons 384, 386, and 390. Condon 386 had four alleles in the population.
TABLE 2.
Major linear B-cell epitopes in the 170-aa binding motif of DBPII
| Peptide | Sequencea |
|---|---|
| 5 | VNNTDTNFH(S/R)DITFR |
| 16 | LIYDAAVEGDLL(L/F)KL |
| 20 | GDLL(L/F)KLNNYRYNKD |
| 45 | NISGT(G/D)E(K/N/Q/T)AQQ(H/R)RKQ |
| 78 | YV(K/R)ELPTEVQKLKEK |
The amino terminus is printed on the left. Polymorphic residues are indicated in bold, and the different alleles identified in the study population are indicated in parentheses. For epitope mapping, synthetic peptide 5 contained a serine (S) in position 10 (codon 308), and for peptides 16 and 20 a leucine (L) in positions 13 and 5, respectively, was incorporated for codon 333. Similarly, synthetic peptide 45 had glycine (G) in position 6 (codon 384), a lysine (K) in position 8 (codon 386), and a histidine (H) in position 12 (codon 390); for peptide 78, a lysine (K) was used in position (codon 447).
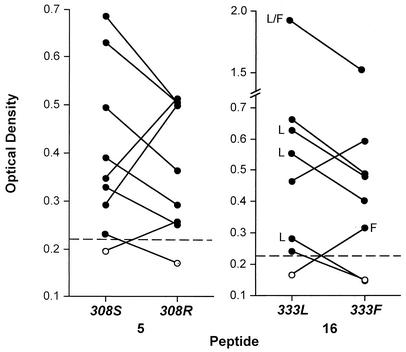
Three epitopes were selected to examine variant-specific binding in greater detail, peptides 5, 16, and 78. The former two peptides also contained T-cell epitopes (26). The locus encoding the polymorphic residue (447) in peptide 78 appears to be under balancing selection (11). We have previously reported differential antibody binding to peptides corresponding to the two different alleles for the linear epitope in peptide 78 (11). To examine whether anti-DBP differentially recognized the two alleles for the linear epitopes in peptides 5 and 16, synthetic peptides were produced corresponding to each allele, as shown in Table 2. Sixty-three of 74 (85%) sera tested responded to peptide 5 (e.g., greater than mean + 3 SD of control sera) (Fig. 3), 56 of which reacted to both peptide variants and 7 which tested positive to one variant but not the other. To evaluate whether some individuals demonstrated different levels of reactivity to each allele, all 74 sera were tested in duplicate to each peptide variant, and the percent difference in OD was calculated for the replicate samples to the same peptide (mean ± SD, 7.0% ± 5.2%) as a control to assess the precision of the ELISA. If the percent difference between the mean OD of the two peptide variants exceeded the mean + 3 SD of the replicate samples [same variant; e.g., (5.2% × 3) + 7% = 22.6%], this was considered a significant difference in levels of serologic reactivity between the two variants. Nine individuals fulfilled this criterion, as shown in Fig. 8 (left panel, S308R).
FIG. 8.
Peptides containing the 308 and 333 residues form linear B-cell epitopes in which substitution by a serine (S) for an arginine (R; 308) or a leucine (L) for a phenylalanine (F; 333) is differentially recognized by antibodies in the same subject (samples connected by lines). Values represent mean ODs of duplicate measurements of serum samples at a 1:10 dilution unless otherwise noted. SD were all <8% of the mean. All OD differences were highly significant based on criterion described in the text. The dashed line represents cutoff values for a positive response, e.g., mean + 3 SD of the unexposed control (see Fig. 3). Only two individuals are the same between the two panels.
A similar analysis was performed for peptide 16. Sixty-two of 74 (84%) sera tested reacted to peptide 16 (e.g., greater than mean + 3 SD of control sera), 58 of these reacted to both peptide variants (Table 2), and 4 tested positive to one variant but not the other. Of the 12 subjects that tested negative to peptide 16, 5 of these subjects reacted to peptide 5. Using the same analysis for peptide 16 as described for peptide 5, eight individuals demonstrated different levels of reactivity between the two variants (Fig. 8, right panel). There was no relationship between age and probability of showing a different response to each variant.
The frequencies of the different S308R and L333F alleles were examined in the study population at the same time serum samples were collected. The frequency of the 333L allele was 0.95, and the frequency of 333F was 0.27 (n = 358) based on RFLP using the restriction enzyme AflII. Since a specific restriction enzyme could not be identified that recognized the different alleles for S308R, direct sequencing of 32 P. vivax isolates for region II of the dbp gene was performed; 308S had a frequency of 0.72 and 308R had a frequency of 0.38 (11). Six of nine (67%) subjects had higher titers to the more common allele for 308 (S), and six of eight (75%) had higher titers for 333 (Fig. 8). Of note, five of eight individuals that demonstrated allele-specific antibodies to peptide 16 were simultaneously infected with P. vivax (Fig. 8, right panel). One sample was infected with at least two parasite strains expressing both alleles. The other four samples had higher antibody titers corresponding to simultaneous infection with parasite strains that expressed the same alleles.
