Abstract
The swine pathogen Actinobacillus pleuropneumoniae possesses a 75-kDa outer membrane protein (OMP), FhuA, the receptor for ferrichrome, a hydroxamate-type siderophore. Polyclonal serum to FhuA reacted with OMP preparations from 12 serotypes of A. pleuropneumoniae under conditions of iron repletion and restriction. Reverse transcription-PCR confirmed that A. pleuropneumoniae fhuA expression is not upregulated in response to low iron levels. An A. pleuropneumoniae fhuA deletion mutant was generated and showed abolishment of ferrichrome uptake.
Actinobacillus pleuropneumoniae is the etiologic agent of porcine pleuropneumonia, a highly infectious disease of pigs which causes economic losses to the swine industry worldwide (7). This bacterium can use hemoglobin, heme-containing compounds (2), porcine transferrin (15), and exogenously supplied siderophores (6) as sources of iron. Under iron-limiting conditions, A. pleuropneumoniae expresses two transferrin binding proteins, TbpA (∼100 kDa) and TbpB (∼60 kDa) (10, 11), both of which have been documented as virulence factors (1). Proximal to the genes tbpA and tbpB and cotranscribed with them is a region that has been identified as exbBD in A. pleuropneumoniae (17); this region is linked to a partial tonB gene which has not been fully sequenced. Although A. pleuropneumoniae can use exogenously supplied siderophores as sources of iron for growth, attempts (5, 6) to identify production of endogenous siderophores have been inconclusive. We first reported (14) the presence of an outer membrane protein (OMP), FhuA, in A. pleuropneumoniae which acts as receptor for ferrichrome, a siderophore belonging to the hydroxamate family. This was the first report of a high-affinity iron acquisition for any siderophore in this organism. Additionally, all four genes of the operon (fhuCDBA) are present in serotypes 1 to 12 of A. pleuropneumoniae (GenBank accession no. AF351135). In this study, we further characterize the receptor FhuA by comparing the sequences of FhuA from different A. pleuropneumoniae serotypes, thereby establishing phylogenetic relationships. Using targeted mutagenesis, we generated a deletion mutant of A. pleuropneumoniae fhuA. We observe that mutating the gene abolishes ferrichrome utilization, showing unequivocally that fhuA encodes the receptor for ferrichrome in A. pleuropneumoniae. We also identify regulation of fhuA that appears to be independent of iron supply.
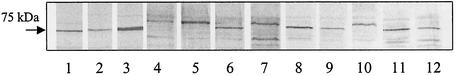
Recombinant FhuA from A. pleuropneumoniae was expressed in Escherichia coli as previously described (14), and the recombinant protein was purified with a QIAexpress purification kit using Ni2+-nitrilotriacetic acid agarose beads (QIAGEN). An antiserum against purified recombinant His6.FhuA was produced in an adult male New Zealand White rabbit following subcutaneous, intramuscular, and intravenous immunizations with purified protein over a 6-week period for a total of seven injections. Ten days after the final injection, the rabbit was anesthetized and blood was obtained through cardiac puncture to yield 50 ml of immune serum. OMPs from strains representing serotypes 1 to 12 of A. pleuropneumoniae were prepared by the method of Hantke (12). The reference strains used were as follows: serotype 1, strain 4074; serotype 2, strain 4226; serotype 3, strain 1421; serotype 4, strain 1462; serotype 5, strain K-17 and field strain 86-4780; serotype 6, strain FEMO; serotype 7, strain WF83; serotype 8, strain 404; serotype 9, strain 13261; serotype 10, strain 13039; serotype 11, strain 56153; and serotype 12, strain 8329/85. Equal amounts of protein from total OMPs of these strains were tested by Western blotting following standard procedures (13) using the aforementioned polyclonal antiserum to recombinant His6.FhuA from serotype 1 at a dilution of 1/10,000. The OMPs were prepared from A. pleuropneumoniae cultures grown both under iron-rich conditions and iron-restricted conditions. The latter ethylene diamine dihydroxyphenyl acetic acid conditions were obtained by the addition of the iron chelator (EDDHA, 50 μM; Sigma). Western blot analyses revealed that the antiserum recognized a minor OMP with an approximate molecular mass of 75 kDa in all strains tested. This strongly suggests that FhuA is expressed in all serotypes and that cross-reactivity exists between serotypes. There appear to be, however, minor variations in the size of this OMP which ranged between 74 and 77 kDa (Fig. 1; Table 1). One major band of 76 kDa was distinguished for serotypes 1, 2, 3, 6, 8, 9, 11, and 12, while a slightly larger band of 77 kDa was identified for serotypes 4, 5, 7, and 10. No increase in the intensities of the bands were observed for all serotypes grown under iron-restricted conditions. Serotype 7 was notable in that two distinct OMPs of 74 and 77 kDa, both of which appeared to be major proteins, reacted with the immune serum. The nucleotide sequence of fhuA from this serotype was determined (see below) and revealed a second translational start site located 138 bp downstream of the first start methionine which might encode a truncated protein with a predicted Mr of 68,000.
