Abstract
The homolactic and catalase-deficient pathogen Streptococcus pneumoniae is not only tolerant to oxygen but requires the activity of its NADH oxidase, Nox, to develop optimal virulence and competence for genetic transformation. In this work, we show that the global regulator RegR is also involved in these traits. Genetic dissection revealed that RegR regulates competence and the expression of virulence factors, including hyaluronidase. In bacteria grown in vitro, RegR represses hyaluronidase. At neutral pH, it increases adherence to A549 epithelial cells, and at alkaline pH, it acts upstream of the CiaRH two-component signaling system to activate competence. These phenotypes are not associated with changes in antibiotic resistance, central metabolism, and carbohydrate utilization. Although the RegR0 (where 0 indicates the loss of the protein) mutation is sufficient to attenuate experimental virulence of strain 23477 in mice, the introduction of an additional hyl0 (where 0 indicates the loss of function) mutation in the RegR0 strain 23302 dramatically reduces its virulence. This indicates that residual virulence of the RegR0 Hyl+ derivative is due to hyaluronidase and supports the dual role of RegR in virulence. This LacI/GalR regulator, not essential for in vitro growth in rich media, is indeed involved in the adaptive response of the pneumococcus via its control of competence, adherence, and virulence.
Genetic transformability is one of the major attributes of the human pathogen Streptococcus pneumoniae. In vitro, this property appears as a consequence of a sequence of events leading to competence development in cultures growing exponentially, under aerobiosis. Competence and virulence are highly regulated, and there is increasing evidence linking the two phenomena at the genetic level. For instance, the ComD histidine kinase, the target for the competence-stimulating peptide encoded by comC (13, 23, 24, 37), is required for virulence (28). Transcription of the comCDE operon is negatively controlled by the two-component system CiaRH (18), which is also involved in virulence (47). In addition, the virulence factor LytA, a choline binding protein showing autolytic activity (8, 20), belongs to a late competence operon (32). Clearly, virulence expression depends on events involved in the competence signaling pathway. Furthermore, mutational alteration of cation transporters and metabolic enzymes decreases both competence and virulence. For instance, mutations affecting the transport of calcium have consequences on competence development, LytA-dependent autolysis, and virulence (4, 48, 49). The ABC transporters encoded by psa (and adc), involved in the uptake of Mn2+ (and possibly Zn2+), are important for growth and competence (15), and the psaA product is an essential virulence factor (10). Mutations of LicD2 and of Nox lead to the alteration of both competence and virulence expression (2, 28, 51, 52). For competence, the NADH oxidase Nox influences the pattern of ComCDE expression and transformability in cultures grown aerobically (17). However, whereas competence is totally inhibited under microaerobiosis (18), the loss of function of Nox partially reduces competence in cultures growing aerobically. This suggests that, in addition to Nox, complementary pathways regulate competence development. By mutational analysis and genetic dissection, we have investigated the role in competence development, virulence, sugar utilization, adherence, and antibiotic susceptibility of RegR, a LacI/GalR homolog. We present evidence suggesting that in the absence of significant effects on growth, central metabolism, sugar utilization, and antibiotic susceptibility, RegR0 (where 0 indicates the loss of the protein) mutations attenuate virulence and lower competence.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All the strains used in this study are listed in Table 1. Escherichia coli was stored and grown under standard conditions (43). For S. pneumoniae, growth media and conditions were used as described by Clavé and Trombe (14). In order to test growth on a panel of sugars, frozen cultures (optical density at 400 nm [OD400], 0.4) of the Cp1015 or Cp8110 strain were thawed and inoculated in 50 volumes of the following medium: CAT (Casitone, 10 g liter−1; tryptone, 5 g liter−1; NaCl, 5 g liter−1; yeast extract, 1 g liter−1), 17 mM K2HPO4, 0.4% bovine serum albumin, 1 mM CaCl2, and 0.2% tested sugar. The medium was adjusted to pH 7 with NaOH.
TABLE 1.
Strains, plasmids, and oligonucleotide primers used in this study
| Bacterial strain, plasmid, or primer | Relevant characteristic(s) | Source or reference |
|---|---|---|
| S. pneumoniae strains | ||
| 23477 | Blood isolate, virulent, serotype 6, Strr | V. Rieux and M.-C. Trombe, unpublished data |
| 23450 | Strr, Nox0, Kmr | 2 |
| 23302 | Product of transformation of 23477 with pTLS34; Strr, RegR0, Kmr | This work |
| 23020 | Product of transformation of 23477 with pVA891-hyl; Strr, Hyl0, Emr | This work |
| 23322 | Product of transformation of 23302 with D39 Δhyl DNA; Strr, RegR0, Hyl0, Kmr, Emr | This work |
| D39 | Blood isolate, virulent, serotype 2, Strr | 3 |
| D39::ΔregR | Product of transformation of D39 with 23302 DNA; RegR0, Kmr | This work |
| D39::Δhyl | Product of transformation of D39 with pVA891-hyl; Hyl0, Emr | 11 |
| D39::ΔregR-Δhyl | Product of transformation of D39::ΔregR with D39::Δhyl DNA; RegR0, Hyl0, Kmr, Emr | This work |
| Cp1015 | Wild type, Strr, HexA0 | 31 |
| Cp1016 | Rifr derivative of Cp1015; rif-23 | Laboratory stocks |
| Cp8302 | Product of transformation of Cp1015 with pTLS34; Strr, HexA0, RegR0, Kmr | This work |
| Cp8110 | Product of transformation of Cp1015 with pTSS10; Strr, HexA0, RegRcds 3K→stopa | This work |
| Cp8056 | Product of transformation of Cp1015 with pN6; Strr, HexA0, Noxcds 71K→stop | 2 |
| Cp8156 | Product of transformation of Cp8056 with pTSS10; Strr, HexA0, RegRcds 3K→stop, Noxcds 71K→stop | This work |
| Cp1800 | Product of transformation of Cp1015 with pPT4; Strr, HexA0, CiaR0, Spr | 18 |
| Cp1810 | Product of transformation of Cp8110 with pPT4; Strr, HexA0, RegRcds 3Q→stop, CiaR0, Spr | This work |
| Cp1020 | Product of transformation of Cp1015 with D39::Δhyl DNA; Strr, HexA0, Hyl0, Emr | This work |
| Cp1120 | Product of transformation of Cp1020 with pTSS10; Strr, HexA0, RegRcds 3K→stop, Hyl0, Emr | This work |
| E. coli strain | ||
| DH5α | supE44Δlac U169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories, 1986 |
| CJ236 | dutI ungI thi-1 relAI/pCJ105 Camr F′ | 27 |
| TG1 | supE hsdΔ5 thi Δ(lac-proAB) F′(traD36 proAB+ laclqlacZΔM15) | 21 |
| Plasmids | ||
| pSP2 | Streptococcal cloning vector; Emr, Tetr | 39 |
| pBluesk− | Ampr, lacZ′; ColE1 origin | 45 |
| pPJl | pUC derivative containing an HincII fragment carrying the kanamycin resistance gene aphA3; Apr, Kmr | 36 |
| pVA891 | Shuttle cloning vector; Emr, Tetr, Camr | 29 |
| pAM239 | pBR322 derivative, ColE1 origin, lacZα selection, Spr | 22 |
| p131 | 5.5-kb BamHI chromosomal fragment of the 15-min region (7 ORFs) of Cp1015 cloned into pSP2 | 34 |
| pBluesk ΔEH | Plasmid derivative of pBluescript− resulting in deletion of the EcoRI-HindIII fragment of the multicloning site | This work |
| pTPS711 | 3.5-kb SpeI-EcoRI chromosomal fragment from Cp1015 containing orfA to regR cloned into λZAP Express; Kmr | R. Palmen, unpublished data |
| pTLS30 | 1-2092 BamHI/BclI fragment of p131 cloned into the BamHI site of pBluesk−ΔEH | This work |
| pTLS34 | Exchange of the 549-1675 HincII fragment of pTLS30 with the aphA3 cassette (HincII fragment) of pPJ1 | This work |
| PTSS9 | 2.3-kb EcoRI/EcoRI fragment of pTPS711 cloned into the EcoRI site of pBluesk− | This work |
| pTSS10 | M13 mutagenesis of pTSS9 by using the mutagenic primer reg7 | This work |
| pPT4 | 0.46-kb KpnI-PstI amplicon containing an internal fragment of ciaR, cloned into pAM239 | 18 |
| pVA891-hyl | 673-bp ClaI-NcoI fragment (from nucleotides 1286 to 1959 of the hyl open reading frame) cloned into the ClaI site of pAV891 | 11 |
| Primers | ||
| PC2 | 5′-AAATTTTCAATATTCCTAGTCGTAACCATC | This work |
| P100 | 5′-ATATTGAAGGGGCTTATTGATCAGCAGAAC | This work |
| reg7 | 5′-ATGGTCAGTTTACTAGTCCAATCTAA | This work |
| SpeI |
CDS, codon stop replacing the wild-type sense codon.