DISCUSSION
Host immune responses to P. vivax DBP are likely to play a key role in acquired immunity to P. vivax because of its essential role for merozoite invasion of erythrocytes. This acquired immunity is likely to result, in part, from generation of antibodies directed to DBP that block its binding to its receptor on red cells. In support of this model, we observed an increase in the proportion and titers of antibodies that recognized rDBP with advancing age, concomitant with a decline in the prevalence and intensity of P. vivax infection. In order to better define potentially protective epitopes in the critical binding motif of DBPII, we identified at least four dominant linear epitopes, two of which corresponded to previously described T-cell epitopes (26). The cumulative antibody levels to these linear epitopes correlated closely to antibody recognition of correctly folded rDBPII (23). This suggests that antibodies directed to some of these linear epitopes also recognize native DBP expressed on the merozoite surface.
Antibody responses to these linear epitopes do not appear to develop with initial vivax malaria infection. Residents of nonendemic areas of malaria who have become infected for the first time with P. vivax develop antibody responses only to the recombinant DBP and not to linear epitopes. This antibody response is lost when rDBP is denatured, indicating initial antibody responses develop to conformational epitopes. A similar pattern occurs in populations in areas endemic for P. vivax in which some children have antibodies that recognize rDBP but not peptides. Most residents of populations in areas endemic for P. vivax, however, recognize linear epitopes of DBP, and the number of epitopes recognized increases with age and, presumably, cumulative exposure. Since a single P. vivax infection does not induce protective immunity (10), these initial antibody responses to immunodominant conformational epitopes may not confer resistance. Repeated P. vivax infection, however, probably results in the release of significant amounts DBP, and along with its partial degradation it stimulates antibody responses to linear epitopes. Partial protection may develop when sufficient antibody titers of high enough affinity are induced to multiple surface-expressed determinants of DBP that contain linear epitopes. This is consistent with early observations of experimental P. vivax infection in humans that at least three to four infections of the same parasite strain were required to achieve protection against disease and a significant reduction in parasitemia (2, 3, 9).
Antibodies that recognize some dominant linear epitopes also bind properly refolded rDBPII. This binding could be partially inhibited by addition of peptides containing these linear epitopes. We also found affinity purification of antibody from immune sera directed to one dominant linear epitope in peptide 5, which also corresponds to a T-cell epitope (26), did not cross-react with other peptides and recognized rDBPII (Fig. 7B). Taken together, these experiments indicate that antibodies to some of these linear epitopes are accessible on the native molecule to naturally acquired antibodies that could block binding of DBP to its receptor, DARC. This was not the case for a linear epitope in peptide 78. It is possible that peptide 78 could be a neodeterminant, produced only with degradation of DBP and not accessible on the native molecule, or it could be part of a conformational epitope. We favor this latter explanation, since a polymorphic residue at codon 447 (position 3 in the linear epitope of peptide 78) occurs on the same haplotype with a polymorphic residue at position 390, both of which are under age-dependent selection (11). This latter residue is also found in peptide 45, which also contains a linear epitope. A combination of both peptides 45 and 78 partially blocked antibody binding (personal observations). To establish that these linear B-cell epitopes are biologically significant, it needs to be demonstrated that these antibodies recognize native DBP in P. vivax schizonts and that these peptide-specific antibodies can inhibit DBPII binding to DARC-positive erythrocytes. These experiments are currently in progress.
Region II of P. vivax DBP, like other parasite molecules important for erythrocyte invasion, such as the MSP119 domain of P. vivax and P. falciparum MSP1, is cysteine rich and, as a consequence, is poorly processed and therefore poorly immunogenic due to its cysteine content (12, 20). This may account for the reduced frequency of antibody and T-cell responses (26) in young children even though many were infected (Fig. 2).
