Abstract
Xenotropic and polytropic murine leukemia viruses (X-MLVs and P-MLVs) cross-interfere to various extents in non-mouse species and in wild Asian mice, suggesting that they might use a common receptor for infection. Consistent with this hypothesis, the susceptibility of some wild mice to X-MLVs has been mapped to the P-MLV receptor locus at the distal end of mouse chromosome 1. In this study, we report the isolation and characterization of a cDNA for the human X-MLV cell surface receptor (X-receptor) by using a human T lymphocyte cDNA library in a retroviral vector. The predicted X-receptor contains 696 amino acids with multiple hydrophobic potential membrane-spanning sequences and with weak homologies to the yeast proteins SYG1, of unknown function, and PHO81, which has been implicated in a system that regulates transport of inorganic phosphate. Expression of the X-receptor in Chinese hamster ovary cells, which are substantially resistant to P-MLVs and to X-MLVs, made them susceptible to both of these virus groups. The mouse homologue of the X-receptor was mapped by hybridization to the distal end of chromosome 1 at the same position as the P-MLV receptor gene Rmc1. These results strongly support the hypothesis that a common gene encodes the receptors for X-MLVs and P-MLVs, with the human X-receptor preferentially mediating X-MLV infections and the homologous protein of inbred mice mediating only P-MLV infections. We propose that X-MLVs and P-MLVs comprise a single family of retroviruses that have coevolved in response to diversification in X-receptor genes of the host.
Retroviral infections are initiated by binding of the viral surface envelope glycoprotein to specific receptor proteins on the host cell membrane. This initial interaction frequently determines the viral host range and, together with interference studies, has been used to classify retroviruses into distinct groups (1). Thus, murine leukemia viruses (MLVs) have been classified into ecotropic, amphotropic, 10A1, xenotropic (X-MLV), and polytropic (P-MLV) host range/interference groups (1–3). Ecotropic MLVs can only infect rats and mice whereas amphotropic and 10A1 MLVs infect a broad range of mammalian species, including humans. X-MLVs are infectious for most mammalian species but are only weakly infectious for wild Asian mice and are unable to infect laboratory strains of mice. In contrast, P-MLVs infect a broad range of mammalian species with variable efficiencies and are exceptionally leukemogenic in mice (4–8). Because they cause changes in mink cell cultures, P-MLVs also have been called mink cell focus-inducing viruses (MCFs) (5, 9).
Cell surface receptors for several of these MLV groups have been functionally cloned and characterized (10–13). Ecotropic MLVs use the cationic amino acid transporter CAT-1 as a receptor (14, 15) whereas amphotropic and 10A1 MLVs use the sodium-dependent phosphate symporter Pit2 (16, 17). In addition, 10A1 MLV can use the sodium-phosphate symporter Pit1 (18, 19), which previously had been identified as the receptor for gibbon ape leukemia virus and feline leukemia virus subgroup B (11, 20).
In contrast, cellular receptors for X-MLVs and P-MLVs have not been identified. Although these viruses have distinct host ranges and pathogenic effects, several lines of evidence have implied that they might use a common receptor in certain cells. In wild Asian mice, which are highly susceptible to P-MLVs and weakly susceptible to X-MLVs, the receptor gene for X-MLV maps at or near the P-MLV receptor gene locus on the distal end of chromosome 1 (Chr1) (21–23). This suggests that a variant of the P-MLV receptor gene in these Asian mice also can be used by X-MLVs. Consistent with this hypothesis, expression of a P-MLV SU glycoprotein in cells from these mice interferes with both P-MLV and X-MLV infections (22, 24–26). Furthermore, in cells from Mus dunni and mink, which are susceptible to both X-MLVs and P-MLVs, interference between the two virus groups is nonreciprocal, with X-MLVs efficiently blocking P-MLV infections and P-MLVs only partially interfering with X-MLVs (27, 28). This evidence has led to the suggestion that P-MLVs and X-MLVs may share a common receptor and that X-MLVs may, in addition, use a unique receptor (27). Alternatively, these viruses might use only a common receptor that is polymorphic in mice and divergent in other mammals. According to this interpretation, nonreciprocal interference would result from unequal affinities of X-MLVs and P-MLVs for their common receptor. This explanation predicts that the degree of interference would depend not only on the cells used but also on the specific X-MLVs and P-MLVs investigated, in agreement with observations (28).
