Abstract
We determined the complete genome sequence of Shigella flexneri serotype 2a strain 2457T (4,599,354 bp). Shigella species cause >1 million deaths per year from dysentery and diarrhea and have a lifestyle that is markedly different from those of closely related bacteria, including Escherichia coli. The genome exhibits the backbone and island mosaic structure of E. coli pathogens, albeit with much less horizontally transferred DNA and lacking 357 genes present in E. coli. The strain is distinctive in its large complement of insertion sequences, with several genomic rearrangements mediated by insertion sequences, 12 cryptic prophages, 372 pseudogenes, and 195 S. flexneri-specific genes. The 2457T genome was also compared with that of a recently sequenced S. flexneri 2a strain, 301. Our data are consistent with Shigella being phylogenetically indistinguishable from E. coli. The S. flexneri-specific regions contain many genes that could encode proteins with roles in virulence. Analysis of these will reveal the genetic basis for aspects of this pathogenic organism's distinctive lifestyle that have yet to be explained.
Shigella is an important human pathogen, responsible for the majority of cases of endemic bacillary dysentery prevalent in developing nations. An estimated 1.1 million deaths and 160 million cases per year are attributed to shigellosis (32). Currently, no vaccine is available that can provide adequate protection against the many different serotypes of Shigella. Existing antimicrobial treatments are becoming compromised due to increased antibiotic resistance, cost of treatment, and continuing poor hygiene and unsanitary conditions in the developing world.
Shigella is pathogenic only for humans. It causes disease by invading the epithelium of the colon, resulting in an intense acute inflammatory response (51). Shigella strains are unusual among enteric bacteria in their ability to gain access to the epithelial cell cytosol, where they replicate and spread directly into adjacent cells. Shigella strains contain a large virulence plasmid that is known to encode genes required and sufficient for invasion of epithelial cells (61). However, chromosomal genes present in “pathogenicity islands” also participate in the pathogenic process directly or contribute to survival in the environments encountered during infection (2, 21, 22, 49, 58, 70). The genetic bases for several aspects of the pathogenic process and intracellular lifestyle of Shigella, including the mechanisms of species specificity, tissue tropism, and restriction of the immune response, are still poorly understood (Table 1) and probably involve chromosomally encoded proteins. In common with other enteric bacteria, Shigella survives the proteases and acids of the intestinal tract by uncertain means. Highly tissue-specific disease results from a very low infectious dose (10 to 100 bacteria) and in the absence of flagellum-based motility. We selected the virulent strain 2457T of Shigella flexneri serotype 2a (33) for sequencing because it has been widely used for genetic research and for clinical challenge studies. Although Shigella spp. have been regarded as distinct from Escherichia coli, as early as 1972, DNA hybridization studies estimated that Shigella and E. coli are taxonomically indistinguishable at the species level (5). Recent work of the Reeves group (34, 56, 57) based on multilocus enzyme electrophoresis and sequencing of a small number of genes places Shigella clearly within the genus Escherichia and arising several times independently. Comparison of the complete S. flexneri genome sequence with that of E. coli K-12 establishes the precise genetic relationship of S. flexneri to E. coli. Given the markedly different lifestyles of intracellular Shigella and extracellular E. coli, the comparison should also reveal important genetic differences expected to underlie pathogenesis, other than the presence or absence of the virulence plasmid.
TABLE 1.
Steps in the pathogenic process of shigellosisa
| Step | Stage of infection or related observationd | Probable molecular mechanism | Candidate ORFs in islandsb | S. flexneri products known to be involved |
|---|---|---|---|---|
| 1 | Penetration through mucus gel to epithelial surface | Activities of mucinases, proteases, and hydrolytic and glycolytic enzymes | Secreted or outer membrane enzymes with putative enzyme activity or of unknown function S3126 (Yp), S1990 (Ph), S0734 (Ph), S3187 (Ec), S2105 (Ec), S3191 (Ec), S3197 (Ec) | PicF (S3178) |
| 2 | M cells translocate bacteria across epithelium | Attachment to M cell surface | Adhesins, fimbriae, hypothetical membrane ORFs S0211 (St), S0213-14 (St), S0215 (Sy), S0217 (St), S2105 (Ec), S3341 (Bc), S3194-5 (Ec), S4048 (Ec), S3197 (Ec), S3229 (Ec) | |
| 3 | Phagocytosis by macrophages; secretion of proinflammatory cytokines | Type III secretion system (plasmid)c | ||
| 4 | Induction of macrophage apoptosis | Specificity of toxicity for particular cell types | Secreted protein S1443 (Me) | IpaB (plasmid) |
| 5 | Binding to basolateral surfaces of colonic epithelial cells | Specificity for human colon; epithelial receptor | Adhesins, fimbriae, hypothetical membrane ORFs S0211 (St), S0213-14 (St), S0215 (Sy), S0217 (St), S2105 (Ec), S3341 (Bc), S3194-5 (Ec), S4048 (Ec), S3197 (Ec), S3229 (Ec) | |
| 6 | S. flexneri-induced uptake into epithelial cells by macropinocytosis | Type III secretion system (plasmid), LPS | ||
| 7 | Lysis of vacuole | Lysis of vacuolar membrane | IpaB, IpaH (plasmid) | |
| 8 | Bacterial replication in cytosol | Metabolic pathways utilized in the cytosol | Nutrient transport proteins S3636-42 (Eo) (PTS sor-like operon); S3114-8 (Eo), S1762 (Sy), S3968 (Eo), S4229 (Cj); metabolic enzymes, hpa operon S4643-55 (Ew) | TonB, CydC, VpsAC, Sit/Iuc/Feo |
| 9 | Actin-based motility | VirG/IcsA (plasmid), DksA | ||
| 10 | Intercellular spread | Interaction with cell-cell junction components | Outer membrane proteins S2105 (Ec), S3197 (Ec), S2341 (St) | |
| 11 | Lysis of double-membrane vacuole | Lysis of two membranes: one from inner face the other from outer face | Lytic secreted or outer membrane proteins S1443 (Me), S2105 (Ec), S2341 (St), S3197 (Ec); regulators of gene expression S2953 (Eo), S2956 (Ec), S4473 (St); S3212 (Vi), S1277 (Ec) | VacJ, plasmid type III secretion effectors, including IpaBC, IcsB, IpgC |
| 12 | Disruption of tight junctions by PMN transmigration and S. flexneri enterotoxins | Bacterial factors induce PMN transmigration and disrupt tight junctions | Secretion of proteins in intestinal lumen S3187 (Ec) | Plasmid type III secretion system; Shet1, Shet2, SigA |
| 13 | S. flexneri passing through open tight junctions | S. flexneri tropism for opened tight junctions | Outer membrane proteins S2105 (Ec), S2341 (St), S3197 (Ec) | |
| 14 | Infected cells secrete cytokines; intense acute inflammatory response | Surface lipoproteins, S0227 (Eo), S3870 (Eo), S3130 (Eo, partial) | LPS, Cld (plasmid) | |
| 15 | Innate immunity prevents systemic spread | Lipoproteins responsible for activity; additional antigens? | Surface lipoproteins S0227 (Eo), S3870 (Eo), S3130 (Eo, partial): surface antigens S0211 (St), S0213-14 (St), S0215 (Sy), S0217 (St), S2341 (St), S3194-S3195 (Ec), S3197 (Ec), S3341 (Bc), S4048 (Ec) | LPS, lipoproteins |
| 16 | Adaptive immune response appears to be T-lymphocyte independent | Mechanism of inhibiting T-lymphocyte response | Secreted or surface proteins S1443 (Me), S2105 (Ec), S2341 (St), S3197 (Ec) | |
| 17 | Chromosomal segments enhance virulence | Additional modulating factors | Regulators of virulence plasmid gene expression S1277 (Ec), S2953 (Eo), S2956 (Ec), S3212 (Vi), S4473 (St) | VirR |
Shown is what is known about the infectious stages of bacterial invasion and spread through the colonic mucosa. Known proteins have been characterized experimentally. Unknown processes are those for which some or all of the genetic determinants are not yet identified and the biochemical or physiological mechanisms remain hidden. Candidate island ORFs encoded in the genome sequence were selected on the basis of homology search results and transmembrane domain predictions and are denoted by the unique identifier (S number) assigned in the annotated 2457T GenBank entry. Other factors that influence virulence, such as the ability to survive passage through the stomach, are also poorly understood, but since they are shared with many other species, they are not included here. PMN, polymorphonuclear leukocyte; LPS, lipopolysaccharide.
