Abstract
Bordetella avium causes an upper respiratory tract disease (bordetellosis) in avian species. Commercially raised turkeys are particularly susceptible. Like other pathogenic members of the genus Bordetella (B. pertussis and B. bronchiseptica) that infect mammals, B. avium binds preferentially to ciliated tracheal epithelial cells and produces similar signs of disease. These similarities prompted us to study bordetellosis in turkeys as a possible nonmammalian model for whooping cough, the exclusively human childhood disease caused by B. pertussis. One impediment to accepting such a host-pathogen model as relevant to the human situation is evidence suggesting that B. avium does not express a number of the factors known to be associated with virulence in the other two Bordetella species. Nevertheless, with signature-tagged mutagenesis, four avirulent mutants that had lesions in genes orthologous to those associated with virulence in B. pertussis and B. bronchiseptica (bvgS, fhaB, fhaC, and fimC) were identified. None of the four B. avium genes had been previously identified as encoding factors associated with virulence, and three of the insertions (in fhaB, bvgS, and fimC) were in genes or gene clusters inferred as being absent or incomplete in B. avium, based upon the lack of DNA sequence similarities in hybridization studies and/or the lack of immunological cross-reactivity of the putative products. We further found that the genotypic arrangements of most of the B. avium orthologues were very similar in all three Bordetella species. In vitro tests, including hemagglutination, tracheal ring binding, and serum sensitivity, helped further define the phenotypes conferred by the mutations. Our findings strengthen the connection between the causative agents and the pathogenesis of bordetellosis in all hosts and may help explain the striking similarities of the histopathologic characteristics of this upper airway disease in avian and mammalian species.
Bordetella avium is the causative agent of bordetellosis, an avian upper respiratory tract disease to which commercially raised turkeys are particularly susceptible (42). As with other pathogenic species of the genus Bordetella (B. pertussis and B. bronchiseptica), B. avium binds preferentially to ciliated tracheal epithelial cells (2, 38, 46). Subsequent death of the ciliated cells is thought to contribute to the clinical signs associated with bordetellosis (e.g., coughing and oculonasal discharge) (42). In addition, infected turkeys are more susceptible to secondary infections with other pathogens, such as Escherichia coli (4, 39, 42).
For B. pertussis and B. bronchiseptica, a number of factors that contribute to virulence in experimental rodent infections have been identified (7, 14, 15, 19, 31, 47). Some of these factors are absent from B. avium (e.g., pertussis toxin and adenylate cyclase) (12), and the absence of other factors has been inferred from the lack of DNA sequence similarities (e.g., the lack of part of the sensory transduction system encoded by bvgA and bvgS) (13). In other instances, B. avium has been shown to have structures and/or activities similar to those in the other pathogenic bordetellae (e.g., fimbriae, dermonecrotic toxin, tracheal cytotoxin, and hemagglutination) (12, 32, 33, 48), but current evidence indicates that B. avium effects the assembly of these structures and the expression of these activities via gene products that have little similarity, at least at the DNA level, to those in the other pathogenic bordetellae (13, 48, 53). Indeed, at the DNA level, there is relatively little to suggest a close association of B. avium with the other two major pathogenic bordetellae (11). Likewise, the avian host is a fairly distant relative of the mammals infected by B. pertussis and B. bronchiseptica. Thus, there is ample reason to accept the view that the factors associated with B. avium virulence differ significantly from those associated with other medically important Bordetella species (49). Nevertheless, the striking similarities of tissue tropism, disease presentation, and pathogenesis produced by all medically important Bordetella species (in their respective hosts) (2, 26, 38) support the suspicion that the bacterial factors producing the characteristic signs of infection are common to all of the bordetellae (35).
In this report, we identified avirulent B. avium mutants that had lesions in four genes (bvgS, fhaB, fhaC, and fimC) whose products are associated with virulence in B. pertussis and B. bronchiseptica. Whereas the genes had limited similarity at the DNA level, they were clearly orthologous when gene size, primary amino acid sequence, and genetic organization were taken into account. Further phenotypic characterization of the mutants in vivo confirmed their profound attenuation, and in vitro assays helped define possible molecular mechanisms responsible for the attenuation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains and plasmids used in this study are listed in Table 1. The B. avium-specific bacteriophage Ba1c1 (40) was also used. Ba1c1 is a clear-plaque derivative of Ba1 that binds to lipopolysaccharide (41). Brain heart infusion (BHI; Difco) and Stainer-Scholte medium were used with previously described B. avium growth conditions (45). Antibiotics were added at previously described concentrations (43). All E. coli strains were grown in Luria broth or agar (29) at 37°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | φ80dlacZ ΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR (rK− mK+) phoA supE44 λ-thi-1 gyrA96 relA1 | Life Technologies |
| MC4100 (λpir) | araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR(λ pir) | 30 |
| S17.1 (λpir) | F−thi pro hsdR recA ΩpRP4 (Amps Tets Kans Tmpr) (λpir) | 18 |
| B. avium | ||
| 197N | parental B. avium strain; Nalr | 45 |
| PAS213 | fimC insertion mutant of B. avium 197N; fimC::mini-Tn5; Kanr Nalr | This study |
| PAS334 | fhaB insertion mutant of B. avium 197N; fhaB::mini-Tn5; Kanr Nalr | This study |
| PAS355 | fhaC insertion mutant of B. avium 197N; fhaC::mini-Tn5; Kanr Nalr | This study |
| PAS356 | bvgS insertion mutant of B. avium 197N; bvgS::mini-Tn5; Kanr Nalr | This study |
| Plasmids | ||
| pUTKm2 | Ampr KanroriR6K oriTRP4 | 10 |
| pLAFR5 | Broad-host-range vector; Tetr | 23 |
| pLAFR5-4a | pLAFR5 containing a 7-kbp segment of B. avium 197N DNA containing the fimB, fimC, fimD, and fhaC genes introduced into the BamHI and HindIII sites; Tetr | This study |
| pLAFR5-5a | pLAFR5 containing a 5-kbp segment of B. avium 197N DNA containing the fimB, fimC, and fimD genes introduced into the BamHI and HindIII sites; Tetr | This study |
STM.
