Abstract
Surface and secreted proteins of schistosomes orchestrate the basic physiologic requirements of a parasitic existence. These proteins are often exposed to host tissues during penetration, migration, feeding, and immune evasion, and they are obvious targets for control strategies. Signal sequence trap (SST) represents a novel approach that selects for cDNAs encoding secreted and surface proteins with N-terminal signal peptides, so we constructed a randomly primed adult Schistosoma mansoni cDNA library fused to a signalless reporter gene encoding placental alkaline phosphatase. The library was used to transfect COS-7 cells, which were then assayed for the presence of reporter at the cell surface. Eighteen S. mansoni cDNA fragments were isolated and sequenced. Expression profiles of the novel clones were determined for different developmental stages; some transcripts were restricted to single-sex adult worms, while others were ubiquitously distributed. Most clones contained signal peptides or signal anchors as determined by the SignalP algorithm. Open reading frames (ORFs) were categorized as follows: (i) previously identified S. mansoni cDNAs encoding proteins of known function; (ii) cDNAs encoding proteins of known function in other organisms but novel for Schistosoma; (iii) S. mansoni expressed sequence tags (ESTs) of unknown function; and (iv) completely novel ORFs without homologues (including ESTs) from any phylum. Clones of particular interest included tetraspanins similar to human cell surface antigens, a protein kinase, and ORFs transcribed in the antisense orientation to previously characterized S. mansoni cDNAs. This is the first report describing the use of SST as a tool for identifying secreted proteins from any pathogenic organism.
Despite significant effort over the past 2 decades, a vaccine to protect against schisotosomiasis is still an elusive goal (22, 26). Current target antigens have generally yielded relatively low levels of protection, and many proteins that showed early promise in murine vaccine trials have proved less efficacious in subsequent trials. There are almost 17,000 Schistosoma mansoni expressed sequence tags (ESTs) in the dbEST database, implying that mRNAs encoding protective antigens have been cloned and sequenced. Moreover, recent efforts at sequencing bacterial artificial chromosome ends have contributed significantly to the pool of genetic information on the parasite. However, many schistosome mRNAs do not share sequence identity with proteins of known function; furthermore, there is a distinct absence of functional screening techniques with which to identify genes of interest from a vaccine development viewpoint.
Vaccine candidate proteins are often secreted by or anchored on the surface of pathogens, and they usually possess N-terminal hydrophobic signal peptides or signal anchors that direct traffic of the protein through the secretory pathway to the cell surface. Proteins that are secreted by or anchored on the surface of intramammalian stages of schistosomes are exposed to host tissues and thus present as potential candidate molecules for the development of new intervention strategies.
Signal peptides are usually 15 to 30 amino acids and consist of a basic N terminus, a hydrophobic center, and a polar C terminus. While they share a similar architecture, high levels of degeneracy make them difficult to identify from primary sequence alone, and they cannot be cloned by degenerative PCR-based methods. Signal sequence trap (SST) is a recently described technique that allows selective cloning of cDNAs that encode open reading frames (ORFs) with an N-terminal signal peptide that directs surface expression of a reporter gene product lacking its endogenous signal peptide. Randomly primed cDNA libraries are ligated into a plasmid vector so that they fuse in-frame with the signalless reporter gene; cDNAs that encode a signal peptide can then restore secretion of the reporter. SST methods have been described with COS cells transfected with a signal peptide-deficient interleukin 6 reporter (29) and Saccharomyces organisms transformed with a signal peptide-deficient invertase reporter (15). Chen and Leder described a simple SST technique that utilized the transmembrane placental alkaline phosphatase (PLAP) protein as a reporter (6). PLAP is heat stable and inactive until transported to the cell surface, and detection of cells expressing surface-derived PLAP (i.e., those containing a cDNA insert with a functional signal peptide) is as simple as staining fixed cells for alkaline phosphatase activity and viewing them with a light microscope.
Given the widespread interest in interactions between host tissues and secreted and surface proteins of pathogens, SST provides an ideal platform for selectively screening parasite transcriptomes for the presence of mRNAs encoding proteins with signal peptides. Indeed, the potential application of SST to parasitology was the focus of a recent review (23), but until now, SST had not been used to clone novel cDNAs from any infectious organism. Here, we employed the PLAP-based peptide signal trap (AP-PST) (6) to identify cDNAs of S. mansoni and showed the utility of this technique in specifically identifying surface and secreted proteins of parasites. We describe the cloning and expression patterns of transcripts that correspond to previously identified schistosome mRNAs, secretory ORFs transcribed antisense to known schistosome mRNAs, ORFs with homology to proteins of known function from other organisms, mRNAs that correspond to S. mansoni ESTs of unknown function, and completely novel mRNAs with no previously identified homologues from any organism. In addition, we obtained full-length cDNA sequence for two tetraspanins that shared sequence identity with host cell surface antigens.
MATERIALS AND METHODS
Parasites.