Species with most similar proteins are in parentheses. Normal letters indicate homologs (i.e., >90% identity over >90% of query and target length). Italic letters indicate matches of <90% but still significant. Bc, Burkholderia cepacia; Cj, Campylobacter; Eo, Escherichia coli O157:H7; Ec, other E. coli pathogens; Ew, E. coli W; Me, Mesorhizobium loti; Sy, S. enterica serovar Typhi; St, S. enterica serovar Typhimurium; Sf, S. flexneri flexneri; Si, Sinorhizobium meliloti; Vi, Vibrio cholerae; Yp, Yersinia pestis.
The type III secretion system includes structural proteins (Mxi and Spa proteins), secreted effector proteins (including IpaBCDA, VirA, MxiC, Spa32, and IpaH), and chaperone proteins.
For steps 15 to 17, these processes, while not sequential steps in infection, are intimately involved in promoting or limiting the progression of infection and are most likely to involve bacterial components yet to be identified.
MATERIALS AND METHODS
Strain.
S. flexneri 2a 2457T was obtained from the Walter Reed Army Institute of Research. The sequenced strain has been redeposited in the American Type Culture Collection under accession no. ATCC 700930.
Genomic DNA preparation, libraries, and sequencing.
Bacteria were grown in Luria-Bertani (LB) medium at 37°C, and genomic DNA was prepared by R. A. Welch at the University of Wisconsin. The genomic DNA was released from bacteria embedded in agarose to prevent shearing during preparation (44). Whole-genome libraries in M13Janus (7) and pBluescript KS− (Stratagene) were prepared by using nebulization to randomly shear genomic DNA extracted from agarose by digestion with Gelase (Epicentre) (44). Random clones were sequenced by Applied Biosystems Prism dye-terminator chemistry, and data were collected with ABI377 and 3700 automated sequencers. Sequence reads (66,219 with an average length of 502 nucleotides [nt]) were assembled by Seqman Genome Edition (DNASTAR). Additional PCRs and sequencing reactions were performed to close gaps, improve coverage, and resolve sequence ambiguities. The final coverage was 7.2X. A whole-genome optical map (38) for restriction enzyme XhoI was prepared to aid the ordering of contigs during assembly and so that the end points and lengths of inversions could be confirmed.
Sequence analysis.
Potential open reading frames (ORFs) were defined by GeneMark.hmm (42) or Genequest (DNASTAR). All predicted proteins larger than 30 amino acids were searched against the nonredundant and local databases. tRNAs were identified with tRNAscan-SE (40). Alternative translation start sites were chosen to conform to the annotated MG1655 sequence. Frameshifts and point mutations were carefully verified for authenticity, and disrupted genes with homologs in K-12 were annotated as “pseudogenes.” Predicted backbone proteins were considered to be orthologs when matches to the corresponding K-12 protein exceeded 90% amino acid identity, alignments included at least 90% of both proteins, and no equivalent match was found elsewhere in the 2457T genome. The protein-level matches were also individually inspected to include genes with lower similarities within colinear regions of the genomes. The genome sequence was compared with that of MG1655 by the modified maximal exact match (MEM) alignment utility that was used for the comparison of EDL933 and K-12 (54). The genomic comparison with strain 301 was performed by a new multigenome comparison tool, Mauve.
Nucleotide sequence accession number.
The complete, annotated sequence was deposited in GenBank under accession no. AE014073.
RESULTS
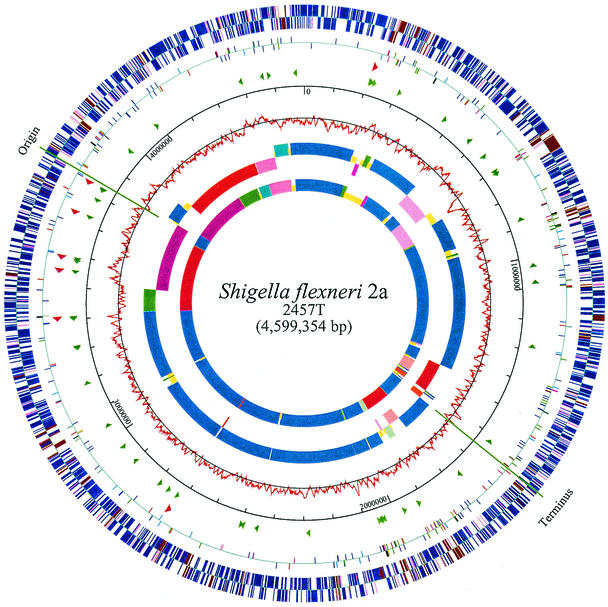
The genome consists of a single circular chromosome of 4,599,354 bp with a G+C content of 50.9%. Features of the genome and its comparison with E. coli K-12 (4) are shown in Fig. 1. Base pair 1 of the chromosome was assigned to correspond with bp 1 in K-12, since the two strains share extensive homology. The origin and terminus of replication were identified within homologous regions. The genome encodes 4,084 predicted genes, with an average size of 873 bp (926 bp if insertion sequences are excluded). The genome is slightly smaller than that of K-12 (4,639,221 bp), and its organization is roughly similar to that described for pathogenic E. coli strain O157:H7 EDL933 (54) and the uropathogen CFT073 (73), with large regions of colinear E. coli backbone punctuated by islands of sequence presumably acquired by horizontal transfer. The number of islands is smaller than those in CFT073 and O157:H7, and a larger proportion of the genome is backbone (82% versus 75% for O157:H7 and CFT073). There are 15 rearrangements >5kb in the genome (inversions and translocations) detected by comparison with K-12 (Fig. 1). Seven rRNA operons are present; their organization was altered from that in K-12 by genomic rearrangements. Ninety-eight tRNA genes include three copies of a novel cluster of four tRNAs (Ile, Arg, Thr, and Gly); only one of these (Gly) is identical to a K-12 tRNA. Each cluster in 2457T is in a prophage region, positioned downstream of the phage Q gene, as in the EDL933 Stx2 phage 933W (55).
FIG. 1.
Circular representation of the S. flexneri 2a 2457T genome and comparison with the E. coli K-12 genome. The outer circle shows the distribution of all ORFs. Blue represents ORFs in backbone regions with sequence identity to K-12, brown represents ORFs in S. flexneri-specific islands, and pink represents IS ORFs. The location outside or inside the axis denotes the direction of transcription. The second circle shows the IS elements; the predominant IS, IS1, is blue. Arrows in the third and fourth circles indicate rRNA (red) and tRNA (green). The fifth circle gives the genome scale in base pairs. The sixth circle shows the C/G skew calculated for each sliding window of 10 kb. In the comparison between S. flexneri 2a 2457T (seventh circle) and E. coli K-12 (the innermost circle), the segments in 2457T that are above (outside) the axis are colinear with K-12, and the segments below (inside) the axis are inverted relative to K-12. Since the difference in K-12 and 2457T genome lengths is small (0.8%) and large segments are homologous, the alignment between circles 7 and 8 is accurate within the limits of resolution for representation of the inversions. Blue represents colinear backbone, various shades of red represent backbone inverted in 2457T relative to K-12, various shades of green represent backbone translocated in 2457T relative to K-12, and yellow represents K-12- or 2457T-specific islands. Segments of the same color and length are homologous. The map was created by GenVision (DNASTAR).
Genome rearrangements.
Large symmetric chromosomal inversions spanning the replication origin and terminus have been observed when closely related bacterial species are compared (10, 13). The architecture of the S. flexneri genome has been affected by multiple large inversions compared to that of the K-12 genome, mostly spanning the axes of the origin and terminus of replication (inner circles in Fig. 1). Additional deletions and unequal crossover events have also taken place, resulting in two replichores of slightly unequal lengths, as found in the genome of Salmonella enterica serovar Typhi strain Ty2 (11). The rearrangement spanning the origin of replication is clearly indicated by the reorganization of the four rRNA operons nearest to it, which have been switched to the other replichore while maintaining their relative locations (shown by a red band in the seventh circle). Figure 1 also shows a smaller segment adjacent to the origin, within the larger inversion, that has reinverted without affecting any rRNA loci (shown by a dark blue band adjacent to the origin in seventh circle). Unlike the inter-replichore inversions reported in Yersinia pestis (10), S. enterica serovar Typhi (11, 39), and E. coli K-12 strain W3110 (28), those in S. flexneri are not associated with rRNA homologies, but instead the insertion sequence (IS) elements that are present at most of the inversion ends most probably mediated the chromosomal recombinations.
ISs.