Signature-tagged mutagenesis (STM) was performed basically as described by Hensel et al. (17) with the following changes. Mini-Tn5 plasmid pUTKm2 containing random DNA tags within the transposon was kindly provided by D. Holden. Twenty-four uniquely tagged plasmids were evaluated for amplification and hybridization efficiencies. Each plasmid was introduced into E. coli strain MC4100 (λpir) by transformation for routine storage and evaluation. For conjugation, the plasmids were introduced into strain S17.1 (λpir) by transformation and subsequently transferred into B. avium by performing 24 conjugation experiments (43) and picking a single exconjugant from each mating. These 24 B. avium mutants comprised what we subsequently refer to as a pool of mutants in that they represent 24 uniquely tagged insertion mutants. Additional mutant pools were obtained by repeating the conjugation procedure and storing the exconjugants in microtiter trays. Prior to turkey inoculations, a pool of mutants was grown (each mutant in a pure culture) in 1.0 ml of BHI containing 40 μg of kanamycin/ml in 48-well cell culture clusters overnight at 37°C. On the following day, 100-μl portions of the cultures were combined, and the resultant mixture (pool) was used to inoculate four BHI agar plates containing kanamycin (40 μg/ml) and nalidixic acid (25 μg/ml). After incubation overnight at 37°C, the bacteria were resuspended in 4 ml of phosphate-buffered saline, and a portion of this suspension was used to isolate input pool chromosomal DNA by the cetyltrimethylammonium bromide (CTAB) protocol (3). The remaining sample was serially diluted and used to infect groups of turkeys.
Ten 1-week-old turkey poults were inoculated with ca. 109 B. avium organisms consisting of approximately equal proportions of the 24 uniquely tagged insertion mutants. At 8 days and at 2 weeks postinoculation, 5 of the 10 birds were removed and their tracheas were swabbed. The birds swabbed at day 8 were marked (with a wing band) and replaced in the brooder (to maintain a constant bird density throughout the experiment), and the remainder were swabbed at day 14. There was little difference in the mutants identified at 8 days and at 14 days. Mutants absent at both 8 days and 14 days are reported here. Tracheal swab samples were placed on BHI agar plates (one plate per poult) containing kanamycin and nalidixic acid and incubated at 37°C overnight. Resultant lawns of bacteria were resuspended in 500 μl of phosphate-buffered saline. Chromosomal DNA was isolated from each recovered pool (10 samples, each representing one poult) by the CTAB protocol (3). Each chromosomal preparation (input and recovered pools) was used as a template for a PCR with primers P2 (5′-dTACCTACAACCTCAAGCT) and P4 (5′-dTACCCATTCTAACCAAGC) (primers that bind to sites common in all tagged transposons) (17). The resultant amplicons were combined into 8-day (five poults) and 14-day (five poults) recovered pools and gel purified (Stratagene). A portion of each mixture (input, 8 day, and 14 day) was reamplified in the presence of digoxigenin, digested with HindIII, and used to probe DNA dot blots. Dot blots comprising the set of 24 original DNA tags (one representing each mutant) were made as recommended by the manufacturer (Bio-Rad).
Genetic analyses of STM mutants.
Cloning and sequence analysis of each mutant was performed as previously described (43). Briefly, B. avium DNA adjacent to the transposon was cloned by taking advantage of the gene encoding neomycin phosphotransferase, conferring kanamycin resistance, within the transposon. Chromosomal DNA from each mutant was digested with BglII or NotI and ligated into pLitmus28 (New England Biolabs) digested with BglII or pBluescript (Stratagene) digested with NotI. The ligation mixture was introduced into competent E. coli DH5α by transformation, and transformants were selected on Luria agar containing 40 μg of kanamycin/ml and 100 μg of ampicillin/ml. The resulting transformants harbored cloned DNA segments containing a portion of the transposon and flanking DNA. The clones were sequenced at the University of North Carolina—Chapel Hill Automated DNA Sequencing Facility by using a model 377 DNA sequencer (Perkin-Elmer, Applied Biosystems Division), an ABI Prism dye terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase FS (Perkin-Elmer, Applied Biosystems Division), and primer P7 (5′-dGCACTTGTGTATAAGAGTCAG) (17) or P6 (5′-dCCTAGGCGCCCAGATCTGAT) (17).
Cloning of the parental bvgA and bvgS genes.