FIG. 1.
OMPs from A. pleuropneumoniae serotypes 1 to 12 immunoblotted with rabbit polyclonal serum to purified recombinant FhuA from serotype 1. Results were similar for OMPs prepared from cultures grown under iron restricted and iron sufficient conditions. Only the relevant portion of the membrane is shown.
TABLE 1.
Characterization of FhuA from reference strains of A. pleuropneumoniae
| A. pleuropneumoniae serotype(s) | Length of predicted OREb (amino acids) | Approx predicted Mr(s) of mature protein (kDa) | Approx Mr(s) of OMP in Western blot (kDa) |
|---|---|---|---|
| 1, 9, 11 | 695 | 74.8 | 76 |
| 10 | 695 | 74.7 | 77 |
| 6 | 694 | 74.7 | 76 |
| 3 | 692 | 74.1 | 76 |
| 2 | 690 | 73.2 | 76 |
| 8 | 690 | 74.0 | 76 |
| 4 | 686 | 72.6 | 77 |
| 5, 5fsa | 684 | 74.3 | 77 |
| 12 | 658 | 70.1 | 76 |
| 7 | 647 | 68.0, 70.4 | 74, 77 |
5fs, field strain of serotype 5 (86-4780) unable to use exogenous ferrichrome for growth.
ORF, open reading frame.
To determine whether a difference in the length of the gene could account for the difference in observed molecular weights of the OMPs in the Western blots, the fhuA gene from A. pleuropneumoniae reference strains representing serotypes 2 to 12 was amplified by PCR. A set of primers (AFor [5′-GATGAGGTGTCGGTGGTT-3′] and ARev [5′-TGTGGCATTGACTTTACG-3′]) based on the mature sequence of fhuA from A. pleuropneumoniae serotype 1 was initially used, and other primers for walking the PCR products were subsequently synthesized as needed. The predicted open reading frames (http://www.ncbi.nlm.nih.gov) deduced from the primary sequences ranged between 647 and 695 amino acids (Table 1) and the predicted molecular masses of the mature protein (http://ca.expasy.org/tools/pi_tool.html) were between 70 and 75 kDa. However, the predicted molecular masses of the mature proteins deduced from the primary sequence differed slightly from the observed molecular masses of the OMPs in the Western blots (Fig. 1; Table 1); variations in electrophoretic mobility could be one plausible explanation. fhuA was also amplified and sequenced from a field strain of serotype 5 (strain 86-780), which cannot grow in the presence of exogenous ferrichrome (6). Genetic analysis of fhuA from this strain showed it to be identical in sequence to the gene in the serotype 5 reference strain indicating that the inability of the field strain to use ferrichrome lies in a gene other than fhuA. The ClustalX algorithm (http://www.clustalw.genome.ad.jp) was utilized to align the amino acid residues predicted from the fhuA primary sequences from A. pleuropneumoniae serotypes 1 to 12 and the field strain of serotype 5. A highly conserved region of six amino acids occurred near the amino terminus where the residues LDEVSV were conserved without exception throughout all test strains. This region constitutes a putative TonB box, a site where TonB-dependent uptake systems interact with the TonB-ExbB-ExbD complex. We proposed (14) a three-dimensional model for FhuA of A. pleuropneumoniae serotype 1 by threading it to the crystal structure of the homologous protein in E. coli (8). Similarly threading the sequences of the remaining 11 serotypes on the three-dimensional model of serotype 1, we observed minor differences in 10 of the 11 surface-exposed loops of these FhuA proteins; conserved regions were in the β-strands of the transmembrane regions. A remarkably conserved region was loop 3, where 33 out of 37 amino acid residues were identical throughout all A. pleuropneumoniae serotypes. This is a key extracellular loop in E. coli involved in ligand recognition and uptake (8) and although loop 3 in A. pleuropneumoniae is four amino acid residues longer than its counterpart in E. coli, conservation of this region in A. pleuropneumoniae implies a similarly significant role.