The S. flexneri chromosome was known to be rich in insertion sequences (45, 53). The IS elements we identified (Table 2) make up 6.7% (309.4 kb) of the chromosome, in contrast to the typical ranges of 0 to ∼4%. The archaeon Sulfolobus solfataricus is a significant exception, because ∼10% of its 2.99-Mb genome is composed of ISs, which is unusual even among archea. In the sequenced E. coli genomes, the IS content is <1.5%, and in Y. pestis, the IS content is ∼3%. The virulence plasmid of S. flexneri also has an extremely high IS content (53% of the plasmid-encoded genes) (69). Of the 284 IS elements in 2457T, 108 are IS1X1 copies. The intact IS1 elements in this genome are typically families with 98 to 100% nucleotide sequence identity. Forty-six IS1 elements still have detectable flanking direct repeats, indicating recent acquisition (20 are full length, 9 bp; 24 are 8 bp; and 2 are 7 bp), and relatively little amelioration has occurred within these IS1 sequences. Comparative genome analysis with E. coli K-12 showed that 156 IS elements are involved in deletions or inversions associated with backbone rearrangements or with presumed horizontal transfer. The arrangements of several nested clusters of IS indicate that at each cluster, one integrated IS has acted as a target for subsequent insertions, resulting in multiple disrupted elements, with only the most recently acquired IS remaining intact.
TABLE 2.
S. flexneri 2457T IS elements
| IS familya | IS name (isotype) | IS size (nt) | No. of IS
|
|||
|---|---|---|---|---|---|---|
| Intact | Broken | Incomplete | Total | |||
| IS1 | IS1X1 | 766 | 105 | 3 | 108 | |
| IS1 | IS1N | 768 | 1 | 1 | ||
| IS2 | IS2 | 1,331 | 29 | 4 | 33 | |
| IS3 | IS3 | 1,258 | 5 | 1 | 6 | |
| IS3 | IS103 | 1,443 | 2 | 1 | 3 | |
| IS3 | IS600 | 1,264 | 35 | 4 | 8 | 47 |
| IS3 | IS629 | 1,310 | 12 | 2 | 14 | |
| IS3 | IS911 | 1,250 | 16 | 2 | 2 | 20 |
| IS3 | ISEhe3 | 1,245 | 3 | 2 | 1 | 6 |
| IS4 | IS4 | 1,426 | 19 | 3 | 22 | |
| IS91 | IS91 | 1,829 | 5 | 1 | 6 | |
| IS91 | IS1294 | 1,689 | 2 | 1 | 3 | |
| IS66 | ISSf13 | 2,729 | 5 | 3 | 2 | 10 |
| IS110 | ISSf14 | 1,451 | 5 | 5 | ||
| Total | 242 | 20 | 22 | 284 | ||
IS families and isotypes were identified in the IS database (43).
Islands.
Comparison of the S. flexneri and K-12 genome sequences revealed 37 islands >1 kb in the S. flexneri backbone that encode at least one gene not related to transposable elements. In contrast, EDL933 and CFT073 both have more than 100 islands >1 kb. The island ORFs show similarity with proteins in a wide range of organisms, including plant and animal pathogens with variety of lifestyles, indicating acquisition from many different sources (Table 3). Eight of the 37 S. flexneri islands encode a putative integrase, and seven islands are located at tRNAs: selC, leuX, aspV, asnT, argW, pheV, and glyU. Only four of the islands at tRNA sites include integrases. Unlike YSH600, a 2a serotype from Japan containing fec and resistance loci at serX (41), 2457T has no island at this site; the fec locus is elsewhere and is not associated with antibiotic resistance. Five of the islands show a cryptic prophage-like organization, and apparently there are two prophages together in two of the islands. Five other islands with few phage genes may also be prophage remnants, for a total of 12 putative prophages. All are cryptic, and the larger ones show mosaic structures that could have been produced by recombination between lambdoid phage genomes. In S. flexneri, the genes responsible for serotype conversion (modification of the basic O antigen via glucosylation and/or O acetylation) are encoded by lysogenic bacteriophages. Although in at least one serotype 2b strain, the type II antigen is encoded by an inducible bacteriophage, SfII (47), in 2457T, the serotype conversion genes (gtrAII, gtrBII or bgt, and gtrII) are part of a cryptic prophage disrupted by multiple IS elements and associated genome rearrangements. One island carries the remnant of an integrated plasmid, including arsenate resistance and plasmid replication genes. Islands lacking phage-like genes are generally bounded by IS elements, which have presumably mediated island integration. The predominance of matches to O157:H7 proteins (Table 3) probably reflects the contents of GenBank rather than suggesting a particularly close relationship between 2457T and O157:H7.
TABLE 3.
ORFs within islands of S. flexneri 2457T categorized by function
| Functional category | No. of ORFs | Species with homologs |
|---|---|---|
| Virulence | 10 | S. flexneri, Y. pestis |
| Adhesin | 7 | S. enterica serovar Typhimurium, other pathogenic E. coli, A. actinomycetemcomitans |
| Regulatory | 5 | E. coli O157:H7, S. enterica serovar Typhi, other pathogenic E. coli |
| Energy metabolism | 31 | other pathogenic E. coli, E. coli O157:H7, S. enterica serovar Typhimurium, C. crescentus, L. monocytogenes, S. enterica serovar Typhi |
| Iron uptake | 12 | S. flexneri, S. boydii, S. enterica serovar Typhimurium, S. enterica serovar Typhi, other pathogenic E. coli |
| Resistance to organic or inorganic chemicals | 7 | other pathogenic E. coli, E. coli O157:H7, C. crescentus, M. loti, A. tumefaciens |
| Transport | 11 | E. coli O157:H7, other pathogenic E. coli, S. enterica serovar Typhi |
| Membrane | 9 | E. coli O157:H7, Y. pestis, K. pneumoniae, C. jejuni, S. enterica serovar Typhi |
| Plasmid replication and transfer functions | 7 | C. crescentus, other pathogenic E. coli |
| DNA replication and transfer functions | 2 | S. flexneri, S. enterica serovar Typhimurium |
| Cell structure | 9 | E. coli O157:H7, other pathogenic E. coli, S. enterica serovar Typhi, S. enterica serovar Typhimurium |
| Biosynthesis | 8 | E. coli O157:H7, other pathogenic E. coli |
| Central intermediary metabolism | 9 | S. flexneri, V. cholerae |
| Conserved unknown | 68 | S. flexneri, nonpathogenic E. coli, other pathogenic E. coli, S. enterica serovar Typhi, P. aeruginosa, X. fastidiosa, S. meliloti |
Plasmids.
S. flexneri was known to harbor a large virulence plasmid, which contains all of the genes required to express the invasive phenotype (61), and two small multicopy plasmids. We sequenced all three plasmids from strain 2457T: pINV-2457T (218 kb), pSf2, and pSf4. We compared the sequence of pINV to those of three S. flexneri virulence plasmids: pWR100 (GenBank accession no. AL391753), pWR501 (AF348706), and pCP301 (AF386526). The results showed that they are all essentially identical, with a few IS element differences and ∼150 single-nucleotide differences distinguishing them. In the course of assembling the genome sequence of S. flexneri 2457T, we also unexpectedly identified a fourth plasmid of 165 kb. This was an S. enterica serovar Typhi R27-like plasmid, which we named “pSf-R27.” The R27 plasmid (62) was thought to be limited to Salmonella, in which it is implicated in the accumulation and spread of antibiotic resistance, but more recently, the similarity noted between R27 and pMT1, the large virulence plasmid of Y. pestis, suggested that there may have been a common ancestral plasmid. Sequence comparison showed that in pSf-R27, Tn10 (carrying tetracycline resistance genes), IS30, and a citrate uptake locus are absent, while the rest of the plasmid is 99.7% identical to R27. PCR was used to screen 142 S. flexneri isolates, including 57 of serotype 2a, for R27 sequences. The sequenced strain, 2457T, was the only strain to give a positive result. 2457T isolates from two other research groups that had obtained the strain from the same source were screened; the plasmid was found in one but not the other. Since 2457T was originally isolated before antibiotic usage had become widespread, it is possible that pSf-R27 may represent a primordial state of the R plasmid subsequently lost from the negative isolate, although we cannot formally exclude the possibility that pSf-R27 was accidentally introduced shortly after the strain was first isolated.
Pseudogenes.
While islands represent insertions into the S. flexneri genome, there are also a large number of gene disruptions and deletions. Disruptions resulted in 372 pseudogenes (8.1% of the genome), caused by several mechanisms, including single-nucleotide indels, point mutations, and IS elements. (IS alone accounts for 27 disruptions and 85 truncations.) Larger IS-mediated deletions and insertions are also seen. In total, 879 genes of K-12 are either absent or are pseudogenes in S. flexneri. Many types of function are missing (Table 4). The missing function is sometimes supplied by a plasmid- or island-encoded gene. The chromosomal fepE is a pseudogene; FepE is a homolog of Cld in K-12, encoding an O-antigen chain-elongation factor. An intact homolog is found on one of the small multicopy S. flexneri plasmids, and this FepE function is required for virulence (23, 65). Similarly, the mhp operon of K-12 is involved in catabolism of small aromatic molecules. Although it is missing from S. flexneri, an alternative system with similar activity is encoded by the hpa locus present on an island. This locus is also found in E. coli C and W and Y. pestis, but not K-12. K-12 genes missing from the S. flexneri backbone are clustered in K-12, suggesting either a single deletion event for each group in S. flexneri or their absence from a common ancestor, with later acquisition by K-12 via horizontal transfer. As an example, the island at tRNA leuX is completely different in K-12, EDL933, CFT073, and 2457T. Clearly, the four strains acquired these islands by distinct events, even if some could have been replacements rather than insertions. Phenotypic tests that have been widely used to distinguish E. coli from S. flexneri are largely explained by pseudogenes, which account for loss of flagellar motility; utilization of mucate, acetate, various sugars, and glycerol; and the requirement for NAD.