A λ12 library was made by partial Tsp509I digestion of B. avium 197N chromosomal DNA ligated into EcoRI-digested λ12 zapII. DNA probes were made by a PCR with primers bvgS-1 (5′-dCAGATAGGCAAACGGCGA) and bvgS-2 (5′-dCGTCCAGATATTGCTGGTGAC), based upon the DNA sequence adjacent to the transposon mutation 20C4 (bvgS). This bvgS probe was used to screen ca. 3,000 λ12 clones by hybridization analysis. Reacting clones were partially sequenced and mapped.
Cloning of the parental fha-fim locus.
Isolation of the fha-fim locus was accomplished in two different ways. Initially, the λ12 library (described above) was screened by using a DNA probe based upon the DNA sequence adjacent to the transposon mutation 8C2 (see Results), which defined the 3′ end of the fimC gene. To gather additional fhaB information, a probe based upon the sequence immediately 3′ to fhaB (from the transposon in mutant 18C4) was used to clone an approximately 7-kbp HindIII chromosomal DNA fragment. This fragment, comprising ca. 6 kbp of fhaB and a partial fimA gene, was then sequenced and mapped. Additional λ library clones, comprising an overlapping set covering the entire region from fhaB to fhaC, were obtained by using DNA probes derived from sequences obtained from the above initial clones. The 5′ end of fhaB, including DNA immediately upstream, was cloned by using BglII to isolate a ca. 9-kbp fragment that overlapped the HindIII clone by 1.7 kbp.
Infectious dose determinations.
A 50% infectious dose (ID50) measurement for each mutant was performed as previously described (45), and the results were analyzed by the method of Reed and Muench (37). Single-dose experiments, in which each of 10 turkeys was inoculated with ca. 107 CFU (41), were used to evaluate the restoration of virulence through complementation.
Erythrocyte agglutination, phage resistance, serum resistance, and tracheal adherence assays.
Erythrocyte agglutination and tracheal adherence assays were performed as previously described (45). Phage resistance was determined by using the B. avium-specific phage Ba1c1 as described by Shelton et al. (41). Serum resistance assays were performed as previously described (43).
Statistical methods.
The standard deviation of the mean was calculated with the aid of the Microsoft Excel STDEV function. The standard error was calculated as the standard deviation divided by the square root of the number of experiments. The statistical significance of mean differences was determined by using Student's t test with the aid of Microsoft Excel statistical analysis software (version 4.0). The mean parental ID50 was calculated by using seven independent determinations.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the two gene clusters identified in this study are AY155575 for the bvg gene cluster and AY155576 for the fha-fim gene cluster.
RESULTS
Identification of B. avium genes orthologous to B. pertussis and B. bronchiseptica genes.
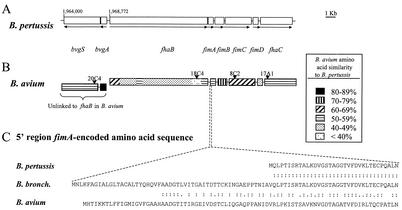
DNA sequence analysis of 10 independent insertion mutants, identified in STM screens as unable to colonize turkey poult tracheas (see Material and Methods), revealed that 4 had lesions in genes similar to those associated with virulence in B. pertussis and B. bronchiseptica (bvgS, fhaB, fhaC, and fimC) (Table 2). None of the four B. avium genes had been previously identified as encoding factors associated with virulence, and several of the insertions (in fhaB, bvgS, and fimC) were in genes or gene clusters inferred as being absent or incomplete in B. avium, based upon the lack of DNA similarity in hybridization studies and/or the lack of immunological cross-reactivity of the putative products (12, 13, 53). DNA sequence comparisons of the completely cloned and sequenced parental genes by using BLASTn (1) revealed at least one reason that some of the genes could have been overlooked: All but one of the genes (bvgS) had very limited DNA similarity (<55% homology) with B. pertussis and B. bronchiseptica genes. At the amino acid level, however, BLASTp comparisons of the primary sequences of the predicted proteins revealed striking similarities among all of the Bordetella species (Fig. 1). Whereas there was considerable variation in the degree of amino acid similarity in certain areas of the predicted gene products, the similarities at the levels of (i) gene size, (ii) overall primary protein structure, and (iii) genetic organization overwhelmingly supported the conclusion that the B. avium genes identified were orthologous to those in B. pertussis and B. bronchiseptica.
TABLE 2.
Genes identified in B. avium by STM
| Insertiona | Strain | Gene disrupted | Gene product structure/functionb |
|---|---|---|---|
| 18C4 | PAS334 | fhaB | Filamentous hemagglutinin (FHA)/attachment |
| 17A1 | PAS355 | fhaC | Accessory protein for FHA/attachment |
| 8C2 | PAS213 | fimC | Pilus chaperone/attachment |
| 20C4 | PAS356 | bvgS | Virulence sensor protein/transcription regulation |
Refer to Fig. 1 for diagram of the insertion site in each gene.
Function refers to those established for B. pertussis and/or B. bronchiseptica as referenced in the text.
FIG. 1.