Adult S. mansoni (Puerto Rican strain; both sexes) worms were perfused from BALB/c mice 7 to 10 weeks after infection with cercariae and washed thoroughly in phosphate-buffered saline (PBS). Schistosome eggs were extracted from mouse livers (harvested from mice infected with the Puerto Rican strain of S. mansoni), as previously described (8). The cercarial stage was collected from infected Biomphalaria glabrata and was kindly provided by Fred Lewis (Biomedical Research Institute, Rockville, Md.)
Cloning and transfection of positive and negative S. mansoni control constructs in the pPST vector.
Before transfecting COS cells with an S. mansoni AP-PST library, we sought to determine whether known schistosome proteins with and without predicted signal peptides (as determined by using the SignalP V2.0 server at http://www.cbs.dtu.dk/services/SignalP-2.0/) could be cloned into the AP-PST vector and direct surface expression of the reporter. CatD (S. mansoni cathepsin D, GenBank accession no. L41346) (2) was predicted to contain an N-terminal signal peptide; schistosome paramyosin (Schistosoma japonicum AF113971) (30) is found on the surface of the tegument (21) but is not predicted to contain a signal peptide and served as a potential negative control. Oligonucleotide primers were designed to amplify the regions encoding the first 60 to 100 amino acids of CatC, CatD, and Pmy fragments, beginning at their translation initiation codons. PCR fragments were directionally cloned into the AP-PST vector (kindly provided by H. Chen and P. Leder, Howard Hughes Medical Institute, Harvard University, Boston, Mass.) by using EcoRI and HindIII restriction enzyme sites incorporated into the primers. AP-PST consists of a modified pcDNA 1.0 backbone with a truncated PLAP sequence (minus signal peptide) next to the multiple cloning site (6). Ligations were electroporated into MC1061/P3 bacterial cells (Invitrogen) and plated onto Luria-Bertani (LB) plates containing ampicillin (25 μg/ml) and tetracycline (10 μg/ml). Colonies were picked and verified for inserts by PCR using the cloning primers, and plasmid mini-preps were prepared from positive colonies with kits (Qiagen). COS cells (106 per 35-mm-diameter well) were transfected with 1.0 μg of each plasmid DNA, as described later.
cDNA library construction.
Construction of a truncated cDNA library from adult S. mansoni and screening and expression assays were performed as described for mouse cDNA (6) with minor modifications. A S. mansoni adult cDNA library was constructed by using a directional random priming strategy (OrientExpress, Novagen). Total RNA from adult worms, eggs, and cercariae was extracted with Trizol (Invitrogen) reagent in accordance with the manufacturer's instructions. Messenger RNA was enriched from total RNA with Ambion Micropoly(A) Pure kit in accordance with the manufacturer's instructions. All RNAs were stored in 0.2 mM EDTA at −80°C. The adult worm AP-PST library was constructed with 4 μg of poly(A)+ RNA. First-strand cDNA for the adult library was synthesized with 5 μg of HindIII-capped random primers in order to produce cDNAs with a smaller average size. Randomly primed cDNAs were purified and size selected by gel electrophoresis and excision of between 300 and 500 bp of DNA with a Qiaquick gel extraction kit (Qiagen). Eluted DNA was cloned directionally into the HindIII- and EcoRI-digested AP-PST vector (6), and the resultant cDNA library, PST-SmA (adult), was electroporated into MC1061/P3 bacterial cells and plated onto LB plates containing ampicillin (25 μg/ml) and tetracycline.
Library screening and staining for PLAP activity.
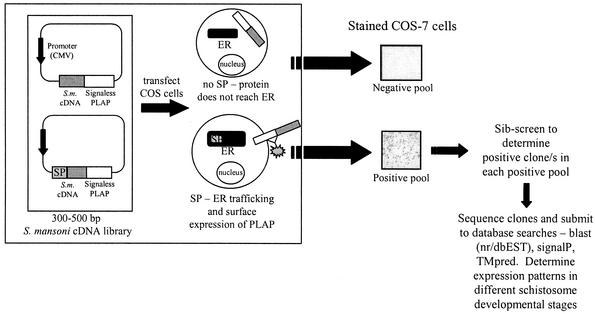
A sib-screen approach was used to screen the library. Fifty-two individual colonies were transferred onto one LB agar plate (containing 40 μg of ampicillin/ml and 10 μg of tetracycline/ml) in a grid format and were designated a pool number. Bacteria from each pool were grown together, and endotoxin-free plasmid DNA was prepared (Qiagen mini preps). One μg of plasmid DNA from each pool was used to transfect 1 × 106 COS-7 cells per 35-mm-diameter well with 6 μl of Genejammer transfection reagent (Stratagene). Cells were incubated at 37°C for 24 h and then fixed in 4% paraformaldehyde (in PBS) for 5 to 10 min. Cells were then washed in three changes of PBS and heated to 65°C for 20 min to inactivate endogenous alkaline phosphatases (note: PLAP is thermostable). Staining was performed by the addition of buffer (100 mM Tris [pH 8.5], 100 mM NaCl, 50 mM MgCl2) containing 1 mg of 4-nitro blue tetrazolium chloride (50× to 75× in 70% dimethylformamide)/ml and 0.1 mg of 5-bromo-4-chloro-3-indolyl-phosphate (100× in 100% dimethylformamide)/ml. Cells were covered in aluminum foil and incubated between 0.5 and 12 h, depending on stain development. Staining was performed at room temperature unless otherwise stated. Cells were then washed in PBS-20 mM EDTA several times and stored in the same buffer at 4°C. When positive pools were detected, pools were divided into 16 subpools and were rescreened in this manner until single positive clones were identified. Pools were judged as positive or negative by comparison to transfections with the positive and negative control plasmids, PST-PLAP and PST, respectively (6). The procedure described here is summarized in Fig. 1.