TABLE 4.
Pseudogenes and E. coli K-12 genes not present in 2457T, categorized by function
| Functional category | No. of pseudogenes
|
||
|---|---|---|---|
| Disrupted/truncated | Missing | Total | |
| Biosynthesis | 13 | 2 | 15 |
| Degradation | 13 | 30 | 43 |
| Metabolism | 29 | 29 | 58 |
| Cell structure | 36 | 27 | 63 |
| Transport | 53 | 71 | 124 |
| Cell processes | 9 | 15 | 24 |
| Regulatory | 26 | 36 | 62 |
| Factor | 10 | 18 | 28 |
| Putative enzymes | 50 | 77 | 127 |
| Hypothetical | 132 | 203 | 335 |
Phylogeny.
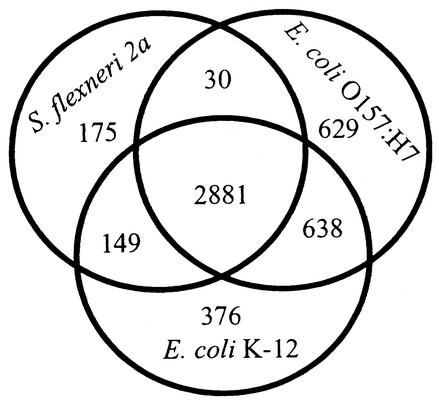
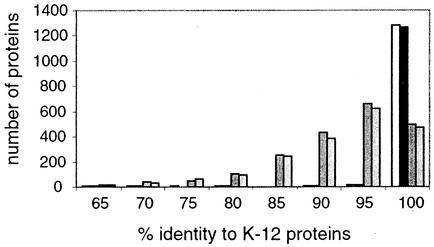
Despite their differences, there persists a high level of similarity among S. flexneri, K-12, and O157:H7. We show in Fig. 2 that the intact proteins shared by all three strains make up by far the largest category. In contrast, few proteins are shared by S. flexneri and O157:H7 but not K-12, demonstrating that the shared colinear backbone is the underlying feature connecting these genomes. The extensive backbone regions we identified in S. flexneri are consistent with phylogenetic reconstructions placing it among the members of the genus Escherichia (56, 57, 71). To examine the predicted proteins on a global scale, we compared backbone proteins in common among S. flexneri, O157:H7, and S. enterica serovars Typhi and Typhimurium (Fig. 3), and these results clearly show that S. flexneri and E. coli are indistinguishable, but quite distinct from the two Salmonella strains, supporting Reeves' suggestion that new nomenclature should be adopted to more accurately reflect the phylogeny (71).
FIG. 2.
Venn diagram showing the distribution of common and unique ORFs among S. flexneri 2a, E. coli K-12, and E. coli O157: H7. Only complete protein-coding ORFs, including hypothetical unknowns, are included. The IS element and phage ORFs, as well as pseudogenes, are excluded.
FIG. 3.
Comparison of backbone proteins. E. coli K-12 proteins with orthologs in all four pathogens (S. flexneri, E. coli O157: H7, and S. enterica serovars Typhi and Typhimurium), were aligned with each pathogen ortholog, and the percentage of identity was calculated. The results are plotted as a histogram. White bars, S. flexneri; black bars, O157: H7; dark gray bars, S. enterica serovar Typhimurium; light gray bars, S. enterica serovar Typhi.
Comparison with S. flexneri strain 301.
At the same time this paper was submitted, the genome sequence of S. flexneri strain 301 was published (25) under GenBank accession no. AE005674. This strain was isolated in 1984 from a patient in China, providing an interesting genome of the same serotype but geographically and temporally separated from 2457T. We compared the genome sequences and annotated features with those of 2457T. The genome of strain 301 is 4,607,203 bp, 7.85 kb larger than 2457T, which is largely accounted for by differences in IS complement, of which strain 301 has 247 complete and 6 partial ISs, whereas 2457T has 242 complete and 42 partial ISs. There are 45 IS loci that are different between the two strains. The genome sequences are very similar, but there are more than 1,400 single-nucleotide differences between them, scattered throughout. We found no evidence in 2457T for the unusual set of three spacer tRNAs (tRNAGlu, as well as tRNAIle and tRNAAla) in the rrnH operon in strain 301, and no example of this type appears in the RNA spacer region database (19). The spacer tRNAs also differ from those in K-12 and 2457T in the rrnA, rrnD, and rrnG operons.
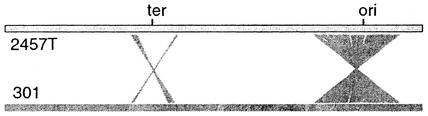
The genome of 2457T shows rearrangements relative to strain 301 (Fig. 4) as well as, and distinct from, those relative to K-12. Around the origin of replication, strain 301 is colinear with K-12, whereas 2457T is not. Around the terminus, a large inversion in 2457T relative to strain 301 was followed by reinversion of most of the DNA within the rearrangement (Fig. 4), leaving two small patches of inverted sequence marking the end points of the initial event. These recombinations were apparently mediated by IS elements.
FIG. 4.
Diagram of differences in genome organization between strains 2457T and 301. Diagonal lines join homologous regions that are not colinear in the two genomes. ter, terminus; ori, origin.
The island contents are similar in the two strains, but some islands show a different organization. Examples are found in the island containing the sitABCD genes and the islands at leuX and thrW. In 2457T, the sit island is integrated at tRNAGly, one of the novel tRNAs, but in strain 301, these tRNAs are distant, due to the rearrangement around the terminus. The thrW island is a serotype-converting prophage that contains several extra unknown genes in strain 301. The leuX island in 2457T contains the cadB pseudogene, which is not present in strain 301. No small multicopy plasmids were reported for strain 301.
The annotated strain 301 genome sequence shows 254 pseudogenes, compared with 372 pseudogenes in 2457T. Some of these differences are due to individual annotation criteria and styles, but 159 are pseudogenes in both strains, of which 42 have unknown functions. Each strain has its own unique set of pseudogenes. Those with known or predicted functions are listed in Table 5: 100 pseudogenes in 2457T and 20 in strain 301. The significance, if any, of the backbone and pseudogene complements of the two strains remains unclear.
TABLE 5.