Single-strand DNA preparation.
Single-strand DNA preparation of the pTSS9 plasmid was performed as previously described by using the M13K07 lytic bacteriophage and E. coli CJ236 as a recipient strain (43). Briefly, a fresh stock of phage was prepared as follows. TG1 cells were grown to an OD595 of 1 in NZY medium (NZ amine, 10 g liter−1; NaCl, 5 g liter−1; Casamino Acid, 1 g liter−1; MgSO4, 2.5 g liter−1; yeast extract, 5 g liter−1). The culture was centrifuged, and the pellet was resuspended to half the original volume in 10 mM MgSO4. Aliquots of this suspension were diluted 1:5 with different dilutions of M13K07 in NZY medium, and the mixture was incubated at room temperature for 20 min. Five milliliters of prewarmed (55°C) top agarose (NZ anime, 10 g liter−1; NaCl, 5 g liter−1; Casamino Acid, 1 g liter−1; MgSO4, 2.5 g liter−1; agarose, 7.5 g liter−1) was then added, and the suspension was poured onto 1.5% agar-NZY plates. After overnight incubation at 37°C, 10 plaques were picked from each plate, resuspended in 10 ml of NZY medium, mixed with 100 μl of TG1, and incubated overnight at 37°C in a shaking water bath. The suspension was then centrifuged, and the supernatant was aliquoted and stored at 4°C (stock solution). The amount of phage was evaluated by titration.
In order to recover single-strand wild-type DNA, 20 μl of the M13K07 stock solution (corresponding to 1010 PFU) was mixed with one colony of strain CJ236 transformed by pTSS9, and the mixture was incubated for 30 min at room temperature. Five hundred microliters of NZY medium plus ampicillin (250 μg ml−1) was added, and the mixture was incubated for 1 h at 37°C. An aliquot of 400 μl was then resuspended in 10 ml of NZY medium plus ampicillin (250 μg ml−1) and kanamycin (80 μg ml−1) and incubated 48 h at 37°C with shaking in a water bath. The culture was centrifuged for 10 min at 3000 × g, and DNA extraction was performed. The supernatant was precipitated with 20% polyethylene glycol 6000 and 3.5 M ammonium acetate for 15 min on ice. After centrifugation, the pellet was resuspended in 200 μl of Tris-EDTA (TE). DNA was purified by sequential extraction with 1 volume of phenol followed by ether. The lower phase was recovered, and DNA was precipitated with 200 μl of 5 M ammonium acetate and 1 ml of ethanol. The precipitated DNA was finally dissolved in 12 μl of TE.
DNA manipulations and mutagenesis.
DNA manipulations were performed as described by Sambrook et al. (43). Oligonucleotides and plasmids are described in Table 1.
M13 mutagenesis was used to construct the Cp8110 strain. Fifty picomoles of mutagenic primer reg7 was phosphorylated with 2 U of T4 kinase in 30 μl of the specific kinase buffer (0.1 M Tris [pH 8], 10 mM MgCl2, 7 mM dithiothreitol, 1 mM ATP) and incubated for 15 min at 37°C. The solution was then warmed for 10 min at 70°C, and 2 μl of the solution was mixed with 200 ng of single-strand pTSS9 DNA, prepared as described above. One microliter of annealing buffer (200 mM Tris [pH 7.4], 1 mM EDTA, 500 mM NaCl) was added, and the volume was made up to 10 μl with sterile distilled water. The solution was covered with paraffin oil to prevent evaporation, boiled for 3 min, and kept standing at 37°C to allow the annealing of the primer to the single-strand DNA. Double-strand DNA was synthesized by the addition of 10 U of T4 DNA ligase, 5 U of T4 DNA polymerase, and 1 μl of synthesis buffer (5 mM deoxynucleoside (triphosphate, 10 mM ATP, 100 mM Tris [pH 7.4], 50 mM MgCl2, 20 mM dithiothreitol) and incubation for 90 min at 37°C. Fifty microliters of TE was then added, and DH5α was transformed, as previously described (43), to recover plasmid pTSS10.
Transformation of S. pneumoniae.
In order to recover isogenic strains differing by a defined mutation, mutated DNA was introduced into the recipient strain by competence-stimulating peptide-induced transformation, according to the protocol described by Echenique et al. (18). For strains constructed from chromosomal DNA, two rounds of transformation were performed. For selection of the CiaR0 and Hyl0 strains and of the RegR0 encapsulated strains, spectinomycin (10 μg ml−1), erythromycin (2 μg ml−1), and kanamycin (40 μg ml−1) were respectively added to the blood agar medium. The screening of the stop codon transformants was performed by restriction analysis of amplicons obtained by direct PCR on the resulting colonies, as previously described (2). The restriction site used to test the P100-PC2 amplicons potentially containing the regR0 mutation was SpeI (Table 1).
The ability of cultures to develop competence in agar medium was determined by a transformation test (18). Briefly, bacteria were grown to an OD400 of 0.1 to 0.2 at 37°C in CTM, pH 7. Volumes of 10 μl of appropriate dilutions of the culture were mixed with 5 μl of Cp1016 transforming DNA encoding the rifampin resistance gene (1 mg ml−1) and diluted in 1 ml of CTM, pH 7.8, plus 1 ml of a 50% (vol/vol) mix of CTM (pH 7.8) and 1.6% agar. This suspension was then poured into petri dishes on a 2-ml layer of the 50% (vol/vol) mix of CTM (pH 7.8) and 1.6% agar. After a 4-h incubation at 37°C, a third layer of medium with or without rifampin (3 μg/ml) was added. The ratio of the number of colonies on plates with antibiotic to the number of colonies on plates without antibiotic was calculated to determine the percentage of transformants obtained. Results were analyzed by the Mann-Whitney U test (one tailed).