Genetic and physical maps of the B. avium genes discovered by STM. (A) Sizes and arrangement of B. pertussis genes (based on data in references 25 and 54). (B) B. avium genes discovered by STM. Locations of insertion mutations that genetically define each gene are indicated by black triangles. The insertions are alphanumerically coded as in Table 2. Each gene is shaded to indicate the approximate degree of similarity to the B. pertussis gene (or region of a gene) at the amino acid sequence level, as denoted by the key. (C) Comparison of the fimA-encoded amino acid sequence of B. avium, truncated FimA of B. pertussis, and complete FimA of B. bronchiseptica (B. bronch.). Additional differences are described in the text. Homology scores (colons indicate identity; periods indicate similarity) were measured by using the PAM 250 scoring matrix in MACAW.
Genotypic analysis of the mutants.
All four B. pertussis genes (bvgS, fhaB, fhaC, and fimC) orthologous to the B. avium genes are closely linked in the arrangement shown in Fig. 1A. We found that B. avium had a similar genetic organization with respect to the linkage of fhaB to the fimABCD-fhaC region (Fig. 1B). Similarly, the B. avium bvgS gene was adjacent to a bvgA-like gene. However, the bvgA-bvgS pair was not immediately 5′ to fhaB. Instead, an open reading frame with similarity to vieA, a gene encoding a sensory regulator in Vibrio cholerae (24), was detected upstream of bvgA, and no evidence of bvg genes were detected upstream of fhaB.
With regard to fim genes, all four genes documented in B. pertussis and B. bronchiseptica (fimABCD) were detected in B. avium. For fimA, the gene encoding the structural subunit of type A fimbriae in B. bronchiseptica, we found an intact open reading frame (Fig. 1C) (the fimA gene is naturally truncated and nonfunctional in B. pertussis) (55). The last gene in the cluster was fhaC, a gene whose product is required for filamentous hemagglutinin (FHA) export and activity in both B. pertussis (54) and B. bronchiseptica (22).
Phenotypic characterization of the mutants.
To confirm that the mutants identified in the STM protocol were in fact attenuated, each mutant was tested individually for its ability to colonize 1-week-old turkey poults. We found that the mutant ID50 values were all significantly higher than those of the parent (Table 3). The mutants were further characterized by in vitro tests that measured the ability of the mutants to (i) agglutinate guinea pig erythrocytes, (ii) bind to tracheal rings from turkey embryos, (iii) resist the normal bactericidal activity of naive turkey serum, and (iv) resist killing by bacteriophage Ba1c1. Like the parent, all of the mutants remained Ba1c1 sensitive (data not shown), suggesting that the lesions did not alter lipopolysaccharide availability for bacteriophage binding (41). In addition, all of the mutants were able to agglutinate erythrocytes at or near parental levels and were indistinguishable from the parent in their levels of serum resistance (Table 3). Further, most of the mutants showed only modest reductions in their ability to bind to tracheal rings in vitro. Whereas the reduced binding was, in most instances, statistically significant; the only mutant that showed a dramatic (>10-fold) decrease in tracheal ring binding was the bvgS mutant (Table 3).
TABLE 3.
In vivo and in vitro properties of STM mutantsa
| Strain | Genotype | ID50 (106)b | % Hagc | Serum resistanced | Tracheal ring bindinge |
|---|---|---|---|---|---|
| 197N | parental | 7 ± 8.7 | (100) | 73 ± 13 | 6.68 ± 2.98 |
| PAS334 | fhaB::miniTn5 | ≥6590 ± 5600* | 100 ± 0 | 103 ± 33 | 7.72 ± 4.03 |
| PAS355 | fhaC::miniTn5 | ≥12400 ± 450** | 88 ± 18 | 75 ± 8 | 2.36 ± 0.90* |
| PAS213 | fimC::miniTn5 | ≥8680 ± 7900* | 75 ± 0 | 75 ± 23 | 0.97 ± 0.45** |
| PAS356 | bvgS::miniTn5 | ≥10500 ± 2500** | 88 ± 18 | 83 ± 27 | 0.52 ± 0.18** |
∗∗, to all values that were significantly different (P < 0.01) from the parental strain using the Student t test. ∗, values that were significantly different (P < 0.05) from the parental strain using the Student's t-test.
ID50 values were determined as previously described (45). Data are averages and standard deviations. Inequality (≥) refers to the fact that no birds were infected at any dose given. The numeric value shown is the lowest possible ID50 achieveable, i.e., it represents the ID50 value calculated if all animals were infected at a dose one order of magnitude higher than the highest dose employed.
Hag, hemagglutination. Overnight cultures were tested for their ability to agglutinate guinea pig erythrocytes in plate agglutination assays (16). Logarithms (base 2) of the reciprocal value of the agglutination titers were compared after normalization to that for the parental strain (100%). Values represent averages ± standard deviation for at least two separate experiments.
Serum resistance values are the percentage of initial inoculum that survives 1 h at 37°C in 50% naive turkey serum as described in the text. Values represent average ± standard deviations for at least two separate experiments.
Tracheal ring binding was carried out as described by Temple et al. (45). Values indicate the percentage of initial inoculum bound to embryonic turkey tracheal rings. Values represent averages ± standard deviations for at least two separate experiments performed in triplicate.