Strain-specific pseudogenes
| ORF identifier | Gene | K-12 homolog | K-12 product |
|---|---|---|---|
| Strain 301 | |||
| SF4079 | metA | b4013 | Homoserine transsuccinylase |
| SF0057 | araA | b0062 | l-Arabinose isomerase |
| SF3734 | bglB | b3721 | Phospho-β-glucosidase B, cryptic oxygen-insensitive NAD(P)H nitroreductase |
| SF0485 | nfnB | b0578 | |
| SF1862 | zwf | b1852 | Glucose-6-phosphate dehydrogenase |
| SF2877 | prfB | b2891 | Peptide chain release factor RF-2 |
| SF1321 | ycjS | b1315 | Putative dehydrogenase |
| SF3214 | yhbX | b3173 | Putative alkaline phosphatase I |
| SF0275 | yafL | b0227 | Putative lipoprotein |
| SF1338, SF1842 | ycjZ | b1328 | Putative transcriptional regulator, LysR type |
| SF1534 | SF1534 | b1696 | Putative AraC-type regulator |
| SF2163 | yegW | b2101 | Putative transcriptional regulator |
| SF2448 | SF2448 | b2382 | Putative AraC-type regulator |
| SF2490 | yfeG | b2437 | Putative AraC-type regulator |
| SF3974 | frvR | b3897 | Putative frv operon regulator |
| SF1368 | SF1368 | b1345 | Putative transposase |
| SF1740 | SF1740 | b1485 | Putative transport protein |
| SF1691 | ydhE | b1663 | Putative transport protein |
| SF2477 | cysW | b2423 | Sulfate transport permease W |
| Strain 2457T | |||
| S4407 | bfr | b3336 | Bacterioferrin, iron storage |
| S1577 | gdhA | b1761 | NADP-specific glutamate dehydrogenase |
| S2171 | cobU | b1993 | Cobinamide kinase/cobinamide phosphate guanylyltransferase |
| S2637 | hemF | b2436 | Coproporphyrinogen III oxidase |
| S3390 | agaB | b3138 | PTS N-acetylgalactosamine-specific IIB component EIIB-AGA |
| S1776 | hdhA | b1619 | NAD-dependent 7α-hydroxysteroid dehydrogenase |
| S4637 | hsdR | b4350 | Host restriction endonuclease R |
| S3489 | degQ | b3234 | Serine endoprotease |
| S4408 | hofD | b3335 | Leader peptidase |
| S4636 | hsdM | b4349 | DNA methylase M |
| S1063 | torS | b0993 | Sensor protein |
| S1894 | narY | b1467 | Cryptic nitrate reductase 2 (β) |
| S2680 | hyfG | b2487 | Hydrogenase 4 subunit |
| S3606 | fdhF | b4079 | Selenopolypeptide subunit of formate dehydrogenase H |
| S4516 | cybC | b4236 | Cytochrome b562 |
| S2957 | rpoS | b2741 | Sigma S (sigma38) factor |
| S0832 | dacC | b0839 | d-Alanyl-d-alanine-carboxypeptidase |
| S4544 | fklB | b4207 | FKBP-type peptidyl-prolyl cis-trans isomerase (rotamase) |
| S4322 | rtcA | b3420 | RNA 3′-terminal phosphate cyclase |
| S1606 | celB | b1737 | PEP-dependent phosphotransferase enzyme II |
| S3990 | glvC | b3683 | PTS system, IIC component |
| S1316 | tpr | b1229 | Protaminelike protein |
| S2859 | intA | b2622 | Prophage CP4-57 integrase |
| S0478 | nfrB | b0569 | Bacteriophage N4 receptor |
| S0349 | yajB | b0404 | Putative glycoprotein |
| S2352 | yohG | b2138 | Putative channel/filament protein |
| S3355 | yhaI | b3104 | Putative cytochrome |
| S0041 | fixB | b0042 | Probable flavoprotein subunit |
| S0364 | yajO | b0419 | Putative NAD(P)H-dependent xylose reductase |
| S0448 | ybbP | b0496 | Putative oxidoreductase |
| S0459 | ylbF | b0520 | Putative carboxylase |
| S0830 | yliI | b0837 | Putative dehydrogenase |
| S1241 | S1241 | b1168 | Putative proteases |
| S1864 | pqqL | b1494 | Putative peptidase |
| S1858 | S1858 | b1498 | Putative sulfatase |
| S1724 | S1724 | b1587 | Putative oxidoreductase, major subunit |
| S1618 | S1618 | b1729 | Kinase (part) |
| S2241 | wcaE | b2055 | Putative glycosyl transferase |
| S2245 | wcaC | b2057 | Putative glycosyl transferase |
| S2484 | elaD | b2269 | Putative sulfatase/phosphatase |
| S2631 | S2631 | b2430 | Putative β-lactamase |
| S2737 | pbpC | b2519 | Putative peptidoglycan enzyme |
| S2869 | S2869 | b2657 | Putative enzyme |
| S3119 | yggC | b2928 | Putative kinase |
| S3321 | ygjH | b3074 | Putative tRNA synthetase |
| S4149 | yiaL | b3576 | Putative lipase/PICK> |
| ORF identifier | Gene | K-12 homolog | K-12 product |
| S4003 | yidX | b3696 | Putative replicase |
| S3846 | ysgA | b3830 | Putative enzyme |
| S3723 | talC | b3946 | Putative transaldolase |
| S4609 | aidB | b4187 | Putative acyl coenzyme A dehydrogenase |
| S2260 | S2260 | b2070 | Putative chaperonin |
| S2576 | S2576 | b2372 | Putative receptor protein |
| S3839 | S3839 | b3837 | Putative histone |
| S1450 | S1450 | b1377 | Putative outer membrane protein |
| S2264 | S2264 | b2074 | Putative membrane protein |
| S2548 | S2548 | b2337 | Putative outer membrane protein |
| S0954 | ycaN | b0900 | Putative transcriptional regulator |
| S2312 | yehI | b2118 | Putative regulator |
| S2684 | hyfR | b2491 | Putative 2-component regulator |
| S4526 | ytfQ | b4227 | Putative transcriptional regulator |
| S4668 | yjjQ | b4365 | Putative regulator |
| S0473 | fimZ | b0535 | Fimbrial regulator, probable signal transducer |
| S0641 | S0641 | b0663 | Putative RNA |
| S0135 | yadC | b0135 | Putative fimbrial protein |
| S1003 | ycbQ | b0938 | Putative fimbrial protein |
| S1098 | ycdV | b1031 | Putative ribosomal protein |
| S1657 | S1657 | b1502 | Putative adhesin, similar to FimH |
| S1506 | yebU | b1835 | Putative nucleolar protein |
| S2295 | yehA | b2108 | Putative type 1 fimbrial protein |
| S2544 | S2544 | b2333 | Putative fimbrial-like protein |
| S2546 | S2546 | b2335 | Putative fimbrial protein |
| S4665 | yjjP | b4364 | Putative structural protein |
| S2087 | fliP | b1948 | Flagellar export protein |
| S2472 | pmrD | b2259 | Polymyxin resistance protein B |
| S0801 | ybiO | b0808 | Putative transport protein |
| S0955 | S0955 | b0899 | Putative transport protein |
| S0998 | ycbM | b0934 | Putative transport system permease |
| S1137 | yceE | b1053 | Putative transport protein |
| S1242 | S1242 | b1169 | Putative ATP-binding component, transport system |
| S1243 | S1243 | b1170 | Putative ATP-binding component, transport system (part) |
| S1436 | ydaH | b1336 | Putative pump protein (transport) |
| S1875 | S1875 | b1483 | Putative ATP-binding component, transport system |
| S1677 | S1677 | b1543 | Putative transport protein |
| S2059 | fliY | b1920 | Putative periplasmic binding transport protein |
| S2895 | S2895 | b2681 | Putative transport protein |
| S4170 | yhiV | b3514 | Putative transport permease |
| S3797 | yihP | b3877 | Putative transport permease |
| S4522 | yjfF | b4231 | Putative transport permease |
| S2311 | molR | b2117 | Molybdate metabolism regulator |
| S1092 | phoH | b1020 | PhoB-dependent, ATP-binding Pho regulon component |
| S2672 | gcvR | b2479 | Transcriptional regulator |
| S2276 | gatR | b2090 | Galactitol utilization operon repressor, fragment 2 |
| S3922 | pssR | b3763 | Regulator of pssA |
| S1455 | feaR | b1384 | Regulator for 2-phenylethylamine catabolism |
| S2032 | flhD | b1892 | Regulator of flagellar biosynthesis, |
| S1158 | flgC | b1074 | Flagellar biosynthesis, cell-proximal portion of basal-body rod |
| S2081 | fliJ | b1942 | Flagellar protein |
| S2569 | dsdX | b2365 | Transport system permease |
| S2038 | araH | b1899 | High-affinity l-arabinose transporter |
| S3764 | rhaT | b3907 | Rhamnose transport |
New genes.
Even though our current knowledge of S. flexneri pathogenesis is detailed in some respects, much remains to be discovered (Table 1). Genome analysis provides important clues to linking processes with specific genes and products. For example, 10 islands may be involved in niche-specific processes or virulence. Some have been analyzed previously and shown to contribute to virulence, including Pic mucinase and ShET1 enterotoxin (2), as well as the aerobactin siderophore genes in SHI-2 at selC (70) (SHI-3 at pheU in S. boydii) (58). Eight other smaller and previously uncharacterized islands contain iron uptake and utilization clusters and putative adhesins. One contains the sit genes, encoding an iron uptake system. With the S. flexneri 2a sequence used as a probe, the sit genes were found in all S. flexneri species and enteroinvasive E. coli, but not in the other pathogenic and nonpathogenic E. coli strains examined (60). Expression of the Sit proteins is induced in the intracellular environment (60). Thus, the Sit system may play an important role in iron sequestration in the intracellular environment of the host. Also encoded in islands are possible specific adhesins, similar to components (LpfA and -C) of long polar fimbriae in S. enterica serovar Typhimurium. Others resemble the Saf proteins of S. enterica serovar Typhimurium.
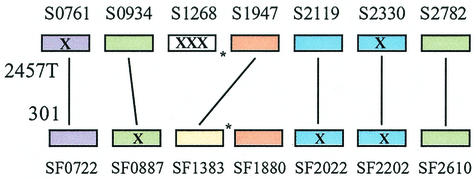
The IpaH proteins encoded on the virulence plasmid of S. flexneri (8, 68) consist of a conserved C-terminal domain and a variable N terminus containing a leucine-rich repeat (LRR). They are secreted by the plasmid type III secretion system, and at least one (IpaH7.8) has been shown to aid S. flexneri in escaping from the macrophage vacuole and is considered to be a virulence factor (16). There are five copies of ipaH on the plasmid, and we found seven more in the 2457T genome, of which four are intact, containing both the LRR domain and the conserved region. The genome sequence of strain 301 (25) also revealed four complete and three incomplete genomic copies. Figure 5 illustrates the differences between the ipaH genes in the two genomes. In both genomes, the incomplete copies are disrupted by insertion sequences or frameshifting mutations. One of the incomplete 2457T copies is highly divergent from all of the other ipaH genes.