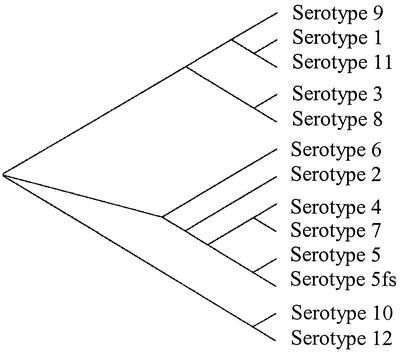
A dendrogram analysis (Fig. 2) assigned FhuA proteins from A. pleuropneumoniae into three branches based on their sequence relatedness. The three main branches of the dendrogram are serotypes 1, 9, 11, 3, and 8; serotypes 6, 2, 4, 7, and 5; and serotypes 10 and 12. Interestingly, this relatedness may be correlated, to some extent, with the serological cross-reactivity that exists between serotypes 1, 9, and 11; 3 and 8; and 4 and 7 due to the structural similarities in the O antigen of each group (3). Taken together, these data could be extrapolated to interpret the common ancestry of different A. pleuropneumoniae serotypes and show their evolution as how they diversified over time.
FIG. 2.
Dendrogram analysis showing sequence relatedness of FhuA from reference strains representing A. pleuropneumoniae serotypes 1 to 12 and a field strain of serotype 5 (5fs).
Strains from different members of the Pasteurellaceae family that are also swine pathogens were assessed for their ability to use ferrichrome as their sole source of iron. These organisms included five strains of Actinobacillus suis (C84, O1/K2; H89-1173, O2/K3; H91-0380, O2/K2; SO4, 01/K1 and VSB-3714), two of Haemophilus parasuis (serotype 5 strain Nagasaki and serotype 6 strain 131) and two of Pasteurella multocida (capsular type A strain 88-761 and capsular type D strain 1703). Zones of bacterial growth were monitored in the area where 5 μl of an aqueous solution of ferrichrome (Sigma) at different concentrations (50 μM, 100 μM, and 1 mM) was spotted on deferrated culture plates (200 μM EDDHA) inoculated with these different bacteria. Results are shown in Table 2. All five strains of A. suis and one strain of H. parasuis were able to grow when ferrichrome was the sole source of iron. Neither strain of P. multocida could grow in the presence of ferrichrome as the sole iron source. This indicates that the ability to use exogenous siderophores is not limited to A. pleuropneumoniae and suggests that a ferrichrome uptake receptor homologous to FhuA of A. pleuropneumoniae could also be present in other porcine Pasteurellaceae. However, when PCR and Southern and Western blot analyses were performed on these strains with primers, probes, and antiserum based on A. pleuropneumoniae sequences, these failed to detect specific individual bands. Sequence variation between species could account for the lack of hybridization signals.
TABLE 2.