To address these issues, we have attempted to clone a human cell surface receptor for X-MLVs. Previous efforts to clone retroviral receptors generally have relied on strategies to stably transfect cells from nonsusceptible species with cDNA or genomic libraries from susceptible species and to select the resulting susceptible cells by infection with a pseudotyped virus that encodes a dominant selectable gene (10–12). Because stable transfectants generally express numerous genes from the library, complex or iterative procedures have been necessary to isolate the receptor clones. Recently, Deng et al. (29) used a retroviral vector library to overcome many of these obstacles and to efficiently clone cDNAs for different coreceptors (BONZO and BOB) that are used by simian immunodeficiency viruses. Retroviral cDNA libraries also have been used to isolate relatively rare genes (30, 31). We, therefore, adapted this approach to successfully clone the X-MLV receptor (X-receptor) from a human T cell cDNA library. The X-receptor protein is predicted to contain 696 amino acids with eight or nine hydrophobic potential membrane-spanning domains. Of interest, the human X-receptor mediates infections by both X-MLVs and P-MLVs. In addition, the human X-receptor cDNA hybridizes strongly with mouse DNA sequences that map to the region of Chr1 that contains the P-MLV receptor locus. These results have important implications for understanding retroviral-receptor coevolution and the roles of receptor polymorphisms in controlling retroviral diseases.
MATERIALS AND METHODS
Cells and Viruses.
Mouse NIH 3T3, human TE671, and Chinese hamster ovary (CHO) cells were used as target cells for infection. TECeB15, TELCeB6 (32), and Phoenix-Eco (Garry Nolan, Stanford University, Stanford, CA) are packaging cell lines producing replication-defective retrovirus. TECeB15 and TELCeB6 cells do not contain retroviral envelope genes and therefore produce noninfectious virus. FLY cells producing replication-defective puro(RD114) pseudotype virus (RD114 virus carrying puromycin resistance gene) were provided by Y. Takeuchi (Institute of Cancer Research, London). CHO cells were maintained in Dulbecco’s modified α medium with 10% fetal bovine serum, and Phoenix-Eco cells were maintained in DMEM with high glucose and 10% fetal bovine serum. All other cell lines were maintained in DMEM with low glucose and 10% fetal bovine serum. X-MLV envelope gene NZB (2) was provided by Jean-Michel Heard (Pasteur Institute, Paris). Replication competent MCF13 (6) and MCF247 (5) viruses were provided by L. Evans (National Institute of Allergy and Infectious Diseases, Rocky Mountain Laboratories, Hamilton, MT).
LacZ(X-MLV) pseudotype virus was generated by transfection (calcium phosphate precipitation, Stratagene) of TELCeB6 cells with FBXsalf vector [xenotropic envelope gene NZB subcloned into the FBsalf retroviral expression vector (32)]. Transfectants were selected with phleomycin (50 μg/ml), and resistant colonies were pooled 2 weeks after addition of selection. LacZ(MCF13) and lacZ(MCF247) viruses were generated by first infecting NIH 3T3 cells with replication-competent MCF13 and MCF247. The resulting cell lines were infected with replication-defective lacZ(A-MLV) pseudotype virus to introduce the lacZ gene. Infection of target cells with lacZ pseudotype virus was carried out as described (33).
Replication-defective xenotropic pseudotype virus carrying the puromycin resistance gene puro(X-MLV) was generated by first transfecting TECeB15 cells with FBXsalf envelope expression vector. Phleomycin-selected transfectants were pooled and then were infected with replication-defective puro(RD114) pseudotype virus to introduce the puromycin resistance gene. Transduced cells were selected with puromycin (1 μg/ml), and the clone expressing the highest titer of puro(X-MLV) was used for infection studies.
Receptor Cloning.