Production of antibodies and Western blot analysis.
Purified recombinant RegR protein was kindly provided by I. Auzat, Centre National de la Recherche Scientifique, Gif sur Yvette, France. Rabbit antiserum directed against RegR was obtained by three successive subcutaneous injections of purified RegR in complete Freund's adjuvant at 3-week intervals. Three weeks after the last injection, blood was recovered and centrifuged and the serum was stored at −80°C. Polyclonal mouse sera directed against CbpA, Hyl, LytA, Ply, PsaA, and PspA were obtained by intraperitoneal immunization with three doses of 10 μg of each protein antigen in alum at 12- to 14-day intervals, and sera were collected from mice 1 week after the third immunization.
For Western immunoblotting, fresh cells were lysed with 0.4% Sarkosyl in 0.02 M Tris (pH 6.9), 0.2% sodium dodecyl sulfate, and 2% glycerol. Cells lysates containing 250 ng of proteins (corresponding to 5 × 106 CFU) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then electroblotted onto nitrocellulose. The transfer efficiency was checked by staining the membrane with a Ponceau solution. After decoloration, the membrane reacted with specific antisera and proteins were detected either by the ECL Western blotting kit (Roche) or by color development with p-nitrophenyl phosphate as the substrate.
Hyaluronidase activity measurement.
The protocol for hyaluronidase activity measurements was derived from that described by Berry et al. (9). Strains were grown in 4 ml of CTM, pH 7, to an OD400 of 0.4. After centrifugation, the bacterial pellet was washed once in phosphate-buffered saline (PBS), resuspended in 400 μl of PBS plus 0.1% deoxycholate, and incubated at 37°C for 10 min to lyse the cells. The cell debris was pelleted by centrifugation, and the supernatant was assayed for the hyaluronidase content as follows. The substrate for the assay was human umbilical cord hyaluronic acid, dissolved in assay buffer (150 mM NaCl, 200 mM sodium acetate [pH 6]) at a concentration of 1 mg ml−1. The supernatant from each strain to be assayed was diluted to 800 μl in the same buffer and then incubated with 60 μl of the substrate for 2 h. The reaction was stopped by the addition of 1,200 μl of stop buffer (2% NaOH, 2.5% cetrimide), and the OD400 was measured by using as control the sample at time 0. One unit was defined as the delta OD400 measured for 5 ml of an initial culture grown to an OD400 of 0.4.
Antibiograms.
Impregnated disks were used as indicated by the manufacturer (Sanofi Diagnostics Pasteur, Marne la Coquette, France). The following antibiotics were tested: penicillin G (10 IU), oxacillin (5 μg), amoxicillin (25 μg), gentamicin (500 μg), netilmicin (30 μg), amikacin (30 μg), minocycline (30 IU), erythromycin (15 IU), clindamycin (2 IU), trimethoprim (1.25 μg), sulfamethoxazole (23.75 μg), nitrofurans (300 μg), rifampin (30 μg), and vancomycin (30 μg).
Adherence to A549 cells.
Bacterial suspensions were plated onto petri dishes containing Trypticase soy agar plus 5% horse blood and grown overnight at 37°C in an atmosphere containing 5% CO2. Cultures were then grown in CAT medium supplemented with 17 mM K2HPO4 and 0.2% glucose and stopped at an OD400 of 0.4. After centrifugation, the bacterial pellet was washed twice with PBS and resuspended in RPMI medium to obtain 3 × 108 to 5 × 108 CFU per ml. Suspensions were finally sonicated (60 W; 60 s; 20 kHz) to avoid clamping of chains.
For adherence, bacterial suspensions were added to the monolayer of A549 cells and the plates were incubated for 60 min at 37°C in an atmosphere containing 5% CO2. Cells were then washed seven times with RPMI medium at 4°C, treated with trypsin, and sonicated (60 W; 45 s; 20 kHz) to dissociate the bacteria. Bacterial viability was determined by plating appropriate dilutions on Trypticase soy agar plus 5% horse blood.
Virulence studies in mice.
For virulence studies in the intraperitoneal model of infection, 6- to 8-week-old male BALB/c mice were obtained from the Central Animal House of Adelaide University and housed at the Medical School Animal House facility. Groups of mice were each infected intraperitoneally with 2.5 × 106 to 2.5 × 107 CFU of strain 23477 or D39 or one of their isogenic derivatives. Survival times were recorded for periods of 10 to 14 days, and median survival times were calculated for each group of mice. Results were analyzed by the Mann-Whitney U test (one tailed).
RESULTS
Genetic organization of a 20.1-kbp region including regR in the S. pneumoniae genome.
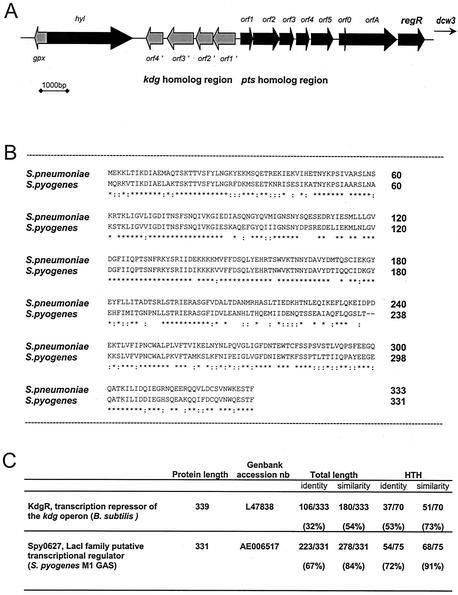
Previous work led to the cloning of a highly conserved 5.5-kb fragment containing genes homologous to those involved in division and cell wall biosynthesis (dcw) (30, 34). At the 5′ end of this fragment, a putative 999-nucleotide gene encoding RegR was identified (GenBank accession number Z79691) (34). Upstream of this open reading frame, a typical extended −10 promoter (6), followed by a ribosome binding site (5), was identified. Identical putative proteins exist in the two recently sequenced S. pneumoniae strains, TIGR4 (The Institute for Genomic Research, Rockville, Md.) and R6 (Eli Lilly and Company, Indianapolis, Ind.). In strain TIGR4, the SP0330 gene encodes a putative 333-amino acid (aa) protein with a single conservative 182H (TIGR4)-to-182Y (Rx) mutation. In R6, the spr0298 gene encodes a 355-aa putative protein, the last 333 aa of which perfectly match the RegR sequence. BLAST search (1) has highlighted significant homology between the 333-aa RegR and transcriptional regulators belonging to the LacI/GalR family. These are characterized by an amino-terminal helix-turn-helix (HTH) motif involved in protein binding to DNA and a carboxy-terminal motif bearing the regulatory function (50). The 1-to-70 amino-terminal part of the 333-aa RegR protein is indeed referenced as “HTH LacI” in the Smart Database (44), and the carboxyl domain is related to “periplasmic binding proteins and the sugar binding domain of the LacI family” in the Pfam database (7). Therefore, the 333-aa product, designated RegR, does show the features of a regulator of the LacI/GalR family. The recent publication of the entire genomes of S. pneumoniae strains TIGR4 and R6 and of the partial chromosomal sequence of strain 19F (16, 25, 46) reveals the inclusion of regR in a 15.5-kb region containing open reading frames encoding putative glutathione peroxidase (gpx), hyaluronidase (hyl), proteins of the 2-keto 3-deoxygluconate metabolism (kdg operon), a phosphotransferase system (PTS) sugar transporter showing homology with N-acetylgalactosamine and mannose PTS (pts operon), and two hypothetical proteins (orf0 and orfA); this 15.5-kbp region precedes the dcw3 region (Fig. 1A). The same genetic organization for kdg, pts, orfA, and regR exists in the Streptococcus pyogenes public genome (19). RegR is indeed an ortholog of Spy0267 of S. pyogenes (GenBank accession number AE006517) (Fig. 1B), KdgR from Clostridium acetobutylicum (GenBank accession number AE007554), and the functionally characterized KdgR from Bacillus subtilis (GenBank accession number L47838). The HTH sequence of these proteins is highly conserved (Fig. 1C). In B. subtilis, kdgR is the first gene of the kdg operon and represses transcription of the operon in galacturonate-free medium (40). As a first attempt to characterize the physiological role of RegR, the effect of its loss-of-function mutation on phenotypes putatively related to the genes of the 20.1-kb region around regR has been investigated. A stop codon mutation has been introduced into the 5′ end of regR by M13 mutagenesis, and the mutated allele has been transferred into the chromosome of the unencapsulated strain Cp1015 to yield strain Cp8110 (see Materials and Methods). Immunoblotting experiments with RegR-specific antiserum have revealed three major reacting proteins in crude extracts from the wild-type strain (∼55, 41, and 34 kDa). A single band was absent in the extracts from strain Cp8110, and it corresponded to the 41-kDa purified RegR (Fig. 2A). Consistently, loss of the RegR protein was expected due to the regR stop codon mutation in this strain.