FIG. 1.
Schematic representation of the AP-PST technique utilized for detection of adult S. mansoni cDNAs encoding ORFs with N-terminal signal peptides. A randomly primed, size-selected S. mansoni cDNA library (adult stage) was constructed in the PST vector (6) and electroporated into MC1061/P3 E. coli. Transformants were patched onto pool plates with grids (52 colonies/plate). DNA from each pool was transfected into COS-7 cells and incubated for 72 h at 37°C. Cells were then fixed and stained for surface alkaline phosphatase activity.
Nucleotide and protein sequence analyses.
The cDNA clones isolated were sequenced from plasmid DNA by using T7 and T3 vector-derived primers and Big Dye sequencing chemistry (ABI) on an ABI 377 automated DNA sequencer. Sequences were compared to those in the GenBank (nonredundant) and dbEST databases with the National Center for Biotechnology Information (NCBI) Blast program. Sequences were also compared to the S. mansoni bacterial artificial chromosome end sequences (http://tigrblast.tigr.org/euk-blast/index.cgi?project=sma1). Signal sequence prediction analysis was performed using the SignalP V2.0 server (http://www.cbs.dtu.dk/services/SignalP-2.0/). SignalP comprises two signal peptide prediction methods, namely, SignalP-NN (based on neural networks and corresponding to SignalP V1.1) and SignalP-HMM (based on hidden Markov models). Internal transmembrane domains were predicted with the TMpred server (http://www.ch.embnet.org/software/TMPRED_form.html).
Expression pattern analysis.
Total RNAs from adult worms, eggs and cercariae were extracted by using Trizol reagent in accordance with the manufacturer's instructions. Single-stranded cDNA was generated by priming 1 to 3 μg of total RNA with 500 ng of oligo(dT)12-18 random primers (Boehringer Mannheim) with Superscript II reverse transcriptase (RT) (Life Technologies). Control reactions (without the addition of RT) were included to monitor for contamination with genomic DNA. Residual RNA was hydrolyzed by the addition of 2 units of Escherichia coli RNase H (Promega) and incubation at 37°C for 20 min. After reverse transcription of the total RNA to single-stranded cDNA, 30 PCR cycles were performed as follows: 94°C for 20 s; 50 to 55°C (depending on primer pairs) for 20 s; and 72°C for 20 s. The sequences of the gene-specific oligonucleotide primers were designed from the 5′ and 3′ ends of each clone and are not reported here but will be made available upon request. Primers targeting a 291-bp region of the S. mansoni gene encoding triosephosphate isomerase were used to amplify the constitutively expressed positive control cDNA (14). RT-PCR amplification products were electrophoresed through 1% agarose gels containing ethidium bromide, and amplicons were viewed with a UV transilluminator.
Rapid amplification of cDNA ends.
Full-length ends of cDNAs were obtained for clones p43F4 and p64A3 by using a GeneRacer kit (Invitrogen). Total RNA from adult worms was extracted as described earlier, and 5 μg was used to make rapid amplification of cDNA ends-ready cDNA, in accordance with the manufacturer's instructions. Sequences were amplified by Touchdown PCR with either the p43F4 or p64A3 gene-specific primers (previously used for RT-PCR analysis) with GeneRacer 3′ or 5′ primers; nested PCRs were performed with nested gene-specific primers and 3′ or 5′ nested GeneRacer primers. Amplification products were electrophoresed through 1% agarose gels containing ethidium bromide, and single bands were gel extracted and either sequenced directly or TA cloned with the TOPO TA cloning kit (Invitrogen). Plasmids were then extracted and inserts were sequenced with vector-derived primers.
Nucleotide sequence accession numbers.