FIG. 5.
Diagram of the organization of ipaH genes in the 2457T and 301 genomes. Lines connect genes in the same positions relative to backbone. X marks genes inactivated by insertion sequences or frameshift mutations. Similar colors denote homology of the N-terminal portions of the encoded proteins. Asterisks show the relative positions of the terminus of chromosomal replication; a chromosomal rearrangement in 2457T spanning the terminus accounts for S1947 having the same flanking backbone regions as SF1383. The figure is not drawn to scale.
DISCUSSION
The genome sequence of S. flexneri offers new candidate genes with potential for involvement in pathogenicity, including predicted proteins similar to virulence factors in other organisms. Among these data, missing links in S. flexneri pathogenesis may be found (Table 1). For example, the molecular mechanisms of species and tissue tropism, including the adhesins potentially specific for the human colonic epithelium, remain hidden. This is due in part to the lack of a suitable animal model. Mice do not become infected following oral inoculation of S. flexneri; therefore, mouse models have been restricted to pulmonary and conjunctival infections, which differ in important respects from colonic infection. Among the island ORFs of S. flexneri are 7 that are similar to adhesins from other pathogenic organisms and 68 that lack significant similarity to proteins of known function, including 9 predicted to encode secreted or membrane proteins, which are therefore strong candidates for mediating direct interactions with host cells. The unique complement of fimbrial adhesins in S. flexneri presumably underlies host specificity, as has been suggested for S. enterica serovar Typhi, another exclusively human pathogen (66). Of particular interest are the ORFs similar to the Salmonella SafABC. As S. enterica serovar Typhimurium is also an intracellular pathogen of intestinal epithelial cells and macrophages, this locus may encode components of an adhesin contributing to host or tissue specificity. In addition, ORFs S3961 and S4048 encode a major type 1 fimbrial subunit and usher protein essentially identical to proteins of enterohemorrhagic E. coli O113:H21, which is pathogenic for humans and cattle.
While a specific host cell receptor may not be the only valid explanation for host specificity, it is consistent with experimental data and in vivo observations. We emphasize that there are clear differences among the consequences of infection of cultured mammalian cells and inoculation of mice or humans. When grown as a nonpolarized and nonconfluent monolayer, cells from a wide variety of hosts and anatomic origins are readily invaded by S. flexneri. When grown as a polarized and confluent monolayer, S. flexneri invades cells only at the basolateral membrane (50). However, in the context of an intact animal host, only cells of the human or monkey colonic mucosa or mouse respiratory epithelium have been shown to be infected by S. flexneri. S. flexneri strains have not been shown to cause intestinal disease in nonprimates, and in mice, S. flexneri strains appear not to invade the colonic mucosa (M. B. Goldberg, unpublished data). Thus, while alternative explanations of S. flexneri species and tissue specificity exist, a specific receptor on polarized primate colonic cells might be involved in the specific invasion of this tissue. In particular, such a receptor might be important to S. flexneri gaining access to the basolateral sides of these cells.
Expression of receptor candidate proteins in nonpathogenic E. coli and screening for adherence to appropriate human tissue (24) might then allow the unique human cellular receptor to be identified (36). From there, the construction of a transgenic mouse model for S. flexneri infection is possible, as reported for Listeria monocytogenes (37), another human-specific intestinal pathogen that causes disease in humans but not mice. An improved animal model will greatly facilitate evaluation of candidate genes with possible roles in virulence.
Experimental evidence suggests that IpaH proteins may play a role in modulating the host response to infection. IpaH7.8 on the invasion plasmid was shown to help S. flexneri escape from macrophage vacuoles (16). Mutations in two ipaH genes on the invasion plasmid induce an exaggerated keratoconjunctivitis response with greater-than-normal inflammation in guinea pig eyes, and IpaH9.8 encoded on the plasmid was shown to translocate to the host nuclei in tissue culture cells (67), but the precise functions of these proteins remain unknown. Unlike the ipaH genes on the invasion plasmid, the genome-encoded ipaH genes are mostly associated with prophage-like islands, reminiscent of the Salmonella lambda-like Gifsy prophages, which encode effector proteins of the YopM/IpaH family (48). Lysogenic conversion with these phages is responsible for much of the diversity of the effector protein repertoires observed among Salmonella spp. (48). The finding that ipaH genes on the plasmid and chromosome may show strain-specific differences in sequences is a novel observation and might suggest that, like in Salmonella, the ipaH gene family might contribute to diversity of effector molecules. This remains to be tested.
IpaH proteins belong to the superfamily of LRR-containing proteins, which includes members from bacteria, plants, and vertebrates (6, 27). The conservation level of these proteins indicates that the LRR probably has structural or functional significance. IpaH-like proteins are found in the animal pathogens Salmonella, Yersinia, and Listeria, as well as the plant pathogens Rhizobium, Bradyrhizobium, and Ralstonia, again often associated with prophage (9, 18, 20, 35). In many host organisms, including plants, receptors involved in recognizing invading pathogens are also LRR proteins: for example, mammalian Toll-like receptors and the NB/LRR family in plants (1, 26). Experimental evidence accumulating from various studies of host-pathogen interactions is beginning to suggest that the bacterial effector proteins might interfere with or modulate the host receptor activity, presumably enabling the pathogen to evade the host's defensive response.
Acquisition of new traits by horizontal transfer has enabled microorganisms to survive in new niches. A complementary loss-of-function mechanism has been proposed (52, 64) by which virulence is enhanced through mutation of ancestral genes encoding factors that interfere with the expression or function of traits necessary for success in the new environment. Acquisition of the virulence plasmid enabled S. flexneri to enter the highly specialized intracellular environment in human intestinal epithelial cells. In this new niche, genes that were required in the intestinal lumen may be deleterious or are no longer beneficial and may accumulate mutations without a selective force to maintain them. Lysine decarboxylase (CadA) produces cadaverine, which inhibits the escape of S. flexneri from the vacuole into the cell cytosol (15, 46). Since S. flexneri replication and spread are dependent upon its access to the cytosol, biosynthesis of cadaverine attenuates virulence. In 2457T, cadA and cadC, which encodes a transcriptional activator of the cad operon, are deleted (entirely absent from the genome). Lack of surface structures such as flagella, fimbriae, and curli in S. flexneri provides the advantage of fewer antigens that can be easily recognized by the host immune system. In 2457T, of 14 dysfunctional genes of flagellar biosynthesis, 11 (fliF, fliJ, fliP, flgC, flgE, flgF, flgK, flgL, flhA, flhB, and cheR) contain frameshifts and 1 (fliA) contains a point mutation, while IS1 elements truncate flhD and flhE.
Although invasion and intercellular spread are well studied (51), many of the signaling and gene expression controls that orchestrate these processes are unknown (Table 1) and might provide new points of therapeutic intervention. Although S. flexneri is an intracellular pathogen, adaptive immunity to S. flexneri may be restricted to B-lymphocyte-dependent humoral responses. Human adaptive immunity is serotype specific, and exposure induces production of specific immunoglobulins (17, 59). In mouse models, adaptive immunity is completely independent of T-lymphocyte function (72). However, the mechanism by which S. flexneri modulates T-lymphocyte responses is unknown. With the sequence known, gene chips could now be used to interrogate expression profiles during infection, identifying all of the genes responding to the various changing conditions of particular interest, including oxidation, temperature shift, and iron depletion, which are specifically induced in the intracellular environment.
The high incidence of shigellosis and the proliferation of drug resistance have spurred serious efforts in vaccine development. Some success has been reported with live attenuated bacteria with mutations in the plasmid gene virG (necessary for intercellular spread), both alone and in combination with chromosomal deletions of aroA (aromatic amino acid synthesis), iuc (aerobactin), set (enterotoxin), or guaBA (purine biosynthesis pathway) (29-31). New candidate genes, when characterized, will provide alternative routes to further attenuation while maintaining antigenicity.
Because of its ability to enter into the cytosol of mammalian cells, S. flexneri strains have been developed as a delivery vehicle of antigens to major histocompatibility complex class I for immunization or of DNA into target cells for gene therapy (3, 12, 14, 63). Again, optimization of these approaches will require sufficient attenuation of the S. flexneri vehicle, specific binding to target cells, and controlled modulation of the immune response.