FIG. 1.
Genome localization of regR and relationship of the RegR product with homologs in S. pyogenes and B. subtilis. (A) Genetic organization of the regR region on the chromosome of S. pneumoniae. The genes encoding putative glutathione peroxidase (gpx), hyaluronidase (hyl), putative proteins of the 2-keto 3-deoxygluconate metabolism (kdg operon, orf1′ to 4′), a putative PTS sugar transporter showing homology with N-acetylgalactosamine and mannose PTS (PTS operon, orf1 to 5), and two hypothetical proteins (orf0 and orfA) are referred to in the TIGR4, S6, and 19F strains, respectively, as SP0313, spr0285, and SPN8189 for gpx; SP0314, spr0286, and SPN8190 for hyl; SP0317 to -0320, spr0287 to -0290, and SPN8195 to -8198 for the kdg operon; SP0321 to -0325, spr0291 to -0295, and SPN8201 to -8203 for the pts operon; and SP0326 and -0327, spr0296 and -0297, and SPN8195 to -8198 for orf0 and orfA. (B) Conservation of the primary structure of RegR in S. pyogenes Spy0627 (AE006517). Stars and colons represent identical and similar amino acids, respectively. (C) The conservation of the HTH motif in both KdgR (L47838) of B. subtilis and Spy0627 of S. pyogenes M1Gas and in RegR of S. pneumoniae has been deduced from the BLAST alignment of KdgR and Spy0627 to RegR. The table indicates percentages of identity and similarity between the protein sequences of RegR and these proteins.
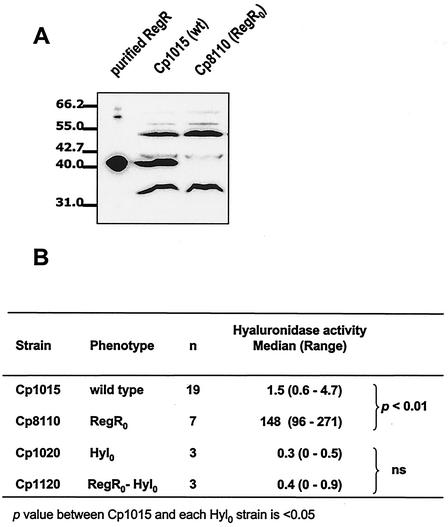
FIG. 2.
Derepression of hyaluronidase due to RegR loss-of-function mutation. (A) Purified RegR and crude extracts from the wild-type (wt) strain Cp1015 and its isogenic derivative Cp8110 were electrophoresed, transferred to nitrocellulose, and immunodetected with the RegR antiserum. The same amounts of extracts were loaded in each lane (see Materials and Methods). (B) Increase in hyaluronidase specific activity due to regR mutation. Crude extracts were prepared and used for the dosage of hyaluronidase activity as described in Materials and Methods. The medians and ranges obtained from n independent experiments are shown. One hyaluronidase unit is defined as the delta OD400 measured for 5 ml of a culture grown to an OD400 of 0.4. ns, not significant.
Impact of regR loss-of-function mutation on sugar utilization, aerobic growth, resistance to β-lactam antibiotics, and hyaluronidase production in vitro.
In vitro, S. pneumoniae grows in a rich medium and requires simple sugars as a carbon source. The homology of RegR with regulators of carbohydrate utilization has prompted us to compare the growth patterns of the wild-type (Cp1015) and RegR0 (Cp8110) strains on different sugars, including sugars susceptible to involvement in the 2-keto-3-deoxygluconate and PTS pathways. We found no obvious differences in growth rates and yields between Cp1015 and Cp8110, irrespective of the carbon source provided (data not shown). Substrates allowing growth of the wild-type strain (mannose, maltose, N-acetylglucosamine, glucose, and lactose) supported the growth of strain Cp8110 as well, while sugars which could not be utilized by the wild-type strain (glucuronic acid, galacturonic acid, hyaluronic acid, and N-acetylgalactosamine) did not support the growth of the RegR0 strain. The growth pattern of strain Cp8110 was not influenced by the oxygen status of the culture in vitro (data not shown). Moreover, when glucose was the carbon source, no specific shift in central metabolism occurred in strain Cp8110 compared to that in strain Cp1015 (data not shown).
In view of the fact that regR lies upstream of the dcw region containing pbpX (the product of which is a β-lactam target), we have examined whether RegR influences β-lactam resistance. Standard antibiograms with impregnated disks were produced for both encapsulated (23477 and 23302) and unencapsulated (Cp1015 and Cp8302) strains by using a series of antibiotics routinely used in hospital strain characterization as controls (Sanofi Diagnostics Pasteur). No significant difference could be observed in the zones of inhibition, indicating similar patterns of antibiotic susceptibility for the wild-type strains and their corresponding RegR0 derivatives (data not shown).
The effect of regR mutation on hyaluronidase expression has been investigated. Hyaluronidase belongs to the family of cell wall-anchored proteins showing the C-terminal LPXTG motif (9, 33); thus, hyaluronidase activity has been determined in culture supernatants and cellular extracts. The absence of RegR was associated with a 90-fold increase in hyaluronidase-specific activity relative to that in the wild-type strain (Fig. 2B). A similar ratio of levels of hyaluronidase activity between the wild-type and the RegR0 strains was detected in culture supernatants (data not shown). This indicates that RegR does reduce the cellular level of hyaluronidase. As a control, it has been shown that extracts from strain Cp1120 (hyl0 regR0 [where 0 indicates the loss of function]) did not contain significant hyaluronidase activity (Fig. 2B). Taken together, these results demonstrate that the high level of hyaluronidase activity in strain Cp8110 was exclusively due to hyl derepression as a result of the RegR loss-of-function mutation.