All novel sequences, including those where the 5′ end extends beyond that of the corresponding ESTs, have been deposited in GenBank under the following accession numbers: p17C5, AF521086; p25D5, AF521087; p28C7, AF521088; p30C2, AF521089; p33F5, AF521090; p43F4, AF521091; p43G5, AF521092; p64A3, AF521093; and p83H3, AF521094. Full-length sequences corresponding to the tetraspanin-encoding clones p64A3 (the full-length sequence is called Sm-tsp-1) and p43F4 (Sm-tsp-2) were deposited in GenBank under accession numbers AF521093 and AF521091, respectively.
RESULTS AND DISCUSSION
As schistosomes mature and penetrate and migrate through the tissues of their intermediate and definitive hosts, morphological changes are accompanied by differential mRNA expression within the various stages. Molecules secreted by, and anchored on, the surfaces of parasites or parasite-infected cells are of interest to most researchers in infectious diseases and present as obvious targets for vaccine and chemotherapeutic intervention strategies. However, despite a plethora of nucleotide sequence information, functional screens for identifying secreted proteins and their genes are laborious, often technically demanding, and usually limited to a particular family of proteins that share common biochemical activities or sequence motifs. SST offers the advantage of targeting all proteins with a signal peptide or signal anchor, and moreover, the AP-PST technique (6) is less labor intensive and does not require the use of specialized equipment such as fluorescence-activated cell sorters. Despite being the topic of a recent review (23), this is the first report to our knowledge of the identification of novel mRNAs from any pathogen of animals by using SST.
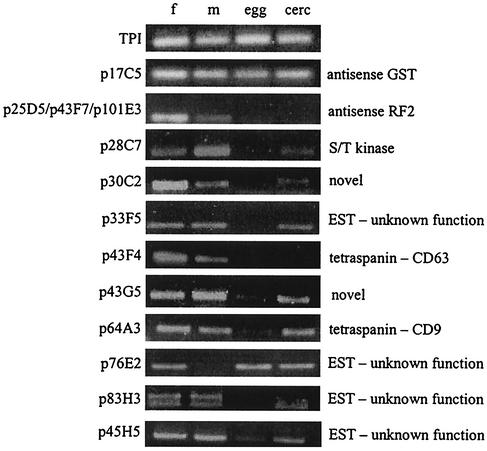
The positive control cDNA encoding the first 100 amino acids of schistosome pre-pro-cathepsin D (2) drove surface expression of the reporter, and PLAP was detected at the cell surface (not shown); the negative control cDNA encoding the first 100 residues of paramyosin did not result in PLAP being detected at the cell surface. The randomly primed S. mansoni cDNA library was then screened, and each clone identified is discussed below in relation to its putative function and expression profile in different developmental stages of S. mansoni. Approximately 5,900 clones of the 300- to 500-bp adult S. mansoni PST-SmA library were screened, and 18 positive clones were obtained. The putative identities and other salient features of each clone are summarized in Table 1. mRNA expression profiles of 10 of the newly described S. mansoni clones were analyzed by RT-PCR and mRNA templates from S. mansoni eggs, cercariae, and adult male and female worms (Fig. 2).
TABLE 1.
S. mansoni cDNA clones isolated from the PST-SmA library by using the alkaline phosphatase peptide signal trap
| Clone(s) | Match(es), % identity, and GenBank accession no. | Kozak sequencee | Putative signal sequenceb | No. of internal transmembrane domains | SignalP-NNc
|
Signal P-HMMd | |||
|---|---|---|---|---|---|---|---|---|---|
| C | S | Y | s | ||||||
| p43F4 | Tetraspanin: human CD63, 39%, and Sm23, 35% | ttaggg_ | MFGACMKNVCLLTTYCILLSILMVAEIAAGIFA IV... | 2 | Y | Y | Y | Y | SP |
| p64A3 | Tetraspanin: bovine CD9, 35%; M81720 | ttcatt_ | MKGCIQCLRVILVVFNFLVVLIGLSVLGFS VY... | 2 | Y | Y | Y | Y | SP |
| p97D2 | S. mansoni eggshell protein EGG2, 100%; M21607 | xxaagc_ | MKQSLTLVFLVAIGYATA HT... | None | Y | Y | Y | Y | SP |
| p25G7, p112E4 | S. mansoni eggshell protein EGG3, 100%; J03982 | tgaaaa_ | MKQSLTLVFLVAIGYATA YT... | None | Y | Y | Y | Y | SP |
| p33F5 | S. mansoni EST,a 100%; AA233973 | cgcatc_ | MKYFICVIITVIIGVALS YS... | None | Y | Y | Y | Y | SP |
| p83H3 | S. mansoni EST,a 100%; BE505109 | tcaatc_ | MNRFFWTVTQCTILLVIICNLNTMKA TS... | None | Y | Y | Y | Y | SP |
| p45H5 | S. mansoni EST, 100%; BG931230 | tcggag_ | MAASHACLDLRALLSVVGLLLASA GR... | None | Y | N | Y | Y | SP |
| p76E2 | S. mansoni EST, 100%; AA559399 | gtgccc_ | MRFPAGTYDELQIPQGSWSELHKQHNKLYNKFFIVSATIATALFAAA FY... | None | N | Y | Y | N | SA |