Growth promotion of different porcine Pasteurellaceae strains by ferrichrome
| Bacterial strain | Ferrichrome use |
|---|---|
| A. pleuropneumoniae | |
| Serotypes 1 to 12 | + |
| Field strain 86-4780 from serotype 5 | − |
| A. suis C84 O1/K2 | + |
| A. suis H89-1173 O2/K3 | + |
| A. suis H91-0380 O2/K2 | + |
| A. suis SO4 O1/K1 | + |
| A. suis VSB-3714 | + |
| H. parasuis serotype 5 | + |
| H. parasuis serotype 6 | − |
| P. multocida A strain 88-761 | − |
| P. multocida D strain 1703 | − |
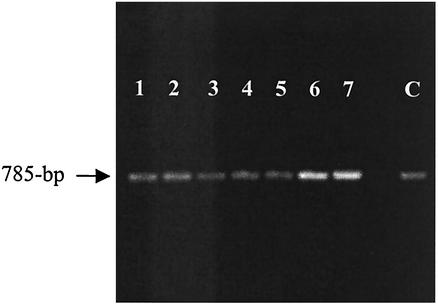
Most high-affinity iron acquisition systems are not expressed constitutively; rather, their expression is up-regulated in response to a decrease in the level of intracellular iron. In A. pleuropneumoniae, the OMP FhuA was expressed under iron-replete conditions in all serotypes as shown by Western blot analysis. This was an indication that unlike other iron uptake genes that are up-regulated in response to a decrease in the level of iron in the environment, fhuA of A. pleuropneumoniae may be regulated differently. To further investigate the effect of iron on the expression of FhuA of A. pleuropneumoniae, we performed reverse transcription (RT)-PCR on total bacterial RNA isolated under different culture conditions of iron repletion, iron restriction, and iron restriction in the presence of a possible second signal such as the ligand ferrichrome. RNA was isolated from A. pleuropneumoniae serotype 1 reference strain 4074 grown under different culture conditions: (i) iron repletion (brain-heart infusion (BHI)-NAD+ broth); (ii) iron restriction (BHI-NAD+ broth supplemented with 50 μM or 100 μM EDDHA); and (iii) iron restriction with the presence of specific iron source (BHI-NAD+ broth supplemented with 50 μM EDDHA and 10 μM ferrichrome). Growth was monitored turbidimetrically, as organisms grown under iron-restricted conditions had a lower growth rate than those grown under iron-replete conditions. Bacteria from all culture conditions were harvested in exponential phase (optical density at 660 nm, ∼0.8). Total bacterial RNA was isolated with an RNeasy Mini Kit (QIAGEN), treated with DNase I (RNase free, fast-performance liquid chromatography purity; Amersham),and purified once more. RT-PCR was performed with the One-Step RT-PCR Kit (QIAGEN) following the recommendations of the manufacturer. Control experiments for DNA contamination were performed in which no reverse transcriptase was added to the RNA samples prior to the PCR step. A primer pair (A3For 5′-TGAAAATGTTCGCTATCG-3′ and 5T7W2.5 5′-GTTTTGACGGGCACGATC-3′) that amplifies an internal fragment of 785 bp in fhuA was employed. As positive control for the RT-PCR, primers (exbDF 5′-CACTTGCTTCTATTGGTGCGGTTG-3′ and RB1 5′-CCAGAACAAGCGACAAGAAACAGC-3′) amplified an 800-bp internal fragment of the tbpB gene in A. pleuropneumoniae, known to be upregulated under iron-restricted conditions (17). The results from the RT-PCR experiments (Fig. 3) showed that the transcript level of fhuA did not increase in response to a decrease in the level of iron, even at higher concentrations of the iron chelator EDDHA (50 or 100 μM) or in the presence of ferrichrome (Fig. 3, lanes 2, 3, and 4). There was no increase in transcripts. These results are in contrast to the expression of genes for transferrin binding proteins in A. pleuropneumoniae, the levels of which are up-regulated in response to diminished levels of iron (Fig. 3, lanes 6 and 7). While fhuA is the first gene transcribed in the fhuACDB operon in E. coli, the gene arrangement in A. pleuropneumoniae is such that fhuA occurs last in the fhuCDBA operon (Fig. 4). RT-PCR was also performed to assess the level of fhuC transcript (primers CFor [5′-GCAATTCGAGCAGGGTAAG-3′] and CRev [5′-CCGGTCGTTTGGTTTCAGG-3′]) and showed no up-regulation in the level of fhuC in response to iron restriction. Results of the RT-PCR experiments confirmed those of the Western blot, indicating that FhuA expression in A. pleuropneumoniae is most likely not regulated by the level of iron in the environment. This phenomenon merits further studies of the promoter region of the fhu operon in A. pleuropneumoniae and does not seem to occur in any other iron uptake system known to date. The prospect of gene regulation at other levels or by other environmental factors is also intriguing. Completing the annotation of the genome of A. pleuropneumoniae and identifying homologies with other iron-regulated OMPs might allow comparison of the nucleotide sequence in the regions upstream of these genes that could potentially be regulated by iron. This will aid in identifying some consensus sequence upstream of iron-regulated genes in A. pleuropneumoniae which seems to be different from the consensus fur box in E. coli.