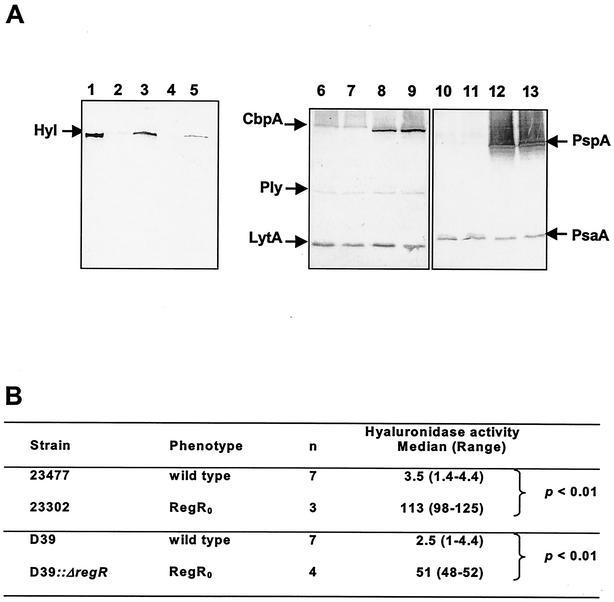
Hyaluronidase is considered to be a potential virulence protein (35). In order to evaluate the role of RegR in the regulation of characterized virulence proteins in S. pneumoniae, encapsulated strains D39 (serotype 2) and 23477 (serotype 6B) have been mutated by the deletion-replacement of regR. An HincII-HincII fragment overlapping regR has been deleted and replaced by the aphA3 cassette from pPJ1 (36) (see Materials and Methods). This yielded the kanamycin-resistant derivatives designated 23302 and D39::ΔregR, respectively (Table 1). Whole-cell lysates from the RegR0 bacteria have been analyzed by Western immunoblotting with polyclonal antisera raised against pneumolysin (Ply), autolysin (LytA), hyaluronidase (Hyl), pneumococcal surface protein A (PspA), pneumococcal surface adhesin A (PsaA), and choline binding protein A (CbpA) (35, 42). In both the RegR0 derivatives 23302 and D39::ΔregR, a marked increase in the intensity of anti-Hyl reactive protein compared to that in the wild type was specifically observed (Fig. 3A). Consistently, hyaluronidase-specific activity in extracts from the RegR0 derivatives 23302 and D39::ΔregR was respectively 30- and 25-fold higher than that in extracts from the corresponding RegR+ strains (Fig. 3B). This shows that RegR represses hyl whatever the genetic background of the strain.
FIG. 3.
Expression of characterized virulence proteins in RegR0 mutants from 23477 and D39. (A) Western blot analysis of cellular extracts from wild-type strains 23477 (lanes 2, 6, and 10) and D39 (lanes 4, 8, and 12) and their regR derivatives 23302 (lanes 3, 7, and 11) and D39::ΔregR (lanes 5, 9, and 13) with specific antisera against hyaluronidase (lanes 1 to 5), a mixture of CbpA, Ply, and LytA (lanes 6 to 9), and a mixture of PsaA and PspA (lanes 10 to 13). Lane 1: purified hyaluronidase. The same amounts of bacterial extracts were loaded in each lane. (B) Increased Hyl activity in 23302 due to regR mutation. Hyl activity in extracts was determined and expressed as described in the legend for Fig. 2.
RegR and hyaluronidase in experimental virulence.
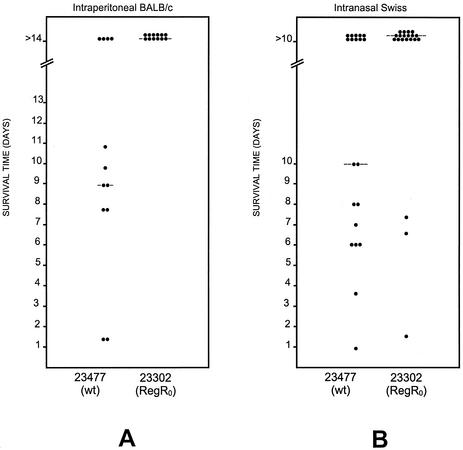
The contribution of hyl to pneumococcal virulence is uncertain. For strain D39, a type 2 capsular serotype, it was demonstrated previously that the hyl0 mutation further reduced the virulence of a Ply− derivative but was silent in the Ply+ isogenic strain in a BALB/c mouse intraperitoneal infection model (11) (see Materials and Methods). Here, the RegR0 mutant gave us a tool to explore the effect of hyl derepression on virulence. Evaluation of the virulence of the RegR0 derivative of D39 after intraperitoneal challenge of BALB/c mice showed that the regR mutation did not affect virulence of the bacteria (data not shown). Surprisingly, strain 23302, the RegR0 derivative of strain 23477, was significantly attenuated in the BALB/c intraperitoneal challenge model. The median survival times of mice challenged with 2.5 × 106 CFU of strains 23477 and 23302 were 9 and >14 days, respectively (P < 0.01) (Fig. 4A). Similarly, the median survival time of Swiss mice challenged intranasally with 108 CFU of 23477 was significantly lower than that of mice challenged with the same dose of 23302 (P < 0.05) (Fig. 4B). In another BALB/c intraperitoneal challenge experiment using 107 CFU, mice challenged with 23302 survived significantly longer than those challenged with 23477; median survival times were 2 days and 1 day, respectively (P < 0.001) (Fig. 5).
FIG. 4.
Effect of regR mutation on virulence. Intraperitoneal (for BALB/c mice) and intranasal (for Swiss mice) challenges were performed with strain 23477 and its isogenic RegR0 derivative 23302 (see Materials and Methods). Inocula were 2.5 × 106 and 108 bacteria for intraperitoneal and intranasal challenges, respectively. Dots represent the number of dead mice at a given time. The bars represent the median survival times. The Mann-Whitney U test (one tailed) was used for analysis. wt, wild type.
FIG. 5.
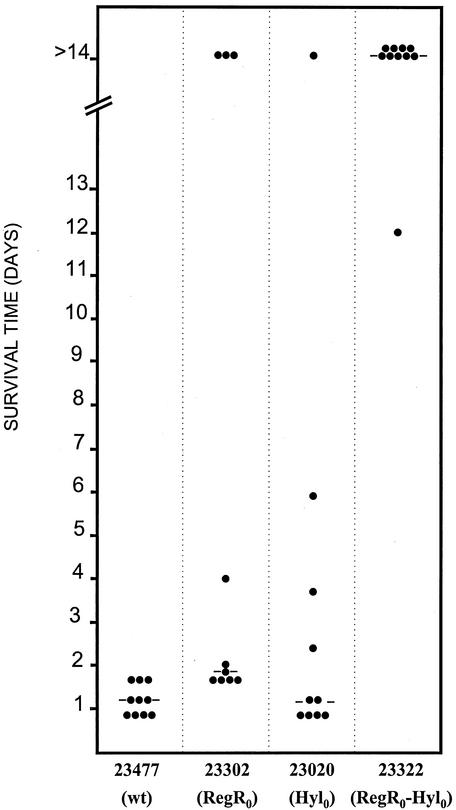
Genetic evidence for the role of both hyaluronidase and RegR in virulence of strain 23477. Bacterial suspensions corresponding to 107 bacteria from cultures of strain 23477 (wild type [wt]) and its isogenic derivatives 23302 (regR0), 23020 (hyl0), and 23322 (regR0 hyl0) were used for intraperitoneal challenge of BALB/c mice. Representation and data analysis were as described in the legend for Fig. 4.