| p43G5 | Unknown | gtgtat_ | MRKMPRFLSIHSGFLHILLS FY... | None | N | N | Y | Y | SA |
| p30C2 | Unknown | atactg_ | MMMIILMVLLSVIRIIIVGLISLVVVKG KL... | None | Y | Y | Y | Y | SP |
| p25D5, p43F7, p101E3 | Antisense S. mansoni ORF-RF2, 100%; M14309 | tccata_ | MIHNSTGASVTASIMAGFVSATFAAFATFATTLTLTATTAITLSIIVAFTTTFSKAVAT VT... | 2 | Y | Y | Y | Y | SA |
| p17C5 | Antisense S. mansoni GST, 100%; M98271 | atattc_ | MFYIFSLTNQLFNTIVFLVCFTHHMMFL RH... | None | N | N | Y | Y | NS |
| p90C8 | S. mansoni actin I, 100%; M80334 | MTQIMFETFNVPAMYVAIQAVLSLYASGRTTG IV... | None | Y | Y | N | N | NS | |
| p28C7 | Human protein kinase DYR2,f 46%; Q92630 | MHKAVIFSFPMIIWLIDMKFS NP... | N/Ag | Y | N | Y | Y | NS | |
| p25C4 | S. mansoni Sm23,f 100%; M34453 | MLLFYCLDNCALA NN... | N/A | N | N | Y | N | NS | |
Clone sequence extends 5′ of the matching EST.
The putative cleavage point of each signal peptide is denoted by a space followed by the first two N-terminal residues of the processed protein.
SignalP-NN is based on neural networks.
SignalP-HMM is based on hidden Markov models. SP, N-terminal signal peptide; SA, signal anchor; NS, nonsecretory.
The Kozak consensus sequence for eukaryotes is GCCRCC_ where R is A or G and _ is an ATG start codon.
Incorrect reading frame drove surface expression.
N/A, not applicable.
FIG. 2.
RT-PCR analysis of mRNA expression patterns in 11 S. mansoni cDNA fragments for adult females (f), adult males (m), egg, and cercariae (cerc). The cDNA clone numbers are indicated on the left side and the identity of the sequence (if known) is on the right. S. mansoni triosephosphate isomerase (TPI) mRNA was used as a constitutively expressed positive control for the RT-PCR.
Tetraspanins.
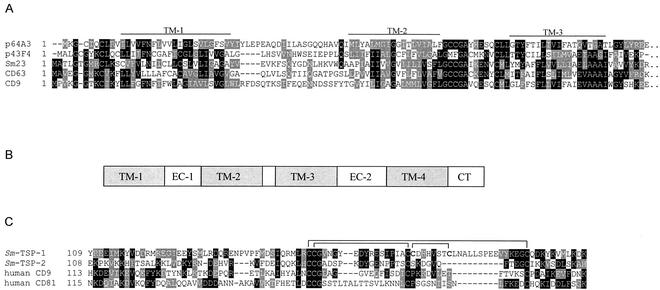
Clones p64A3 and p43F4 belonged to the tetraspanin family of membrane-spanning receptors and shared 35% identity with human CD9 (p64A3) and 39% identity with CD63 (p43F4). Tetraspanins traverse the cell membrane four times and play multiple roles in mammalian cell signaling, notably interactions between immune effector cells and their ligands (13). Neither p64A3 nor p43F4 had been previously identified from the 17,000 ESTs from S. mansoni in dbEST. Both transcripts were detected in adult males and females and cercariae but only weakly (p64A3) or not at all (p43F4) in eggs (Fig. 2). Other tetraspanins have been reported from schistosomes, and members of this protein family include the tegumental vaccine candidate Sm23 (7, 18) (also identified in this study as clone p25C4) and the Sj25 family from S. japonicum (10). Outside parasitology, tetraspanins have gained notoriety due to their expression on the surface of human leukocytes; CD81 and CD9 are expressed on the surfaces of cell types, including B and T lymphocytes, where they mediate a wide range of biological processes (19). Figure 3A shows the similarity between the two new S. mansoni tetraspanins identified in this study and other members of the superfamily, including Sm23 and mammalian cell surface antigens. The SST clones identified did not extend past transmembrane domain 3 in the 3′ direction, so we identified the remaining 5′ untranslated region and 3′ coding sequence and 3′ untranslated region of these mRNAs using rapid amplification of cDNA ends. Tetraspanins bind to their ligands via extracellular loop 2 (EC2), situated between transmembrane domains 3 and 4. The EC2 of CD81 binds to various mammalian ligands and is also a receptor for hepatitis C envelope glycoprotein (24). Further analysis of the schistosome tetraspanin ORFs encoded by these new mRNAs identified the entire ORFs of p64A3 (the full-length cDNA is now called Sm-tsp-1) and p43F4 (Sm-tsp-2), including the EC2 regions. The EC2 of Sm-TSP-1 contained six Cys residues that likely form disulfide bridges, as shown in Fig. 3B. Most tetraspanins have six cysteines (27), but some of the well-characterized family members, including CD81 and CD9, have four cysteines in their EC2 domains and form two disulfide bridges. Sm-TSP-2 is most similar to group I tetraspanins, including CD9, CD81, and Sm23, and contains four Cys residues in its EC2 region (Fig. 3B).