For the fimC mutant, previous studies carefully documented the polarity of at least one fimC insertion mutation upon fhaC expression in B. bronchiseptica (54). Because of this finding, we took the precaution of examining our fimC lesion in a cis-trans test for complementation. Recombinant plasmids containing the fimB, fimC, and fimD genes, either with or without fhaC, were tested in in vivo complementation experiments (turkey colonization). Complementation of both the fimC and the fhaC mutations required the fhaC gene product (Table 4). This result (i) confirmed that fhaC was important for virulence; (ii) indicated that the fhaC insertion was not polar for downstream genes (at least those relevant to our in vitro assay); and (iii) revealed that, as in B. bronchiseptica, the fimC lesion was polar for fhaC expression. The last point indicated that the fimC insertion could produce avirulence, entirely or partly, through a polar effect on fhaC expression.
TABLE 4.
Complementation of fimC and fhaC mutations
| Strain | Plasmid | Relevant plasmid genotype or property | No. of turkeys colonized/ no. testeda |
|---|---|---|---|
| PAS213 (197N [fimC::Tn5]) | None | 0/10 | |
| pLAFR5 | Cloning vector | 0/10 | |
| pLAFR5-4a | fimB fimC fimD fhaC | 10/10 | |
| pLAFR5-5a | fimB fimC fimD | 1/10 | |
| PAS355 (197N [fhaC::Tn5]) | None | 0/10 | |
| pLAFR5 | Cloning vector | 0/10 | |
| pLAFR5-4a | fimB fimC fimD fhaC | 9/9 | |
| pLAFR5-5a | fimB fimC fimD | 0/10 |
Number of turkeys colonized after 2 weeks/number inoculated with ∼107 CFU of the strain indicated as described in the text.
DISCUSSION
The use of experimental models to understand the disease pathogenesis of exclusively human infectious agents presents distinct challenges. At best, an acceptable match between host and pathogen is achieved so that the model host develops some signs of disease similar to those in humans and the model microorganism possesses some of the properties of the normal infectious agent (e.g., mice and Salmonella enterica serovar Typhimurium, humans and S. enterica serovar Typhi). Most recently, emphasis has been placed on the development of model infections in which less complex and less tactile hosts can be used in lieu of mammals. Such emphasis has given rise to a variety of infectious models, including plants (36), nematodes (8), insects (9), and fish (34). Whereas the genetic tractability of these hosts provides a powerful impetus for their examination, we are unaware of any nonmammalian model that mimics human disease as well as the domestic turkey with bordetellosis mimics human whooping cough. Our present work provides support for the genetic similarity of the two agents causing these diseases. Perhaps the most useful of our findings was our observation that several of the B. pertussis factors (whose role in virulence has remained uncertain due to the impediment of accurately modeling the disease) were found to be important for virulence when an entirely different Bordetella species was used in a natural infection of a distantly related host.
With regard to the specific genes identified in the STM screen, there is complete agreement that the sensory transducing products of the bvgA and bvgS genes (BvgA and BvgS) are required for virulence in B. pertussis and B. bronchiseptica (6, 51). For B. avium, we found that our bvgS mutant was attenuated in vivo, and in one of the in vitro tests of virulence, the bvgS mutant was the most defective, binding tracheal rings at 1/10 the level of the parent. DNA sequencing of regions adjacent to bvgS revealed the 5′ presence of a bvgA homologue but did not confirm the 3′ presence of a bvgR homologue (28)—although the sequence of the 3′ region was limited (ca. 50 bp). Our unpublished observations indicate that a bvgA insertion mutant, like the bvgS mutant, is completely avirulent (consistent with the model of the cooperative interaction between the bvgA and bvgS products in controlling the expression of a number of genes involved in virulence) (50). For B. avium, the arrangement of bvgA and bvgS (with respect to each other) was identical to that found in both B. bronchiseptica and B. pertussis. Unlike in B. pertussis, however, the B. avium bvgA and bvgS genes were unlinked to fhaB (44). We suspect that the putative B. avium bvgA and bvgS gene products function analogously to their counterparts in B. pertussis and B. bronchiseptica. The prototypic example of a gene regulated by the bvgA and bvgS gene products in B. pertussis is fhaB (44). We do not know whether fhaB is transcriptionally regulated by BvgA or BvgS in B. avium; however, the available data do not contradict this idea, since B. avium mutants with insertions in either bvgS or fhaB were attenuated.
Two of our four mutants had lesions in genes (fhaB and fhaC) whose counterparts in B. pertussis and B. bronchiseptica are required for the expression of FHA. In these two species, the product of the fhaC gene (FhaC) is required for the stability of the fhaB gene product (FHA) (54). A definitive role for FHA in the progression of a natural infection with bordetellae is best characterized for B. bronchiseptica, as FHA appears to play no agreed-upon role in virulence in B. pertussis (25, 52). Cotter et al. (7) found that B. bronchiseptica fhaB mutants were less able to colonize the tracheal epithelium of rats. Our results were similar in that our fhaB mutant showed a dramatic increase in ID50 but it was still capable of binding embryonic turkey tracheal rings in vitro, suggesting a role for B. avium FHA distinct from simple tracheal cell adherence. It is likely that the putative B. avium FHA is not a cell-associated hemagglutinin (i.e., required for agglutination when bacteria are mixed with erythrocytes). This supposition is based upon two observations. First, both B. avium fhaB and fhaC mutants were hemagglutination positive when bacteria were mixed with erythrocytes (unpublished results). Second, we have mapped 25 independently isolated B. avium insertion mutations that confer a hemagglutination-negative phenotype, and none of them maps to the fhaB gene (unpublished results).