Derepression of hyaluronidase in vitro was associated with attenuation of strain 23302. In order to investigate the importance of hyaluronidase in the residual virulence of this strain, a 23302 derivative carrying the hyl insertion mutation was obtained and named 23322 (see Materials and Methods and Table 1). A combination of both hyl and regR mutations culminated in strong attenuation of strain 23322. No mouse death had occurred 10 days after intraperitoneal infection with strain 23322 (107 CFU/mouse), and the overall survival rate of mice was significantly higher than that for mice infected with either 23302 (P < 0.01) or 23020 (P < 0.001) (Fig. 5). This demonstrates that hyaluronidase is important for virulence expression in vivo. It is possible that in the regR+ strain, virulence attenuation due to the Hyl defect is masked by the expression of complementary virulence factors positively regulated by RegR. Examination of colonies on agar plates indicated that the isogenic encapsulated wild-type 23477 and RegR0 23302 strains show comparable mucoid phenotypes (data not shown), suggesting that the effect of RegR on virulence does not involve gross modulation of capsule biosynthesis. When in parallel experiments the regR hyl D39 double mutant strain was used, no virulence attenuation was measured (data not shown), indicating that in the S2 strain these mutations were silent with regard to experimental virulence. Taken together, these results highlight the importance of the bacterial genetic background on the impact of a given virulence factor in experimental virulence and show the complexity of virulence factor regulation in S. pneumoniae.
The first step in virulence expression is host colonization. The role of RegR in the initial stages of host colonization has been evaluated by measurements of adherence to alveolar epithelial (A549) cells (see Materials and Methods). Bacteria from cultures at pH 8.0 from strain Cp1015 and its RegR0 derivative Cp8110 did not show any difference in their adherence properties (50 CFU/100 cells). However, when bacteria were grown at pH 7.0, a net twofold increase in adherence was specifically observed for the wild-type strain. The number of adherent bacteria from Cp1015 shifted to 100 CFU/100 cells, and it remained at 50 CFU/100 cells for Cp8110. This indicates that RegR activates the adherence of bacteria grown at pH 7. Impact on adherence might account partly for the positive effect of RegR on virulence; in any case, it reveals a role for RegR in the bacterial response to pH.
RegR is an activator of competence, acting upstream of CiaRH in addition to Nox.
As adherence experiments revealed a pH-dependent role for RegR, we have investigated its effect on competence, a response highly controlled by the pH of the culture. Indeed, in alkaline medium, competence has been shown to develop in cultures growing exponentially, whereas its expression is reduced and appears later when the pH of the medium is neutral (12). In in situ transformation tests in soft agar medium (see Materials and Methods) with bacteria from early exponential cultures (OD400 of 0.05 to 0.1), transformants from the wild-type strain were selectively obtained when the test was performed at pH 8.0 and aerobically. Transformants from the RegR0 mutant strain Cp8110 were also selectively obtained at pH 8.0 from aerobically grown bacteria, indicating that competence repression at pH 7.0 and under microaerobiosis is not due to RegR (Table 2). However, reduced levels of transformant recovery for strain Cp8110 compared to levels of recovery for the wild type (P < 0.01) show indeed that the LacI/GalR family regulator RegR is required for optimal competence development in alkaline medium. The effect of the regR mutation was abolished by the insertion mutation in ciaR (Table 2), indicating that RegR acts upstream of the competence signaling pathway including CiaRH and ComDE (18). A similar conclusion had been obtained previously for the NADH oxidase Nox (2). In order to determine the relative influence of these two functions on competence development, strain Cp8156 (RegR0 Nox0) has been constructed by transformation of Cp8056 with pTSS10 carrying the regR0 mutation (see Materials and Methods). Strain Cp8156 consistently gave significantly lower yields of transformants than did strain Cp8110 (RegR0). For strains Cp8056 and Cp8156, counts were too low to reveal any significant difference between the RegR0 Nox0 and Nox0 strains by the Mann-Whitney U test (Table 2). It is therefore not possible to discriminate between schemes involving cumulative effects of Nox and RegR and epistasis of nox to the regR mutation. It has been verified by Western blot analysis that the regR mutation did not influence the cellular level of Nox in bacteria growing aerobically (data not shown). In any case, the data presented highlight the role of both Nox and RegR upstream of CiaRH in the positive regulation of competence in cultures growing aerobically in alkaline medium.
TABLE 2.
Influence of RegR on competence expressiona
| Strain | Phenotype | Median % of transformants (range) | No. of expts | P value compared to Cp1015b |
|---|---|---|---|---|
| Cp1015 | Wild type | 2.42 (0.6-5.2) | 19 | |
| Cp8110 | RegR0 | 0.27 (0-2.5) | 13 | <0.01 |
| Cp8056 | Nox0 | 0.03 (0-0.2) | 7 | <0.01 |
| Cp8156 | RegR0 Nox0 | 0.02 (0-0.07) | 6 | <0.01 |
| Cp1800 | CiaR0 | 1.93 (1.1-6.8) | 8 | NS |
| Cp1810 | CiaR0 RegR0 | 5.82 (0.8-10.1) | 4 | NS |
Bacteria from exponential cultures (OD400 = 0.1) were mixed with DNA from strain Cp1016 carrying the Rifr mutation and plated in transformation medium as described by Echenique et al. (18) and in Materials and Methods. The percentage of rifampin-resistant colonies in the population was calculated and values were compared by using the Mann-Whitney U test.
NS, not significant.
DISCUSSION
Our work focused on the global role of RegR in the physiology of S. pneumoniae. Phenotypes associated with the loss-of-function mutation in regR were identified in both in vitro and in vivo studies. In vitro, RegR lowers the cellular level and specific activity of hyaluronidase, irrespective of the genetic background, increases adherence of bacteria grown at neutral pH, and positively regulates competence. For competence, regulation by RegR occurs upstream of the signaling route through CiaRH and ComDE. In a mouse model of infection, attenuation of virulence, due to the regR deletion, depends on the genetic background of the bacteria. Mutational analysis and genetic dissection in the S2 genetic background did not reveal any role for RegR and for hyaluronidase in virulence. However, this strategy made it possible to establish the function of the global regulator RegR and of hyaluronidase in the virulence of strain 23477, a 6B capsular serotype blood isolate.