The dominant linear epitopes identified contained polymorphic residues, and approximately 10 to 15% of partially immune individuals had antibodies that differentially recognized peptides corresponding to the two different alleles (Fig. 8). This indicates that these polymorphic residues can affect antibody binding, suggesting that they arose by immune selection. Most individuals, however, recognized peptides corresponding to both alleles. This is not unexpected, since both strains exist in the population and individuals are likely to have been exposed to both. Indeed 18% of P. vivax-positive samples came from people who were infected simultaneously with both strains. Concurrent infection with a particular parasite strain appeared to boost strain-specific immunity, as suggested by the correlation of infection by a particular parasite strain with a corresponding higher peptide-specific antibody response to that strain. Although it would be expected that young individuals with less exposure should be more likely to recognize one allele and not the other, this was not observed. This lack of an association with age may reflect the inability of a cross-sectional analysis to adequately reflect prior exposure to parasites with the same alleles (e.g., at the time of study the 333L allele was much more common than the 333F allele).
No difference in antibody response was observed between individuals with any concurrent P. vivax infection and those without detectable infection. Active malaria infection should boost antibody responses. This lack of association between infection and elevated antibody levels probably occurs because most individuals are consistently parasitemic during a high-transmission time of year, although often below levels of detection. This was suggested by longitudinal studies showing that individuals who were negative at one time point were often found to be parasitemic 1 month later (personal observations). There was also no association between antibody levels and concurrent P. falciparum infection, suggesting that falciparum infection does not suppress (or boost) the short-term antibody response to P. vivax DBP. Although we found no concordance in antibody responses with rDBPII and region II of EBA-175, the P. falciparum homologue to P. vivax DBPII (17), in the same individual or antibody reactivity to rDBP from sera of Duffy-negative P. falciparum-infected subjects from Kenya (personal observations), it is possible that some cross-reactivity may occur between P. falciparum and P. vivax linear epitopes. These experiments are currently in progress.
In conclusion, the present study demonstrates that immune responses in an endemic population develop to both linear and conformational B-cell epitopes in the critical binding region of DBPII. The challenge will be to determine whether protective epitopes can be identified which transcend this diversity such that a recombinant vaccine can provide protection to diverse parasite strains.
Acknowledgments
The Department of Veteran's Affairs Medical Service supported this work.
We appreciate the cooperation and curiosity of the residents of Liksul and neighboring villages for helping us to perform these studies and for the support and encouragement of John Reeder.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Ball, J. M., N. L. Henry, R. C. Montelaro, and M. J. Newman. 1994. A versatile synthetic peptide-based ELISA for identifying antibody epitopes. J. Immunol. Methods 171:37-44. [DOI] [PubMed] [Google Scholar]
- 2.Boyd, M. F. 1942. Criteria of immunity and susceptibility in naturally induced vivax malaria infections. Am. J. Trop. Med. Hyg. 22:217. [Google Scholar]
- 3.Boyd, M. F., S. Stratman-Thomas, and K. Warren. 1933. Studies on benign terian malaria. I. On the occurrence of acquired tolerance to Plasmodium vivax. Am. J. Trop. Med. 17:55. [Google Scholar]
- 4.Burkot, T. R., P. M. Graves, R. Paru, R. A. Wirtz, and P. F. Heywood. 1988. Human malaria transmission studies in the Anopheles punctulatus complex in Papua New Guinea: sporozoite rates, inoculation rates, and sporozoite densities. Am. J. Trop. Med. Hyg. 39:135-144. [DOI] [PubMed] [Google Scholar]
- 5.Cattani, J. A., J. L. Tulloch, H. Vrbova, D. Jolley, F. D. Gibson, J. S. Moir, P. F. Heywood, M. P. Alpers, A. Stevenson, and R. Clancy. 1986. The epidemiology of malaria in a population surrounding Madang, Papua New Guinea. Am. J. Trop. Med. Hyg. 35:3-15. [DOI] [PubMed] [Google Scholar]
- 6.Chitnis, C. E., and M. J. Blackman. 2000. Host cell invasion by malaria parasites. Parasitol. Today 16:411-415. [DOI] [PubMed] [Google Scholar]