FIG. 3.
(A) Alignment of the tetraspanin family members, bovine CD9, and human CD63 with the ORFs from S. mansoni clones p43F4 and p64A3, identified by AP-PST. Black boxes denote identical amino acids; gray boxes indicate similarity. Transmembrane domains TM-1 to TM-3 are marked by a line spanning the region in reference to the CD9 sequence. (B) Schematic arrangement of domains in tetraspanin family members. EC, extracellular loop; CT, cytoplasmic tail. (C) Alignment of the regions spanning the ligand binding domains (extracellular loop 2) of clones Sm-TSP-1 (corresponds to clone p64A3), Sm-TSP-2 (corresponds to p43F4), and other tetraspanin family members. Lines denote the known (CD81) and putative (all other sequences) disulfide bond linkages.
Sm23 is expressed in the tegument of S. mansoni, but its ligand has yet to be determined; Sm-TSP-1 and -2 might also be exposed on the surface of the parasite, where they could acquire host-derived ligands. Indeed, interruption of the binding of tetraspanins to their ligands, by antibody-mediated mechanisms or with synthetic peptidomimetics, might be an attractive future control strategy for schistosomiasis.
Other clones corresponding to known S. mansoni genes.
Three clones were identified that shared 100% sequence identity with S. mansoni eggshell proteins. p97D2 encoded the EGG2 ORF (5, 16), and both p25G7 and p43F7 encoded the ORF of EGG3 (25). Sex-specific gene products, notably those involved in reproduction, are attractive targets for antihelminth intervention strategies (3), and S. mansoni egg- and uterus-derived proteins are worthy of further investigation. Clone p90C8 encoded S. mansoni actin I, but translation began at Met-121 of the actin I ORF; this stretch of residues from the actin I protein (Met-121 to Asp-189) gained a positive C and S score in the SignalP-NN program. However, it clearly does not represent the initiator Met of actin but rather an internal hydrophobic tract of residues. Although actin is not a secreted protein, this provides further proof of the concept that the AP-PST technique can identify internal transmembrane or other hydrophobic motifs that contain an N-terminal Met.
Extending the 5′ ends of truncated ESTs.
Many of the S. mansoni ESTs in the public domain are not full length due to incomplete reverse transcription during cDNA library construction, resulting in premature truncation at the 5′ ends of mRNAs. As we have shown here, SST complements random shotgun cloning by permitting alignment and stitching of 5′ SST clones with partially overlapping ESTs that have truncated 5′ termini. Two clones, p33F5 and p83H3, had regions of 100% identity at their 3′ ends with the 5′ ends of S. mansoni ESTs that encoded proteins of unknown function. These ESTs were truncated at their 5′ ends, and as such, signal peptides were not identified from the existing ORFs. These clones were expressed in adult worms and cercariae but not in eggs (Fig. 2). Alignment of these two SST clones with their corresponding ESTs extended the 5′ ends of these cDNA consensus sequences so that the ORF of each cDNA contained a signal peptide including an initiator Met and 5′ untranslated region.
Secreted and surface proteins of unknown function.
Numerous clones contained signal peptide-encoding ORFs, but they did not share homology with proteins of known function. Four clones corresponded to S. mansoni ESTs of unknown function (p33F5, p45H5, p76E2, and p83H3), and two clones (p30C2 and p43G5) were completely novel and did not share similarity with any sequences in the public databases, including dbEST. p76E2 showed a restricted expression pattern and was detected in eggs, cercariae, and female worms but not in male worms (Fig. 2). Of the novel clones that did not correspond to ESTs, p30C2 was expressed in cercariae and adult worms (notably higher levels in female worms) but not in eggs, while p43G5 was expressed in all stages (Fig. 2). Future work will involve localization of these novel clones at both the mRNA and protein levels to determine if any are expressed in the tegument and might therefore warrant further investigation as potential vaccine candidates.
ORFs transcribed antisense to known S. mansoni mRNAs.