Knowledge of all the proteins encoded in the 2457T genome provides the entire repertoire of surface proteins that are potential vaccine targets, and candidates found to be adequately antigenic could therefore be used singly or in combination, engineered for expression from recombinant constructs, or even used directly in DNA vaccines. The sequence will also facilitate identification of many of the corresponding vaccine candidate genes in other S. flexneri serotypes, both type specific or in common. Comparison with the genome of nonpathogenic E. coli will reveal factors that, like cadaverine, block or limit survival of S. flexneri in host tissue. Thus, functions no longer active (pseudogenes) in S. flexneri but expressed in nonpathogenic E. coli may lead to the development of novel S. flexneri-specific therapies by virtue of a suppressive effect on bacterial growth or tissue invasion. These genome-driven research activities will serve as starting points for a new phase of vaccine and molecular pathogenicity investigation.
Acknowledgments
We thank the members of the University of Wisconsin genomics team for expert technical assistance.
This work was supported by Public Health Service grants AI-44387 to F.R.B and AI-43562 to M.B.G.
J.W., M.B.G., and V.B. contributed equally to this work.
Editor: J. T. Barbieri
Footnotes
Paper no. 3603 from the Laboratory of Genetics.
REFERENCES
- 1.Aderem, A., and R. J. Ulevitch. 2000. Toll-like receptors in the induction of the innate immune response. Nature 406:782-787. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hasani, K., I. R. Henderson, H. Sakellaris, K. Rajakumar, T. Grant, J. P. Nataro, R. Robins-Browne, and B. Adler. 2000. The sigA gene which is borne on the she pathogenicity island of Shigella flexneri 2a encodes an exported cytopathic protease involved in intestinal fluid accumulation. Infect. Immun. 68:2457-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, R. J., M. F. Pasetti, M. B. Sztein, M. M. Levine, and F. R. Noriega. 2000. ΔguaBA attenuated Shigella flexneri2a strain CVD 1204 as a Shigella vaccine and as a live mucosal delivery system for fragment C of tetanus toxin. Vaccine 18:2193-2202. [DOI] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Brenner, D. J., G. R. Fanning, A. G. Steigerwalt, I. Ørskov, and F. Ørskov. 1972. Polynucleotide sequence relatedness among three groups of pathogenic Escherichia coli strains. Infect. Immun. 6:308-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchanan, S. G., and N. J. Gay. 1996. Structural and functional diversity in the leucine-rich repeat family of proteins. Prog. Biophys. Mol. Biol. 65:1-44. [DOI] [PubMed] [Google Scholar]
- 7.Burland, V., D. L. Daniels, G. Plunkett III, and F. R. Blattner. 1993. Genome sequencing on both strands: the Janus strategy. Nucleic Acids Res. 21:3385-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buysse, J. M., A. B. Hartman, N. Strockbine, and M. Venkatesan. 1995. Genetic polymorphism of the ipaH multicopy antigen gene in Shigella spps. and enteroinvasive Escherichia coli. Microb. Pathog. 19:335-349. [DOI] [PubMed] [Google Scholar]
- 9.Cossart, P., J. Pizarro-Cerda, and M. Lecuit. 2003. Invasion of mammalian cells by Listeria monocytogenes: functional mimicry to subvert cellular functions. Trends Cell Biol. 13:23-31. [DOI] [PubMed] [Google Scholar]
- 10.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng, W., S.-R. Liou, G. Plunkett III, G. F. Mayhew, D. J. Rose, V. Burland, V. Kodoyianni, D. C. Schwartz, and F. R. Blattner. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devico, A. L., T. R. Fouts, M. T. Shata, R. Kamin-Lewis, G. K. Lewis, and D. M. Hone. 2002. Development of an oral prime-boost strategy to elicit broadly neutralizing antibodies against HIV-1. Vaccine 20:1968-1974. [DOI] [PubMed] [Google Scholar]
- 13.Eisen, J. A., J. F. Heidelberg, O. White, and S. L. Salzberg. 2000. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 1(6):0011.1-0011.9. [Online.] http://genomebiology.com. [DOI] [PMC free article] [PubMed]
- 14.Fennelly, G. J., S. A. Khan, M. A. Abadi, T. F. Wild, and B. R. Bloom. 1999. Mucosal DNA vaccine immunization against measles with a highly attenuated Shigella flexneri vector. J. Immunol. 162:1603-1610. [PubMed] [Google Scholar]
- 15.Fernandez, I. M., M. Silva, R. Schuch, W. A. Walker, A. M. Siber, A. T. Maurelli, and B. A. McCormick. 2001. Cadaverine prevents the escape of Shigella flexneri from the phagolysosome: a connection between bacterial dissemination and neutrophil transepithelial signaling. J. Infect. Dis. 184:743-753. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-Prada, C. M., D. L. Hoover, B. D. Tall, A. B. Hartman, J. Kopelowitz, and M. M. Venkatesan. 2000. Shigella flexneri IpaH7.8 facilitates escape of virulent bacteria from the endocytic vacuoles of mouse and human macrophages. Infect. Immun. 68:3608-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Formal, S. B., E. V. Oaks, R. E. Olsen, M. Wingfield-Eggleston, P. J. Snoy, and J. P. Cogan. 1991. Effect of prior infection with virulent Shigella flexneri 2a on the resistance of monkeys to subsequent infection with Shigella sonnei. J. Infect. Dis. 164:533-537. [DOI] [PubMed] [Google Scholar]
- 18.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Martinez, J., I. Bescos, J. J. Rodriguez-Sala, and F. Rodriguez-Valera. 2001. RISSC: a novel database for ribosomal 16S-23S RNA genes spacer regions. Nucleic Acids Res. 29:178-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grosdent, N., I. Maridonneau-Parini, M.-P. Sory, and G. R. Cornelis. 2002. Role of Yops and adhesins in resistance of Yersinia enterocolitica to phagocytosis. Infect. Immun. 70:4165-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson, I. R., J. Czeczulin, C. Eslava, F. Noriega, and J. P. Nataro. 1999. Characterization of Pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun. 67:5587-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong, M., Y. Gleason, E. E. Wyckoff, and S. M. Payne. 1998. Identification of two Shigella flexneri chromosomal loci involved in intercellular spreading. Infect. Immun. 66:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong, M., and S. M. Payne. 1997. Effect of mutations in Shigella flexneri chromosomal and plasmid-encoded lipopolysaccharide genes on invasion and serum resistance. Mol. Microbiol. 24:779-791. [DOI] [PubMed] [Google Scholar]
- 24.Isberg, R. R., D. L. Voorhis, and S. Falkow. 1987. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell 50:769-778. [DOI] [PubMed] [Google Scholar]
- 25.Jin, Q., Z. Yuan, J. Xu, Y. Wang, Y. Shen, W. Lu, J. Wang, H. Liu, J. Yang, F. Yang, X. Zhang, J. Zhang, G. Yang, H. Wu, D. Qu, J. Dong, L. Sun, Y. Xue, A. Zhao, Y. Gao, J. Zhu, B. Kan, K. Ding, S. Chen, H. Cheng, Z. Yao, B. He, R. Chen, D. Ma, B. Qiang, Y. Wen, Y. Hou, and J. Yu. 2002. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 30:4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kjemtrup, S., Z. Nimchuk, and J. L. Dangl. 2000. Effector proteins of phytopathogenic bacteria: bifunctional signals in virulence and host recognition. Curr. Opin. Microbiol. 3:73-78. [DOI] [PubMed] [Google Scholar]
- 27.Kobe, B., and A. V. Kajava. 2001. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11:725-732. [DOI] [PubMed] [Google Scholar]
- 28.Kohara, Y., K. Akiyama, and K. Isono. 1987. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell 50:495-508. [DOI] [PubMed] [Google Scholar]
- 29.Kotloff, K. L., F. Noriega, G. A. Losonsky, M. B. Sztein, S. S. Wasserman, J. P. Nataro, and M. M. Levine. 1996. Safety, immunogenicity, and transmissibility in humans of CVD 1203, a live oral Shigella flexneri 2a vaccine candidate attenuated by deletions in aroA and virG. Infect. Immun. 64:4542-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotloff, K. L., F. R. Noriega, T. Samandari, M. B. Sztein, G. A. Losonsky, J. P. Nataro, W. D. Picking, E. M. Barry, and M. M. Levine. 2000. Shigella flexneri 2a strain CVD 1207, with specific deletions in virG, sen, set, and guaBA, is highly attenuated in humans. Infect. Immun. 68:1034-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kotloff, K. L., D. N. Taylor, M. B. Sztein, S. S. Wasserman, G. A. Losonsky, J. P. Nataro, M. Venkatesan, A. Hartman, W. D. Picking, D. E. Katz, J. D. Campbell, M. M. Levine, and T. L. Hale. 2002. Phase I evaluation of ΔvirG Shigella sonnei live, attenuated, oral vaccine strain WRSS1 in healthy adults. Infect. Immun. 70:2016-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotloff, K. L., J. P. Winickoff, B. Ivanoff, J. D. Clemens, D. L. Swerdlow, P. J. Sansonetti, G. K. Adak, and M. M. Levine. 1999. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull. W. H. O. 77:651-666. [PMC free article] [PubMed] [Google Scholar]