FIG. 3.
RT-PCR with RNA from A. pleuropneumoniae serotype 1 cells grown under iron-replete conditions (lanes1 and 5); under iron-restricted conditions with 50 μM EDDHA (lanes 2 and 6), under iron-restricted conditions with 100 μM EDDHA (lanes 3 and 7), and under iron-restricted conditions (50 μM EDDHA) in the presence of 10 μM ferrichrome (lane 4) or with total DNA (lane C). Lanes 1 to 4 and lane C employed a primer pair (A3For, 5T7W2.5) that amplifies a 785-bp internal fragment in fhuA; lanes 5 to 7 employed a primer pair (ExbDF, RB1) that amplifies and internal fragment of the same size in tbpB of A. pleuropneumoniae.
FIG. 4.
Organization of the genes fhuC, fhuD, fhuB, and fhuA in the fhu operon of A. pleuropneumoniae serotype 1 strain 4074. Cm, chloramphenicol cassette that interrupts the fhuA gene in the mutant strain DG02.
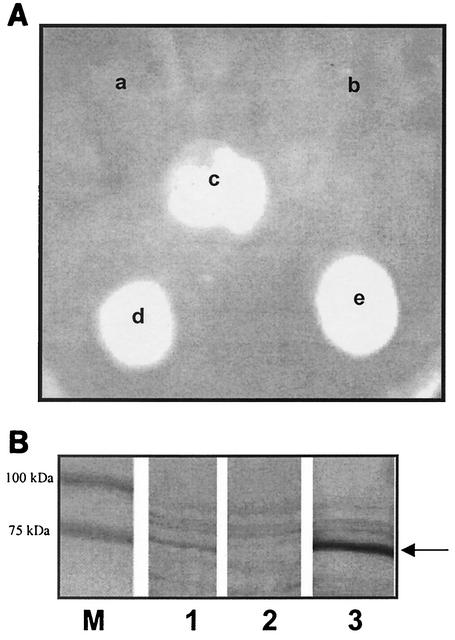
In order to construct a deletion mutant of A. pleuropneumoniae fhuA, a single-step transconjugation system using a sucrose sensitivity marker (vector pBMK1) designed by Oswald and colleagues (16) was utilized with some modifications. We used inverse PCR to delete 1,015 bp of the 2,088-bp coding sequence of the fhuA gene and subsequently cloned in that region an A. pleuropneumoniae-derived promoter (omlA) that is transcriptionally fused to a chloramphenicol acetyltransferase (cat) gene. To facilitate allelic exchange, additional upstream and downstream sequences to fhuA were also included in the construct. The 12.2-kb plasmid having pBMK1 vector for backbone was transformed into diaminopimelic acid auxotrophic E. coli β2155 [thrB1004pro thi hsdS lacZΔM15 (F′lacZΔM15lacIq traD36 proA+ proB+) Δdap::erm(Ermr)recA::RP4-2-tet(Tcr)Mu-km(Kmr)λpir] cells as the donor for transconjugation into A. pleuropneumoniae. We followed a filter mating technique described by Dehio and Meyer (4) and adapted by Oswald et al. (16) which allows for two consecutive crossing over events to be monitored. After transconjugation, Cmr A. pleuropneumoniae colonies were analyzed for the presence of a single crossover event by PCR using primers that amplified both wild type and ΔfhuA as well as chloramphenicol, kanamycin, and sucrose cassettes. The presence of plasmid cointegrates was also confirmed by Southern blot. Counterselection of transformants for a second crossover was carried out on NAD+-supplemented Mueller-Hinton agar plates with 10% sucrose and chloramphenicol (1 μg/ml). After 6 days of incubation at 30°C, single sucrose resistant colonies were isolated. One clone, DG02, that had undergone a successful second crossover, was subsequently isolated and analyzed by growth promotion assays for assessment of phenotype. While this clone was unable to grow in deferrated media with either ferrichrome or ferricrocin as the sole source of iron (Fig. 5A [a and b]), growth was still possible with other sources of iron such as porcine hemoglobin, hemin and transferrin (Fig. 5A) indicating that iron uptake from these sources remained intact. PCR and Southern blot analyses verified the genotype of the A. pleuropneumoniae ΔfhuA strain DG02 (Fig. 4) as all anticipated PCR products with various primer combinations were present at the expected sizes. Southern blot analyses revealed that each of the parent strain and the mutant strain DG02 demonstrated only one differential hybridization signal when probed with fhuA, indicating the existence of a single copy of fhuA in