The predicted B. avium FHA protein had an extended (72-amino-acid) signal sequence and the attachment motifs (RGD, RRARR, and CRD), secretion motifs (NPNL and NPNG), and proline-rich region described for B. pertussis (22). However, there was no evidence of the repeat regions (R1 and R2), the heparin-binding site, or the cleavage site (22). Further biochemical characterization and site-directed mutagenesis of B. avium FHA are required to establish the relationship of the numerous interesting biochemical features of FHA (brought out in studies of B. pertussis) to the pathogenesis of avian bordetellosis.
With regard to the B. avium fimC mutant, an early study by Mooi et al. (32) indicated that B. avium produces fimbriae. This result is supported by our unpublished electron microscopic observations and those of others (G. Luginbuhl, personal communication). Previous studies documented the requirement of fimC for piliation in B. pertussis (55). Electron microscopic examination of our fimC mutant reveled no piliated cells. However, relatively few examples of our parental strain were piliated (at least when grown under our standard laboratory conditions), weakening somewhat our ability to draw substantive conclusions as to the complete lack of piliation in the fimC mutants (unpublished observations). Whereas our present genetic evidence indicated that fimC may not be required for virulence and a fimC mutant emerged in the STM screen by virtue of its polar effect on fhaC expression, the fimC mutation did direct us to the unexpected and very dramatic similarity in the organization of the fha-fim regions in all medically important bordetellae.
Another interesting feature of the fim characterization was our finding of an intact fimA gene in B. avium. In B. pertussis, the fimA gene is truncated and the major pilin subunit for each of the different antigenic types of fimbriae is supplied by a gene unlinked to the fim cluster (55). In B. bronchiseptica, the fimA gene is intact and makes a product (5). However, available evidence suggests that the actual amount of pili made by this subunit in B. bronchiseptica is modest compared to the amounts made by other fimbrial subunits that are, as in B. pertussis, encoded elsewhere on the chromosome. In keeping with this theme, one report has indicated that B. avium produces pili with a subunit molecular mass of 13.1 kDa (20)—a molecular mass inconsistent with the 20.6-kDa product predicted from the fimA sequence.
In B. bronchiseptica, careful studies (27) have revealed that fimbrial production is required for rat tracheal colonization but not for nasal colonization. These results suggest a circumscribed role for B. bronchiseptica fimbriae in the infectious process. Our B. avium fimC mutant showed significantly reduced tracheal ring binding compared to the fhaC mutant (Table 3). The presence of this distinguishing phenotype indicates that the fimC lesion has effects beyond just a polar effect on fhaC expression. This may permit a more refined genetic analysis that will better define a role for fimC in pathogenesis.
The last gene in the cluster, fhaC, encodes a product required for FHA stability in B. pertussis and B. bronchiseptica (21, 22). Our finding that both fhaB and fhaC insertion mutants were avirulent is consistent with an interactive role. Indeed, in all virulence measurements that we performed (including tracheal ring binding), the two mutants were phenotypically indistinguishable when an appropriate statistical test was applied.
More work is required to rigorously confirm a role for each of the gene products described here in virulence. For example, replacing the insertions with well-defined in-frame deletion mutations should aid further studies of the role of these gene products in virulence by minimizing polarity. Such studies can now be approached with more assurance that at least some of the molecular pathogenic mechanisms of the bordetellae are likely to be quite similar, as the features of airway histopathologic characteristics have long suggested.
Acknowledgments
We thank Craig Altier and Scott Stibitz for critical reading of the manuscript. We thank Denarra Nevels for technical assistance.
This work was supported in part by grants from the U.S. Department of Agriculture, the National Institutes of Health, and the State of North Carolina and by a Wellcome Trust Programme Grant.