- 33.LaBrec, E. H., H. Schneider, T. J. Magnani, and S. B. Formal. 1964. Epithelial cell penetration as an essential step in the pathogenesis of bacillary dysentery. J. Bacteriol. 88:1503-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lan, R., B. Lumb, D. Ryan, and P. R. Reeves. 2001. Molecular evolution of large virulence plasmid in Shigella clones and enteroinvasive Escherichia coli. Infect. Immun. 69:6303-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavie, M., E. Shillington, C. Eguiluz, N. Grimsley, and C. Boucher. 2002. PopP1, a new member of the YopJ/AvrRxv family of type III effector proteins, acts as a host-specificity factor and modulates aggressiveness of Ralstonia solanacearum. Mol. Plant-Microbe Interact. 15:1058-1068. [DOI] [PubMed] [Google Scholar]
- 36.Lecuit, M., S. Dramsi, C. Gottardi, M. Fedor-Chaiken, B. Gumbiner, and P. Cossart. 1999. A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 18:3956-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lecuit, M., S. Vandormael-Pournin, J. Lefort, M. Huerre, P. Gounon, C. Dupuy, C. Babinet, and P. Cossart. 2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292:1722-1725. [DOI] [PubMed] [Google Scholar]
- 38.Lim, A., E. T. Dimalanta, K. D. Potamousis, G. Yen, J. Apodoca, C. Tao, J. Lin, R. Qi, J. Skiadas, A. Ramanathan, N. T. Perna, G. Plunkett III, V. Burland, B. Mau, J. Hackett, F. R. Blattner, T. S. Anantharaman, B. Mishra, and D. C. Schwartz. 2001. Shotgun optical maps of the whole Escherichia coli O157:H7 genome. Genome Res. 11:1584-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, S. L., and K. E. Sanderson. 1995. Rearrangements in the genome of the bacterium Salmonella typhi. Proc. Natl. Acad. Sci. USA 92:1018-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luck, S. N., S. A. Turner, K. Rajakumar, H. Sakellaris, and B. Adler. 2001. Ferric dicitrate transport system (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes. Infect. Immun. 69:6012-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lukashin, A. V., and M. Borodovsky. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahillon, J., H. A. Kirkpatrick, H. L. Kijenski, C. A. Bloch, C. K. Rode, G. F. Mayhew, D. J. Rose, G. Plunkett III, V. Burland, and F. R. Blattner. 1998. Subdivision of the Escherichia coli K-12 genome for sequencing: manipulation and DNA sequence of transposable elements introducing unique restriction sites. Gene 223:47-54. [DOI] [PubMed] [Google Scholar]
- 45.Matsutani, S., and E. Ohtsubo. 1993. Distribution of the Shigella sonnei insertion elements in Enterobacteriaceae. Gene 127:111-115. [DOI] [PubMed] [Google Scholar]
- 46.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 95:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mavris, M., P. A. Manning, and R. Morona. 1997. Mechanism of bacteriophage SfII-mediated serotype conversion in Shigella flexneri. Mol. Microbiol. 26:939-950. [DOI] [PubMed] [Google Scholar]
- 48.Mirold, S., W. Rabsch, H. Tschape, and W. D. Hardt. 2001. Transfer of the Salmonella type III effector SopE between unrelated phage families. J. Mol. Biol. 312:7-16. [DOI] [PubMed] [Google Scholar]
- 49.Mogull, S. A., L. J. Runyen-Janecky, M. Hong, and S. M. Payne. 2001. dksA is required for intercellular spread of Shigella flexneri via an RpoS-independent mechanism. Infect. Immun. 69:5742-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mounier, J., T. Vasselon, R. Hellio, M. Lesourd, and P. J. Sansonetti. 1992. Shigella flexneri enters human colonic Caco-2 epithelial cells through the basolateral pole. Infect. Immun. 60:237-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nhieu, G. T., and P. J. Sansonetti. 1999. Mechanism of Shigella entry into epithelial cells. Curr. Opin. Microbiol. 2:51-55. [DOI] [PubMed] [Google Scholar]
- 52.Ochman, H., and N. A. Moran. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096-1099. [DOI] [PubMed] [Google Scholar]
- 53.Ohtsubo, H., K. Nyman, W. Doroszkiewicz, and E. Ohtsubo. 1981. Multiple copies of iso-insertion sequences of IS1 in Shigella dysenteriae chromosome. Nature 292:640-643. [DOI] [PubMed] [Google Scholar]
- 54.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 55.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pupo, G. M., D. K. Karaolis, R. Lan, and P. R. Reeves. 1997. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect. Immun. 65:2685-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pupo, G. M., R. Lan, and P. R. Reeves. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. USA 97:10567-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Purdy, G. E., and S. M. Payne. 2001. The SHI-3 iron transport island of Shigella boydii 0-1392 carries the genes for aerobactin synthesis and transport. J. Bacteriol. 183:4176-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robin, G., D. Cohen, N. Orr, I. Markus, R. Slepon, S. Ashkenazi, and Y. Keisari. 1997. Characterization and quantitative analysis of serum IgG class and subclass response to Shigella sonnei and Shigella flexneri 2a lipopolysaccharide following natural Shigella infection. J. Infect. Dis. 175:1128-1133. [DOI] [PubMed] [Google Scholar]
- 60.Runyen-Janecky, L. J., and S. M. Payne. 2002. Identification of chromosomal Shigella flexneri genes induced by the eukaryotic intracellular environment. Infect. Immun. 70:4379-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sansonetti, P. J., D. J. Kopecko, and S. B. Formal. 1982. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect. Immun. 35:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sherburne, C. K., T. D. Lawley, M. W. Gilmour, F. R. Blattner, V. Burland, E. Grotbeck, D. J. Rose, and D. E. Taylor. 2000. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 28:2177-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sizemore, D. R., A. A. Branstrom, and J. C. Sadoff. 1995. Attenuated Shigella as a DNA delivery vehicle for DNA-mediated immunization. Science 270:299-302. [DOI] [PubMed] [Google Scholar]
- 64.Sokurenko, E. V., D. L. Hasty, and D. E. Dykhuizen. 1999. Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 7:191-195. [DOI] [PubMed] [Google Scholar]
- 65.Stevenson, G., A. Kessler, and P. R. Reeves. 1995. A plasmid-borne O-antigen chain length determinant and its relationship to other chain length determinants. FEMS Microbiol. Lett. 125:23-30. [DOI] [PubMed] [Google Scholar]
- 66.Townsend, S. M., N. E. Kramer, R. Edwards, S. Baker, N. Hamlin, M. Simmonds, K. Stevens, S. Maloy, J. Parkhill, G. Dougan, and A. J. Bäumler. 2001. Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect. Immun. 69:2894-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Toyotome, T., T. Suzuki, A. Kuwae, T. Nonaka, H. Fukuda, S. Imajoh-Ohmi, T. Toyofuku, M. Hori, and C. Sasakawa. 2001. Shigella protein IpaH9.8 is secreted from bacteria within mammalian cells and transported to the nucleus. J. Biol. Chem. 276:32071-32079. [DOI] [PubMed] [Google Scholar]
- 68.Venkatesan, M. M., J. M. Buysse, and A. B. Hartman. 1991. Sequence variation in two ipaH genes of Shigella flexneri 5 and homology to the LRG-like family of proteins. Mol. Microbiol. 5:2435-2445. [DOI] [PubMed] [Google Scholar]
- 69.Venkatesan, M. M., M. B. Goldberg, D. J. Rose, E. J. Grotbeck, V. Burland, and F. R. Blattner. 2001. Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect. Immun. 69:3271-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vokes, S. A., S. A. Reeves, A. G. Torres, and S. M. Payne. 1999. The aerobactin iron transport system genes in Shigella flexneri are present within a pathogenicity island. Mol. Microbiol. 33:63-73. [DOI] [PubMed] [Google Scholar]
- 71.Wang, L., W. Qu, and P. R. Reeves. 2001. Sequence analysis of four Shigella boydii O-antigen loci: implication for Escherichia coli and Shigella relationships. Infect. Immun. 69:6923-6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Way, S. S., A. C. Borczuk, and M. B. Goldberg. 1999. Thymic independence of adaptive immunity to the intracellular pathogen Shigella flexneri serotype 2a. Infect. Immun. 67:3970-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S.-R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. T. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of a uropathogenic strain of Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]