the chromosome of A. pleuropneumoniae serotype 1. In the fhu operon of A. pleuropneumoniae, fhuA is the last gene which is transcribed. Analysis of sequence information 100 bp downstream of fhuA reveals a partial gene in a region displaying similarity with a hypothetical protein from Neisseria meningitidis NMA0986 (Fig. 4) (13), a gene not pertinent to iron uptake which is transcribed in the opposite direction. Hence, polar effects due to mutating fhuA of A. pleuropneumoniae are not thought to play a role on downstream genes. Western blot using a rabbit polyclonal serum raised against A. pleuropneumoniae recombinant FhuA revealed that the OMP FhuA was no longer expressed in DG02 (Fig. 5B, lane 2) whereas the 75-kDa OMP of the parent strain reacted with the antibody. Additionally, the fhuA gene was also cloned into the shuttle vector pJF224-XN (9) and introduced into the mutant strain DG02 by electroporation, in an attempt to complement the ferrichrome negative phenotype. Results of the complementation experiment demonstrated that fhuA was able to restore the phenotype of the mutant strain as successful transformants were selected in deferrated medium supplemented with ferrichrome as the sole source of iron. The 75-kDa OMP FhuA from the complemented strain DG02c reacted with the polyclonal serum to recombinant FhuA (Fig. 5B, lane 3). Collectively, these data establish the role of fhuA as the receptor for the uptake of ferric hydroxamate in A. pleuropneumoniae. It would be interesting to test the virulence of A. pleuropneumoniae ΔfhuA strain DG02 in pigs and compare it with the virulence of the parent strain in an experimental setup similar to the one carried out for TbpA and TbpB mutants (1). Based on experiments conducted in the present study, we conclude that while FhuA of A. pleuropneumoniae acts as the receptor for ferrichrome and looks similar among different serotypes, it is regulated differently from the homologous proteins in other species.
FIG. 5.
(A) Growth promotion assay for strain DG02 (A. pleuropneumoniae ΔfhuA) on defferated media using the following exogenous sources of iron: ferrichrome (a), ferricrocin (b), porcine transferrin (c), hemoglobin (d), and hemin (e). (B) OMP preparations from A. pleuropneumoniae serotype 1 strain 4074 (lane 1), A. pleuropneumoniae ΔfhuA strain DG02 (lane 2), and complemented A. pleuropneumoniae ΔfhuA strain DG02c (lane 3) immunoblotted with rabbit polyclonal serum to purified recombinant A. pleuropneumoniae FhuA from serotype 1. M represents a prestained protein standard marker. The arrow indicates the position of FhuA.
Acknowledgments
This work was supported in part by the Canadian Research Network on Bacterial Pathogens of Swine, a network of the Natural Sciences and Engineering Research Council (NSERC) of Canada and industrial partners. Financial support was also provided by Fonds pour la formation de chercheurs et l'aide à la recherche (99-ER-0214 and 2002-ER-71900) and Strategic Grant 224192 from NSERC to M.J. and J.W.C.
We thank G. Gerlach at the Institut für Mikrobiologie und Tierseuchen in Hannover, Germany, for the generous donation of the E. coli β2155 strain and vector pBMK1; J. MacInnes at the Ontario Veterinary College in Guelph, Ontario, Canada, for the A. suis strains; and K. R. Mittal for the H. parasuis strains. J. Labrie assisted with the growth promotion assays, and D. Niven and F. Bahrami assisted with the RT-PCR experiments.