Editor: D. L. Burns
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Arp, L. H., and N. F. Cheville. 1984. Tracheal lesions in young turkeys infected with Bordetella avium. Am. J. Vet. Res. 45:2196-2201. [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Barnes, H. J., and M. S. Hofstad. 1978. Factors involved in respiratory disease of turkeys in Iowa. J. Am. Vet. Med. Assoc. 173:889-897. [Google Scholar]
- 5.Boschwitz, J. S., H. G. J. van der Heide, F. R. Mooi, and D. A. Relman. 1997. Bordetella bronchiseptica expresses the fimbrial structural gene fimA. J. Bacteriol. 179:7882-7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotter, P. A., and J. F. Miller. 1994. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect. Immun. 62:3381-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter, P. A., M. H. Yuk, S. Mattoo, B. J. Akerley, J. Boschwitz, D. A. Relman, and J. F. Miller. 1998. Filamentous hemagglutinin of Bordetella bronchiseptica is required for efficient establishment of tracheal colonization. Infect. Immun. 66:5921-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darby, C., L. L. Cosma, J. H. Thomas, and C. Manoil. 1999. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:15202-15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Argenio, D. A., L. A. Gallagher, C. A. Berg, and C. Manoil. 2001. Drosophila as a model host for Pseudomonas aeruginosa infection. J. Bacteriol. 183:1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLey, J., P. Seger, K. Kersters, W. Mannheim, and A. Lievens. 1986. Intra and intergeneric similarities of the Bordetella ribosomal ribonucleic acid cistrons: proposal for a new family, Alcaligenaceae. Int. J. Syst. Bacteriol. 36:405-414. [Google Scholar]
- 11.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gentry-Weeks, C. R., B. T. Cookson, W. E. Goldman, R. B. Rimler, S. B. Porter, and R. Curtiss III. 1988. Dermonecrotic toxin and tracheal cytotoxin, putative virulence factors of Bordetella avium. Infect. Immun. 56:1698-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gentry-Weeks, C. R., D. L. Provence, J. M. Keith, and R. Curtiss III. 1991. Isolation and characterization of Bordetella avium phase variants. Infect. Immun. 59:4026-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geuijen, C. A., R. J. Willems, M. Bongaerts, J. Top, H. Gielen, and F. R. Mooi. 1997. Role of the Bordetella pertussis minor fimbrial subunit, FimD, in colonization of the mouse respiratory tract. Infect. Immun. 65:4222-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodwin, M. S., and A. A. Weiss. 1990. Adenylate cyclase toxin is critical for colonization and pertussis toxin is critical for lethal infection by Bordetella pertussis in infant mice. Infect. Immun. 58:3445-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris, S. L., P. A. Spears, E. A. Havell, T. S. Hamrick, J. R. Horton, and P. E. Orndorff. 2001. Isolation and characterization of Escherichia coli type 1 pilus mutants that have altered binding specificities. J. Bacteriol. 183:4099-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hensel, M., J. E. Shea, C. Gleeson, M. D. Hjones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann, A., T. Thimm, M. Dröge, E. R. B. Moore, J. C. Munch, and C. C. Tebbel. 1998. Intergeneric transfer of conjugative and mobilizable plasmids harbored by Escherichia coli in the gut of the soil microarthropod Folsomia candida (Collembola). Appl. Environ. Microbiol. 64:2652-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa, H., and W. Sato. 1997. Role of Bordetella bronchiseptica sialic acid-binding hemagglutinin as a putative colonization factor. J. Vet. Med. Sci. 59:43-44. [DOI] [PubMed] [Google Scholar]
- 20.Jackwood, M. W., and Y. M. Saif. 1987. Pili of Bordetella avium: expression, characterization, and role in in vitro adherence. Avian Dis. 31:277-286. [PubMed] [Google Scholar]
- 21.Jacob-Dubuisson, F., C. El-Hamel, N. Saint, S. Guedin, E. Willery, G. Molle, and C. Locht. 1999. Channel formation by FhaC, the outer membrane protein involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem. 274:37731-37735. [DOI] [PubMed] [Google Scholar]
- 22.Jacob-Dubuisson, F., T. Kehoe, E. Willery, N. Reveneau, C. Locht, and D. A. Relman. 2000. Molecular characterization of Bordetella bronchiseptica filamentous hemagglutinin and its secretion machinery. Microbiology 146:1211-1221. [DOI] [PubMed] [Google Scholar]
- 23.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 24.Lee, S. H., M. J. Angelichio, J. J. Mekalanos, and A. Camilli. 1998. Nucleotide sequence and spatiotemporal expression of the Vibrio cholerae vieSAB genes during infection. J. Bacteriol. 180:2298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Locht, C., P. Bertin, F. D. Menozzi, and G. Renauld. 1993. The filamentous hemagglutinin, a multifaceted adhesion produced by virulent Bordetella spp. Mol. Microbiol. 9:653-660. [DOI] [PubMed] [Google Scholar]
- 26.Mallory, F. B., and A. A. Horner. 1913. Pertussis: the histological lesion in the respiratory tract. J. Med. Res. 27:115-123. [PMC free article] [PubMed] [Google Scholar]
- 27.Mattoo, S., J. F. Miller, and P. A. Cotter. 2000. Role of Bordetella bronchiseptica fimbriae in tracheal colonization and development of a humoral immune response. Infect. Immun. 68:2024-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merkel, T. J., C. Barros, and S. Stibitz. 1998. Characterization of the bvgR locus of Bordetella pertussis. J. Bacteriol. 180:1682-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Miller, V. L., and J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mooi, F. R., W. H. Jansen, H. Brunings, H. Gielen, H. G. van der Heide, H. C. Walvoort, and P. A. Guinee. 1992. Construction and analysis of Bordetella pertussis mutants defective in the production of fimbriae. Microb. Pathog. 12:127-135. [DOI] [PubMed] [Google Scholar]
- 32.Mooi, F. R., H. G. J. van der Heide, A. R. ter Avest, K. G. Welinder, I. Livey, B. A. M. van der Zeijst, and W. Gaastra. 1987. Characterization of fimbrial subunits from Bordetella species. Microb. Pathog. 2:473-484. [DOI] [PubMed] [Google Scholar]
- 33.Moore, K. M., M. W. Jackwood, T. P. Brown, and D. W. Dreesen. 1994. Bordetella avium hemagglutination and motility mutants: isolation, characterization and pathogenicity. Avian Dis. 38:50-58. [PubMed] [Google Scholar]
- 34.Neely, M. N., J. D. Pfeifer, and M. Caparon. 2002. Streptococcus-zebra fish model of bacterial pathogenesis. Infect. Immun. 70:3904-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orndorff, P. E. 1991. Bacterial virulence, p. 640-658. In A. Balows, H. G. Truper, M. Dworkin, W. Harner, and H.-K. Schleifer (ed.), The prokaryotes, 2nd ed., vol. I. Springer-Verlag, New York, N.Y.