- 7.Chitnis, C. E., A. Chaudhuri, R. Horuk, A. O. Pogo, and L. H. Miller. 1996. The domain on the Duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. J. Exp. Med. 184:1531-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chitnis, C. E., and L. H. Miller. 1994. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J. Exp. Med. 180:497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciuca, M., M. Chelarescu, and A. Sofletea. 1955. Studies on immunity in malaria. Ed. Acad. Republique Roumaine, Bucharest 14:61-77. [Google Scholar]
- 10.Clyde, D. F. 1975. Immunization of man against falciparum and vivax malaria by use of attenuated sporozoites. Am. J. Trop. Med. Hyg. 24:397-401. [DOI] [PubMed] [Google Scholar]
- 11.Cole-Tobian, J., A. Cortes, M. Baisor, W. Kastens, J. Xainli, M. Bockarie, J. Adams, and C. L. King. 2002. Age-acquired immunity to a Plasmodium vivax invasion ligand, the duffy binding protein. J. Infect. Dis. 186:531-540. [DOI] [PubMed] [Google Scholar]
- 12.Egan, A., M. Waterfall, M. Pinder, A. Holder, and E. Riley. 1997. Characterization of human T- and B-cell epitopes in the C terminus of Plasmodium falciparum merozoite surface protein 1: evidence for poor T-cell recognition of polypeptides with numerous disulfide bonds. Infect. Immun. 65:3024-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraser, T., P. Michon, J. W. Barnwell, A. R. Noe, F. Al-Yaman, D. C. Kaslow, and J. H. Adams. 1997. Expression and serologic activity of a soluble recombinant Plasmodium vivax Duffy binding protein. Infect. Immun. 65:2772-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horuk, R., C. E. Chitnis, W. C. Darbonne, T. J. Colby, A. Rybicki, T. J. Hadley, and L. H. Miller. 1993. A receptor for the malarial parasite Plasmodium vivax: the erythrocyte chemokine receptor. Science 261:1182-1184. [DOI] [PubMed] [Google Scholar]
- 15.Mendis, K., B. J. Sina, P. Marchesini, and R. Carter. 2001. The neglected burden of Plasmodium vivax malaria. Am. J. Trop. Med. Hyg. 64:97-106. [DOI] [PubMed] [Google Scholar]
- 16.Michon, P., T. Fraser, and J. H. Adams. 2000. Naturally acquired and vaccine-elicited antibodies block erythrocyte cytoadherence of the Plasmodium vivax Duffy binding protein. Infect. Immun. 68:3164-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michon, P., J. R. Stevens, O. Kaneko, and J. H. Adams. 2002. Evolutionary relationships of conserved cysteine-rich motifs in adhesive molecules of malaria parasites. Mol. Biol. Evol. 19:1128-1142. [DOI] [PubMed] [Google Scholar]
- 18.Michon, P. A., M. Arevalo-Herrera, T. Fraser, S. Herrera, and J. H. Adams. 1998. Serologic responses to recombinant Plasmodium vivax Duffy binding protein in a Colombian village. Am. J. Trop. Med. Hyg. 59:597-599. [DOI] [PubMed] [Google Scholar]
- 19.Miller, L. H., S. J. Mason, D. F. Clyde, and M. H. McGinniss. 1976. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group. N. Engl. J. Med. 295:302-304. [DOI] [PubMed] [Google Scholar]
- 20.Quin, S. J., E. M. Seixas, C. A. Cross, M. Berg, V. Lindo, B. Stockinger, and J. Langhorne. 2001. Low CD4+ T cell responses to the C-terminal region of the malaria merozoite surface protein-1 may be attributed to processing within distinct MHC class II pathways. Eur. J. Immunol. 31:72-81. [DOI] [PubMed] [Google Scholar]
- 21.Ranjan, A., and C. E. Chitnis. 1999. Mapping regions containing binding residues within functional domains of Plasmodium vivax and Plasmodium knowlesi erythrocyte-binding proteins. Proc. Natl. Acad. Sci. USA 96:14067-14072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh, A. P., S. K. Puri, and C. E. Chitnis. 2002. Antibodies raised against receptor-binding domain of Plasmodium knowlesi Duffy binding protein inhibit erythrocyte invasion. Mol. Biochem. Parasitol. 121:21-31. [DOI] [PubMed] [Google Scholar]
- 23.Singh, S., K. Pandey, R. Chattopadhayay, S. S. Yazdani, A. Lynn, A. Bharadwaj, A. Ranjan, and C. Chitnis. 2001. Biochemical, biophysical, and functional characterization of bacterially expressed and refolded receptor binding domain of Plasmodium vivax Duffy-binding protein. J. Biol. Chem. 276:17111-17116. [DOI] [PubMed] [Google Scholar]
- 24.Tsuboi, T., S. H. Kappe, F. Al-Yaman, M. D. Prickett, M. Alpers, and J. H. Adams. 1994. Natural variation within the principal adhesion domain of the Plasmodium vivax duffy binding protein. Infect. Immun. 62:5581-5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xainli, J., J. H. Adams, and C. L. King. 2000. The erythrocyte binding motif of Plasmodium vivax duffy binding protein is highly polymorphic and functionally conserved in isolates from Papua New Guinea. Mol. Biochem. Parasitol. 111:253-260. [DOI] [PubMed] [Google Scholar]
- 26.Xainli, J., M. Baisor, W. Kastens, M. Bockarie, J. H. Adams, and C. L. King. 2002. Age-dependent cellular immune responses to Plasmodium vivax duffy binding protein in humans. J. Immunol. 169:3200-3207. [DOI] [PubMed] [Google Scholar]