Clones p17C5, p25D5, p43F7, and p101E3 were 100% identical to S. mansoni nucleotide sequences. Intriguingly, however, closer inspection of these clones revealed that their ORFs were transcribed in the antisense orientation to known S. mansoni mRNAs. For example, the nucleotide sequences of clones p25D5, p43F7, and p101E3 (5′-3′) were identical to the reverse complement (antisense) of the female-specific S. mansoni ORF-RF2 transcript (4). It is interesting that ORF-RF2 mRNA was specifically expressed in female worms (4); however we detected expression of all three antisense transcripts in both female and male worms (Fig. 2). Taq DNA polymerase generated double-stranded cDNA during the RT-PCRs; as a result, the antisense fragments generated might have derived from the second strand. Proper confirmation of the existence of transcribed antisense mRNAs requires a more detailed investigation by using single-stranded probes for Northern blotting and sequencing of the antisense genes to identify exon-intron boundaries. Antisense transcription is not uncommon in eukaryotes. Antisense reading frames occur commonly in the Trypanosoma genome (20), and an antisense ORF encoding a CC chemokine-like protein was detected in the causative agent of river blindness Onchocerca volvulus (9). This is the first report, however, of potential antisense transcripts from schistosomes, and we are further exploring this area of research.
Concluding remarks.
We have shown here that SST is a powerful tool for detecting mRNAs that encode secreted and transmembrane proteins from parasites. Previous reports in the literature have focused on application of SST to mammalian tissues (6, 28, 29), plants (12, 17), and insects (1, 11). This is the first time, to our knowledge, that SST has been used to successfully identify novel mRNAs from any animal pathogen. Furthermore, this is the first report describing recognition and successful trafficking of schistosome signal peptides in mammalian cells. Future work will involve localization of the mRNAs and their protein products to specific tissues within S. mansoni. Clones that are detected in the tegument will become the focus of further research to enhance our understanding of the host-parasite interactions as well as provide new potential vaccine antigens against schistosomiasis.
Acknowledgments
We sincerely thank Hsiuchen Chen and Philip Leder of The Howard Hughes Medical Institute, Harvard Medical School, for provision of the AP-PST vector and associated clones. We also thank Mary Duke of QIMR for maintenance of the schistosome life cycle and Fred Lewis of the Biomedical Research Institute for provision of schistosomes and infected snails. We are grateful to Malcolm Jones of QIMR for technical assistance and helpful discussions.
This work was funded by a block grant awarded to QIMR by the National Health and Medical Research Council of Australia (NHMRC). A.L. was supported by a Howard Florey Centenary Research Fellowship from the NHMRC.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Arca, B., F. Lombardo, M. de Lara Capurro, A. della Torre, G. Dimopoulos, A. A. James, and M. Coluzzi. 1999. Trapping cDNAs encoding secreted proteins from the salivary glands of the malaria vector Anopheles gambiae. Proc. Natl. Acad. Sci. USA 96:1516-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, M. M., S. A. Harrop, J. P. Dalton, B. H. Kalinna, D. P. McManus, and P. J. Brindley. 1995. Cloning and characterization of the Schistosoma japonicum aspartic proteinase involved in hemoglobin degradation. J. Biol. Chem. 270:24496-24501. [DOI] [PubMed] [Google Scholar]
- 3.Boag, P. R., S. E. Newton, and R. B. Gasser. 2001. Molecular aspects of sexual development and reproduction in nematodes and schistosomes. Adv. Parasitol. 50:153-198. [DOI] [PubMed] [Google Scholar]
- 4.Bobek, L., D. M. Rekosh, H. van Keulen, and P. T. LoVerde. 1986. Characterization of a female-specific cDNA derived from a developmentally regulated mRNA in the human blood fluke Schistosoma mansoni. Proc. Natl. Acad. Sci. USA 83:5544-5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobek, L. A., D. M. Rekosh, and P. T. LoVerde. 1988. Small gene family encoding an eggshell (chorion) protein of the human parasite Schistosoma mansoni. Mol. Cell. Biol. 8:3008-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, H., and P. Leder. 1999. A new signal sequence trap using alkaline phosphatase as a reporter. Nucleic Acids Res. 27:1219-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Da'dara, A. A., P. J. Skelly, M. M. Wang, and D. A. Harn. 2001. Immunization with plasmid DNA encoding the integral membrane protein, Sm23, elicits a protective immune response against schistosome infection in mice. Vaccine 20:359-369. [DOI] [PubMed] [Google Scholar]