We used genetic dissection to highlight the opposite effects of RegR on the regulation of Hyl and on the regulation of putative complementary virulence factors. The combination of an insertion mutation in hyl and a deletion-replacement mutation in regR, leading to the loss of both proteins in strain 23322, severely reduced the bacterial virulence compared to that of each single mutant strain 23302 and 23020. This allows us to propose that in the RegR0 derivative (23302), virulence might be attributed to hyl derepression, while the virulence of the Hyl0 RegR+ strain (23020) might be due to the expression of uncharacterized virulence factors. In strain 23322, defects in hyaluronidase due to the hyl mutation and in unknown virulence factors due to the regR mutation culminate in a strong attenuation of the bacteria. Thus, RegR might be considered as a modulator of the expression of virulence factors, adjusting their production to specific infection sites and thus optimizing the infection process. Indeed, a site-specific role of hyaluronidase has been proposed by others. When clinical strains from otitis media and meningitis cases were compared to commensal isolates for their hyaluronidase production, both the frequencies of producing strains and the levels of production varied according to the isolate. The incidence of Hyl-producing strains was 100% for cerebrospinal fluid (CSF) isolates, 84.6% for otitis media isolates, and 11.5% for commensal strains (26). High producers were found among the CSF isolates, and low producers were found among strains from otitis media cases and from the throat. In experimental virulence studies, the addition of purified hyaluronidase to a type 6A clinical isolate from CSF was required to cause meningitis in mice (53). Moreover, for strain 19F, attenuation due to a hyl mutation was specifically observed in an intranasal model but not in an intraperitoneal challenge model, leading to the proposal that hyaluronidase could be involved in the colonization of the host (38). Thus, it is possible that hyaluronidase is temporarily involved in the infection strategy of S. pneumoniae, in combination with other virulence determinants positively regulated by RegR. The regulatory role of RegR (acting negatively on Hyl and positively on complementary virulence factors) might be a result of the direct activation of specific genes, or it might be indirect via the repression of repressors. More work is needed to characterize the functions positively regulated by RegR. Importantly, such studies might reveal specific ligands and accessory proteins involved in the control of this virulence modulator.
Adherence studies using A549 pulmonary epithelial cells revealed that RegR increases the adherence of bacteria grown at neutral pH. This trait might be important for the colonization of the host. In any case, it reveals the contribution of RegR to the bacterial response to the pH of the culture. However, RegR is not involved in competence repression at pH 7; rather, genetic dissection revealed a positive role for RegR in addition to Nox in competence development at alkaline pH, upstream of the signaling route involving CiaRH and ComDE. It is thus proposed that, in the presence of oxygen, pH-dependent metabolic pathways adjust the cytoplasmic concentration of RegR ligand, culminating in the activation of competence and adherence. RegR might thus be considered as a port of entry for metabolic signals in the global adaptive network of S. pneumoniae.
The LacI/GalR family consists of transcriptional regulators involved in metabolic regulation. For S. pneumoniae, the characteristics of MalR, a negative regulator of the maltose operon, fit the canonical model described for other species (41). The high level of similarity between RegR and KdgR, a repressor of hexuronate utilization via the modified Entner-Doudoroff pathway in B. subtilis (40), supports a role for RegR in this metabolism. However, simple growth studies did not reveal specific carbohydrate utilization or shifts in central metabolism in vitro and in rich media due to RegR deficiency.
In conclusion, we have exploited the observation of different phenotypes in different models and experimental approaches to demonstrate the physiological impact of a global regulator of the LacI/GalR family in S. pneumoniae. The pleiotropic effect of the regR0 mutation demonstrates the role of this regulator in the repression of the degradative enzyme hyaluronidase, with impact on experimental virulence for the S6 strain 23477. The physiological effect of RegR concerns both competence and virulence expression. For competence, RegR acts upstream of the signaling cascade involving CiaRH and ComDE, in addition to the NADH oxidase Nox. Although the effect of RegR was not observed with strain D39, the findings in this work indicate its role both in vivo and in vitro via its fine tuning of the expression of virulence and competence genes, culminating in the adjusted response of the bacteria to specific environmental conditions.
Acknowledgments
This work was supported by Université Paul Sabatier, Toulouse, France, the National Health and Medical Research Council of Australia, Aventis Pharma, France, and the Association pour la Recherche sur le Cancer (ARC), France.
We gratefully acknowledge Didier Fournier for his contribution to the M13 mutagenesis success and Nick Lindley for the analysis of metabolic fluxes and for helpful discussion. We sincerely thank Isabelle Auzat for purifying the RegR protein and Gérard Chabanon for providing the antibiogram disks. We thank Delphine Dos Santos and Saliha Mimar for their skillful technical assistance.
Editor: A. D. O'Brien
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auzat, I., S. Chapuy-Regaud, G. Le Bras, D. Dos Santos, A. D. Ogunniyi, I. Le Thomas, J. R. Garel, J. C. Paton, and M. C. Trombe. 1999. The NADH oxidase of Streptococcus pneumoniae: its involvement in competence and virulence. Mol. Microbiol. 34:1018-1028. [DOI] [PubMed] [Google Scholar]
- 3.Avery, O. T., C. M. McLeod, and M. McCarthy. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. J. Exp. Med. 79:137-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azoulay-Dupuis, E., V. Rieux, C. Rivier, and M. C. Trombe. 1988. Pleiotropic mutations alter the kinetics of calcium transport, competence regulation, autolysis and experimental virulence in Streptococcus pneumoniae. Res. Microbiol. 149:5-13. [DOI] [PubMed] [Google Scholar]
- 5.Bacot, C. M., and R. H. Reeves. 1991. Novel tRNA gene organization in the 16S-23S intergenic spacer of the Streptococcus pneumoniae rRNA gene cluster. J. Bacteriol. 173:4234-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barne, K. A., J. A. Bown, S. J. Busby, and S. D. Minchin. 1997. Region 2.5 of the Escherichia coli RNA polymerase sigma 70 subunit is responsible for the recognition of the ‘extended-10’ motif at promoters. EMBO J. 16:4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. Sonnhammer. 2000. The Pfam protein families database. Nucleic Acids Res. 28:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berry, A. M., R. A. Lock, D. Hansman, and J. C. Paton. 1989. Contribution of autolysin to virulence of Streptococcus pneumoniae. Infect. Immun. 57:2324-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berry, A. M., R. A. Lock, S. M. Thomas, D. P. Rajan, D. Hansman, and J. C. Paton. 1994. Cloning and nucleotide sequence of the Streptococcus pneumoniae hyaluronidase gene and purification of the enzyme from recombinant Escherichia coli. Infect. Immun. 62:1101-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berry, A. M., and J. C. Paton. 1996. Sequence heterogeneity of PsaA, a 37-kDa putative adhesin essential for virulence of Streptococcus pneumoniae. Infect. Immun. 64:5255-5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berry, A. M., and J. C. Paton. 2000. Additive attenuation of virulence of Streptococcus pneumoniae by mutation of the genes encoding pneumolysin and other putative pneumococcal virulence proteins. Infect. Immun. 68:133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J. D., and D. A. Morrison. 1987. Modulation of competence for genetic transformation in Streptococcus pneumoniae. J. Gen. Microbiol. 133:1959-1967. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, Q., E. A. Campbell, A. M. Naughton, S. Johnson, and H. R. Masure. 1997. The com locus controls genetic transformation in Streptococcus pneumoniae. Mol. Microbiol. 23:683-692. [DOI] [PubMed] [Google Scholar]
- 14.Clavé, C., and M. C. Trombe. 1989. DNA uptake in competent Streptococcus pneumoniae requires ATP and is regulated by cytoplasmic pH. FEMS Microbiol. Lett. 53:113-118. [DOI] [PubMed] [Google Scholar]
- 15.Dintilhac, A., G. Alloing, C. Granadel, and J. P. Claverys. 1997. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol. Microbiol. 25:727-739. [DOI] [PubMed] [Google Scholar]
- 16.Dopazo, J., A. Mendoza, J. Herrero, F. Caldara, Y. Humbert, L. Friedli, M. Guerrier, E. Grand-Schenk, C. Gandin, M. de Francesco, A. Polissi, G. Buell, G. Feger, E. Garcia, M. Peitsch, and J. F. Garcia-Bustos. 2001. Annotated draft genomic sequence from a Streptococcus pneumoniae type 19F clinical isolate. Microb. Drug Resist. 7:99-125. [DOI] [PubMed] [Google Scholar]
- 17.Echenique, J. R., and M. C. Trombe. 2001. Competence modulation by the NADH oxidase of Streptococcus pneumoniae involves signal transduction. J. Bacteriol. 183:768-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Echenique, J. R., S. Chapuy-Regaud, and M. C. Trombe. 2000. Competence regulation by oxygen in Streptococcus pneumoniae: involvement of ciaRH and comCDE. Mol. Microbiol. 36:688-696. [DOI] [PubMed] [Google Scholar]
- 19.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia, P., J. L. Garcia, E. Garcia, and R. Lopez. 1986. Nucleotide sequence and expression of the pneumococcal autolysin gene from its own promoter in Escherichia coli. Gene 43:265-272. [DOI] [PubMed] [Google Scholar]
- 21.Gibson, J. F., R. K. Poole, M. N. Hughes, and J. F. Rees. 1984. Filamentous growth of Escherichia coli K12 elicited by dimeric, mixed-valence complexes of ruthenium. Arch. Microbiol. 139:265-271. [DOI] [PubMed] [Google Scholar]
- 22.Gil, D. 1990. Elaboration et caractérisation d'un nouveau type de vecteur de clonage à nombre de copies régulable. University thesis. Université Paul Sabatier, Toulouse, France.