Editor: F. C. Fang
REFERENCES
- 1.Baltes, N., I. Hennig-Pauka, and G. F. Gerlach. 2002. Both transferrin binding proteins are virulence factors in Actinobacillus pleuropneumoniae serotype 7 infection. FEMS Microbiol. Lett. 209:283-287. [DOI] [PubMed] [Google Scholar]
- 2.Bélanger, M., C. Bégin, and M. Jacques. 1995. Lipopolysaccharides of Actinobacillus pleuropneumoniae bind pig hemoglobin. Infect. Immun. 63:656-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beynon, L. M., D. W. Griffith, J. C. Richards, and M. B. Perry. 1992. Characterization of the lipopolysaccharide O antigens of Actinobacillus pleuropneumoniae serotypes 9 and 11: antigenic relationships among serotypes 9, 11, and 1. J. Bacteriol. 174:5324-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dehio, C., and M. Meyer. 1997. Maintenance of broad-host-range incompatibility group P and Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal transfer from Escherichia coli. J. Bacteriol. 179:538-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deneer, H. G., and A. A. Potter. 1989. Effect of iron restriction on the outer membrane proteins of Actinobacillus (Haemophilus) pleuropneumoniae. Infect. Immun. 57:798-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diarra, M. S., J. A. Dolence, E. K. Dolence, I. Darwish, M. J. Miller, F. Malouin, and M. Jacques. 1996. Growth of Actinobacillus pleuropneumoniae is promoted by exogenous hydroxamate and catecholate siderophores. Appl. Environ. Microbiol. 62:853-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenwick, B., and S. Henry. 1994. Porcine pleuropneumonia. J. Am. Vet. Med. Assoc. 204:1334-1340. [PubMed] [Google Scholar]
- 8.Ferguson, A. D., E. Hofmann, J. W. Coulton, K. Diederichs, and W. Welte. 1998. Siderophore-mediated iron transport: crystal structure of the FhuA with bound lipopolysaccharide. Science 282:2215-2220. [DOI] [PubMed] [Google Scholar]
- 9.Frey, J. 1992. Construction of a broad host range shuttle vector for gene cloning and expression in Actinobacillus pleuropneumoniae and other Pasteurellaceae. Res. Microbiol. 143:263-269. [DOI] [PubMed] [Google Scholar]
- 10.Gerlach, G. F., C. Anderson, A. A. Potter, S. Klashinsky, and P. J. Willson. 1992. Cloning and expression of a transferrin-binding protein from Actinobacillus pleuropneumoniae. Infect. Immun. 60:892-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez, G. C., R.-H. Yu, P. R. J. Rosteck, and A. B. Schryvers. 1995. Sequence, genetic analysis, and expression of Actinobacillus pleuropneumoniae transferrin receptor genes. Microbiology 141:2405-2416. [DOI] [PubMed] [Google Scholar]
- 12.Hantke, K. 1981. Regulation of ferric iron transport in Escherichia coli K-12: isolation of a constitutive mutant. Mol. Gen. Genet. 182:288-292. [DOI] [PubMed] [Google Scholar]
- 13.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Mikael, L. G., P. Pawelek, J. Labrie, M. Sirois, J. W. Coulton, and M. Jacques. 2002. Molecular cloning and characterization of the ferric hydroxamate uptake (fhu) operon in Actinobacillus pleuropneumoniae. Microbiology 148:2869-2882. [DOI] [PubMed] [Google Scholar]
- 15.Niven, D. F., J. Donga, and F. S. Archibald. 1989. Responses of Haemophilus pleuropneumoniae to iron restriction; changes in the outer membrane protein profile and the removal of iron from porcine transferrin. Mol. Microbiol. 3:1083-1089. [DOI] [PubMed] [Google Scholar]
- 16.Oswald, W., T. Walaiporn, G. Ohrt, and G. F. Gerlach. 1999. A single-step transconjugation system for the introduction of unmarked deletions into Actinobacillus pleuropneumoniae serotype 7 using a sucrose sensitivity marker. FEMS Microbiol. Lett. 179:153-160. [DOI] [PubMed] [Google Scholar]
- 17.Tonpitak, W., S. Thiede, W. Oswald, N. Baltes, and G. F. Gerlach. 2000. Actinobacillus pleuropneumoniae iron transport: a set of exbBD genes is transcriptionally linked to the tbpB gene and required for utilization of transferrin-bound iron. Infect. Immun. 68:1164-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]