- 36.Rahme, L. G., M. W. Tan, L. Le, S. M. Wong, R. G. Tompkins, S. B. Calderwood, and F. M. Ausubel. 1997. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 94:13245-13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent end points. Am. J. Hyg. 27:293-299. [Google Scholar]
- 38.Rhea, L. J. 1915. The comparative pathology of the tracheal and bronchial lesions produced in man by B. pertussis (whooping cough) and those produce in dogs by B. bronchiseptica (canine distemper). J. Med. Res. 32:471-474. [PMC free article] [PubMed] [Google Scholar]
- 39.Saif, Y. M., P. D. Moorhead, R. N. Dearth, and D. J. Jackwood. 1980. Observations on Alcaligenes faecalis infection in turkeys. Avian Dis. 24:665-684. [PubMed] [Google Scholar]
- 40.Shelton, C. B., D. M. Miyamoto, D. R. Crosslin, J. L. Casey, L. M. Temple, and P. E. Orndorff. 2000. Discovery, purification, and characterization of a temperate transducing bacteriophage, Ba1, for Bordetella avium. J. Bacteriol. 182:6130-6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shelton, C. B., L. M. Temple, and P. E. Orndorff. 2002. Use of bacteriophage Ba1 to identify properties associated with Bordetella avium virulence. Infect. Immun. 70:1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skeeles, J. K., and L. H. Arp. 1997. Bordetellosis (turkey coryza), p. 275-288. In B. W. Calnek, H. J. Barnes, C. W. Beard, L. R. McDougal, and Y. M. Saif (ed.), Diseases of poultry. Iowa State University Press, Ames.
- 43.Spears, P. A., L. M. Temple, and P. E. Orndorff. 2000. A role for lipopolysaccharide in turkey tracheal colonization by Bordetella avium as demonstrated in vivo and in vitro. Mol. Microbiol. 36:1425-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stibitz, S., A. A. Weiss, and S. Falkow. 1988. Genetic analysis of a region of the Bordetella pertussis chromosome encoding filamentous hemagglutinin and the pleiotropic regulatory locus vir. J. Bacteriol. 170:2904-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Temple, L. M., A. A. Weiss, K. E. Walker, H. J. Barnes, V. L. Christensen, D. M. Miyamoto, C. B. Shelton, and P. E. Orndorff. 1998. Bordetella avium virulence measured in vivo and in vitro. Infect. Immun. 66:5244-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toumanen, E. I., and J. O. Hendley. 1983. Adherence of Bordetella pertussis to human respiratory epithelial cells. J. Infect. Dis. 148:125-130. [DOI] [PubMed] [Google Scholar]
- 47.van den Berg, B. M., H. Beekhuizen, R. J. Willems, F. R. Mooi, and R. van Furth. 1999. Role of Bordetella pertussis virulence factors in adherence to epithelial cell lines derived from the human respiratory tract. Infect. Immun. 67:1056-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker, K. E., and A. A. Weiss. 1994. Characterization of the dermonecrotic toxin in members of the genus Bordetella. Infect. Immun. 62:3817-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiss, A. A. 1991. The genus Bordetella, p. 2530-2543. In A. Balows, H. G. Truper, M. Dworkin, W. Harner, and H.-K. Schleifer (ed.), The prokaryotes, 2nd ed., vol. I. Springer-Verlag, New York, N.Y.
- 50.Weiss, A. A., and S. Falkow. 1984. Genetic analysis of phase change in Bordetella pertussis. Infect. Immun. 43:263-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiss, A. A., E. Hewlett, G. A. Meyers, and S. Falkow. 1983. Tn5-induced mutations affecting virulence factors of Bordetella pertussis. Infect. Immun. 42:33-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiss, A. A., E. L. Hewlett, G. A. Meyers, and S. Falkow. 1984. Pertussis toxin and extra-cytoplasmic adenylate cyclase as virulence factors of Bordetella pertussis. J. Infect. Dis. 150:219-222. [DOI] [PubMed] [Google Scholar]
- 53.Willems, R. J., C. Geuijen, H. G. van der Heide, M. Matheson, A. Robinson, L. F. Versluis, R. Ebberink, J. Theelen, and F. R. Mooi. 1993. Isolation of a putative fimbrial adhesin from Bordetella pertussis and the identification of its gene. Mol. Microbiol. 9:623-634. [DOI] [PubMed] [Google Scholar]
- 54.Willems, R. J., C. Geuijen, H. G. van der Heide, G. Renauld, P. Bertin, W. M. van den Akker, C. Locht, and F. R. Mooi. 1994. Mutational analysis of the Bordetella pertussis fim/fha gene cluster: identification of a gene with sequence similarities to haemolysin accessory genes involved in export of FHA. Mol. Microbiol. 11:337-347. [DOI] [PubMed] [Google Scholar]
- 55.Willems, R. J. L., C. Geuijen, H. G. J. van der Heide, and F. R. Mooi. 1992. Characterization of a Bordetella pertussis fimbrial gene cluster which is located directly downstream of the filamentous hemagglutinin gene. Mol. Microbiol. 6:2661-2671. [DOI] [PubMed] [Google Scholar]