- 23.Havarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Håvarstein, L. S., P. Gaustad, I. F. Nes, and D. A. Morrison. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863-869. [DOI] [PubMed] [Google Scholar]
- 25.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kostyukova, N. N., M. O. Volkova, V. V. Ivanova, and A. S. Kvetnaya. 1995. A study of pathogenic factors of Streptococcus pneumoniae strains causing meningitis. FEMS Immunol. Med. Microbiol. 10:133-137. [DOI] [PubMed] [Google Scholar]
- 27.Kunkel, T. A., J. D. Roberts, and R. A. Zakour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 28.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 29.Macrina, F. L., R. P. Evans, J. A. Tobian, D. L. Hartley, D. B. Clewell, and K. R. Jones. 1983. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene 25:145-150. [DOI] [PubMed] [Google Scholar]
- 30.Massidda, O., D. Anderluzzi, L. Friedli, and G. Feger. 1998. Unconventional organization of the division and cell wall gene cluster of Streptococcus pneumoniae. Microbiology 144:3069-3078. [DOI] [PubMed] [Google Scholar]
- 31.Morrison, D. A., M. C. Trombe, M. K. Hayden, G. A. Waszak, and J. D. Chen. 1984. Isolation of transformation-deficient Streptococcus pneumoniae mutants defective in control of competence using insertion-duplication mutagenesis with the erythromycin resistance determinant of pAM beta 1. J. Bacteriol. 159:870-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mortier-Barrière, I., A. de Saizieu, J. P. Claverys, and B. Martin. 1998. Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pneumoniae. Mol. Microbiol. 27:159-170. [DOI] [PubMed] [Google Scholar]
- 33.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmen, R., A. D. Ogunniyi, P. Berroy, S. Larpin, J. C. Paton, and M. C. Trombe. 1999. Insertional mutation of orfD of the DCW cluster of Streptococcus pneumoniae attenuates virulence. Microb. Pathog. 27:337-348. [DOI] [PubMed] [Google Scholar]
- 35.Paton, J. C., P. W. Andrew, G. J. Boulnois, and T. J. Mitchell. 1993. Molecular analysis of the pathogenicity of Streptococcus pneumoniae: the role of pneumococcal proteins. Annu. Rev. Microbiol. 47:89-115. [DOI] [PubMed] [Google Scholar]
- 36.Peeters, B. P., J. H. de Boer, S. Bron, and G. Venema. 1988. Structural plasmid instability in Bacillus subtilis: effect of direct and inverted repeats. Mol. Gen. Genet. 212:450-458. [DOI] [PubMed] [Google Scholar]
- 37.Pestova, E. V., L. S. Havarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853-862. [DOI] [PubMed] [Google Scholar]
- 38.Polissi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prats, H., B. Martin, P. Pognonec, A. C. Burger, and J. P. Claverys. 1985. A plasmid vector allowing positive selection of recombinant plasmids in Streptococcus pneumoniae. Gene 39:41-48. [DOI] [PubMed] [Google Scholar]
- 40.Pujic, P., R. Dervyn, A. Sorokin, and S. D. Erlich. 1998. The kdgRKAT operon of Bacillus subtilis: detection of the transcript and regulation by the kdgR and ccpA genes. Microbiology 144:3111-3118. [DOI] [PubMed] [Google Scholar]
- 41.Puyet, A., A. M. Ibanez, and M. Espinosa. 1993. Characterization of the Streptococcus pneumoniae maltosaccharide regulator MalR, a member of the LacI-GalR family of repressors displaying distinctive genetic features. J. Biol. Chem. 268:25402-25408. [PubMed] [Google Scholar]
- 42.Rosenow, C., P. Ryan, J. N. Weiser, S. Johnson, P. Fontan, A. Ortqvist, and H. R. Masure. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25:819-829. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Schultz, J., R. R. Copley, T. Doerks, C. P. Ponting, and P. Bork. 2000. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28:231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Short, J. M., J. M. Fernandez, J. A. Sorge, and W. D. Huse. 1988. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 16:7583-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 47.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. G. Wallis, D. J. Brown, D. J. Holmes, M. Rosenberg, and K. R. Burnham. 2000. A genomic analysis of two-component signal transduction systems in Streptococcus pneumoniae. Mol. Microbiol. 35:566-576. [DOI] [PubMed] [Google Scholar]
- 48.Trombe, M. C. 1993. Characterization of a calcium porter of Streptococcus pneumoniae involved in calcium regulation of growth and competence. J. Gen. Microbiol. 139:433-439. [DOI] [PubMed] [Google Scholar]
- 49.Trombe, M. C., V. Rieux, and F. Baille. 1994. Mutations which alter the kinetics of calcium transport alter the regulation of competence in Streptococcus pneumoniae. J. Bacteriol. 176:1992-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weickert, M. J., and S. Adhya. 1992. A family of bacterial regulators homologous to Gal and Lac repressors. J. Biol. Chem. 267:15869-15874. [PubMed] [Google Scholar]
- 51.Yu, J., A. P. Bryant, A. Marra, M. A. Lonetto, K. A. Ingraham, A. F. Chalker, D. J. Holmes, D. Holden, M. Rosenberg, and D. McDevitt. 2001. Characterization of the Streptococcus pneumoniae NADH oxidase that is required for infection. Microbiology 147:431-438. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, J. R., I. Idanpaan-Heikkila, W. Fischer, and E. I. Tuomanen. 1999. Pneumococcal licD2 gene is involved in phosphorylcholine metabolism. Mol. Microbiol. 31:1477-1488. [DOI] [PubMed] [Google Scholar]
- 53.Zwijnenburg, P. J., T. van der Poll, S. Florquin, S. J. van Deventer, J. J. Roord, and A. M. van Furth. 2001. Experimental pneumococcal meningitis in mice: a model of intranasal infection. J. Infect. Dis. 183:1143-1146. [DOI] [PubMed] [Google Scholar]