- 8.Dalton, J. P., S. R. Day, A. C. Drew, and P. J. Brindley. 1997. A method for the isolation of schistosome eggs and miracidia free of contaminating host tissues. Parasitology 115:29-32. [DOI] [PubMed] [Google Scholar]
- 9.Erttmann, K. D., D. W. Buttner, and M. Y. Gallin. 1995. A putative protein related to human chemokines encoded antisense to the cDNA of an Onchocerca volvulus antigen. Trop. Med. Parasitol. 46:123-130. [PubMed] [Google Scholar]
- 10.Fan, J., and P. J. Brindley. 1998. Characterization of cDNAs encoding a new family of tetraspanins from schistosomes—the Sj25 family. Gene 219:1-8. [DOI] [PubMed] [Google Scholar]
- 11.Goo, J. H., Y. Ahn, O. K. Park, and W. J. Park. 1999. Selection of Drosophila genes encoding secreted and membrane proteins. Mol. Cells 9:564-568. [PubMed] [Google Scholar]
- 12.Goo, J. H., A. R. Park, W. J. Park, and O. K. Park. 1999. Selection of Arabidopsis genes encoding secreted and plasma membrane proteins. Plant Mol. Biol. 41:415-423. [DOI] [PubMed] [Google Scholar]
- 13.Hemler, M. E. 2001. Specific tetraspanin functions. J. Cell Biol. 155:1103-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hooker, C. W., and P. J. Brindley. 1996. Cloning and characterisation of strain-specific transcripts encoding triosephosphate isomerase, a candidate vaccine antigen from Schistosoma japonicum. Mol. Biochem. Parasitol. 82:265-269. [DOI] [PubMed] [Google Scholar]
- 15.Klein, R. D., Q. Gu, A. Goddard, and A. Rosenthal. 1996. Selection for genes encoding secreted proteins and receptors. Proc. Natl. Acad. Sci. USA 93:7108-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koster, B., H. Dargatz, J. Schroder, J. Hirzmann, C. Haarmann, P. Symmons, and W. Kunz. 1988. Identification and localisation of the products of a putative eggshell precursor gene in the vitellarium of Schistosoma mansoni. Mol. Biochem. Parasitol. 31:183-198. [DOI] [PubMed] [Google Scholar]
- 17.Kristoffersen, P., T. Teichmann, R. Stracke, and K. Palme. 1996. Signal sequence trap to clone cDNAs encoding secreted or membrane-associated plant proteins. Anal. Biochem. 243:127-132. [DOI] [PubMed] [Google Scholar]
- 18.Lee, K. W., K. A. Shalaby, A. M. Medhat, H. Shi, Q. Yang, A. M. Karim, and P. T. LoVerde. 1995. Schistosoma mansoni: characterization of the gene encoding Sm23, an integral membrane protein. Exp. Parasitol. 80:155-158. [DOI] [PubMed] [Google Scholar]
- 19.Levy, S., S. C. Todd, and H. T. Maecker. 1998. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol. 16:89-109. [DOI] [PubMed] [Google Scholar]
- 20.Liniger, M., K. Bodenmuller, E. Pays, S. Gallati, and I. Roditi. 2001. Overlapping sense and antisense transcription units in Trypanosoma brucei. Mol. Microbiol. 40:869-878. [DOI] [PubMed] [Google Scholar]
- 21.Loukas, A., M. K. Jones, L. T. King, P. J. Brindley, and D. P. McManus. 2001. Receptor for Fc on the surfaces of schistosomes. Infect. Immun. 69:3646-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McManus, D. P. 1999. The search for a vaccine against schistosomiasis—a difficult path but an achievable goal. Immunol. Rev. 171:149-161. [DOI] [PubMed] [Google Scholar]
- 23.Nene, V., and R. Bishop. 2001. Trapping parasite secretory proteins in baker's yeast. Trends Parasitol. 17:407-409. [DOI] [PubMed] [Google Scholar]
- 24.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 25.Rodrigues, V., M. Chaudhri, M. Knight, H. Meadows, A. E. Chambers, W. R. Taylor, C. Kelly, and A. J. Simpson. 1989. Predicted structure of a major Schistosoma mansoni eggshell protein. Mol. Biochem. Parasitol. 32:7-13. [DOI] [PubMed] [Google Scholar]
- 26.Ross, A. G., P. B. Bartley, A. C. Sleigh, G. R. Olds, Y. Li, G. M. Williams, and D. P. McManus. 2002. Schistosomiasis. N. Engl. J. Med. 346:1212-1220. [DOI] [PubMed] [Google Scholar]
- 27.Seigneuret, M., A. Delaguillaumie, C. Lagaudriere-Gesbert, and H. Conjeaud. 2001. Structure of the tetraspanin main extracellular domain. A partially conserved fold with a structurally variable domain insertion. J. Biol. Chem. 276:40055-40064. [DOI] [PubMed] [Google Scholar]
- 28.Skarnes, W. C., J. E. Moss, S. M. Hurtley, and R. S. Beddington. 1995. Capturing genes encoding membrane and secreted proteins important for mouse development. Proc. Natl. Acad. Sci. USA 92:6592-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tashiro, K., H. Tada, R. Heilker, M. Shirozu, T. Nakano, and T. Honjo. 1993. Signal sequence trap: a cloning strategy for secreted proteins and type I membrane proteins. Science 261:600-603. [DOI] [PubMed] [Google Scholar]
- 30.Yang, W., G. J. Waine, D. G. Sculley, X. Liu, and D. P. McManus. 1992. Cloning and partial nucleotide sequence of Schistosoma japonicum paramyosin: a potential vaccine candidate against schistosomiasis. Int. J. Parasitol. 22:1187-1191. [DOI] [PubMed] [Google Scholar]