A human T lymphocyte cDNA library, cloned into the retroviral vector pBabe-X (31), was generously provided by R. Sutton (Baylor College of Medicine, Houston, TX). Approximately 10 μg of retroviral plasmid library DNA was transfected into Phoenix-Eco packaging cells (2 × 106 cells in a 100-mm tissue culture plate) by using Qiagen SuperFect transfection reagent (Qiagen, Valencia, CA). Two days after transfection, the viral supernatant was filtered and added with 8 μg/ml polybrene to 10 100-mm tissue culture dishes, each containing 5 × 105 NIH 3T3 cells. After 16 h of incubation, the viral supernatant was replaced with fresh medium. The following day, the transduced NIH 3T3 cells were transferred to 150-mm tissue culture plates and were incubated with 10 ml of puro(X-MLV) supernatant for 16 h. The cells then were incubated with a fresh 10 ml of puro(X-MLV) supernatant for a further 4 h, after which the medium was replaced. The next day, cells in each plate were transferred to 2 × 150-mm plates, and puromycin was added at 5 μg/ml. Selection medium was replaced every 2 days until resistant colonies had appeared. Resistant colonies then were tested for susceptibility to lacZ(X-MLV) pseudotype.
Isolation of Receptor cDNA and Expression in Mammalian Cells.
The transduced cDNA was recovered by subjecting 250 ng of genomic DNA, isolated from lacZ(X-MLV) sensitive NIH 3T3 clones, to PCR amplification by using the Expand PCR kit from Boehringer Mannheim. The 4.5-kilobase (kb) X-MLV receptor cDNA X3 was amplified by using primers complementary to pBabe-X vector sequences flanking the cDNA insert (upstream primer, 5′-GATCCCAGTGTGCTGGAAAG-3′; downstream primer, 5′-GGTGGGGTCTTTCATTCC-3′). The PCR was run for 30 cycles with 54°C annealing temperature for 1.5 minutes, and with an extension temperature of 68°C for 7 min. The amplified DNA then was cloned into the pCR2.1TOPO vector (Invitrogen), and the subsequent plasmid was named pTOPO-X3. The DNA sequence was determined by the Microbiology and Molecular Immunology Core Facility on the PE/ABD 377 sequencer by using dye terminator cycle sequencing chemistry (Applied Biosystems, Foster City, CA). The accession number for the X3 cDNA sequence is AF089744.
NIH 3T3 and CHO cells expressing human X-MLV receptor were generated by transfection of the 4.5-kb receptor cDNA subcloned into pcDNA3.1(−) mammalian expression vector (Invitrogen). The receptor cDNA was isolated from pTOPO-X3 by XhoI-HindIII digestion and was cloned into XhoI–HindIII-cut pcDNA3.1. Transfectants were selected with G418 (1 mg/ml), and resistant cells were analyzed for sensitivity to lacZ(X-MLV).
Northern Blots.
Multiple tissue Northern blots containing ≈2 μg Poly(A)+ RNA from various human tissues were obtained from CLONTECH. The blots were probed with full length X3 cDNA that was labeled with 32P by using random primer extension system from New England Nuclear life science products.
Genetic Mapping.
The X3 cDNA isolated from pTOPO-X3 was used as hybridization probe to map the corresponding gene on the mouse linkage map by Southern blot analysis of the progeny of two sets of genetic crosses: (NFS/N or C58/J × Mus musculus musculus) × M. musculus musculus and (NFS/N × Mus spretus) × M. spretus or C84/J [C. A. Kozak and C. E. Buckler, Mouse Genome Database, Release 3.2, Mouse Genome Informatics, The Jackson Laboratory (http://www.informatics.jax.org)]. Progeny of these crosses have been typed for >1,200 loci distributed over the 19 autosomes and the X chromosome. Recombinational distances were determined according to Green (34), and loci were ordered by minimizing the number of recombinants.
RESULTS
Isolation of the 4.5-kb Xenotropic MLV Receptor (X-Receptor) cDNA.
To clone the receptor for X-MLV, we used a human T lymphocyte cDNA library subcloned into pBabe-X retroviral vector (see Materials and Methods). Forty-eight hours after transducing the library into NIH 3T3 fibroblasts, which are naturally resistant to X-MLVs, the cells were challenged with a replication-defective xenotropic pseudotype virus that encodes the dominant selectable gene for puromycin resistance. Selection in the presence of 5 μg/ml puromycin yielded seven resistant colonies. Only one of these (X3) was highly sensitive to lacZ(X-MLV) pseudotype virus whereas parental NIH 3T3 cells were resistant to infection (Fig. 1 A and B). Because NIH 3T3 cells are susceptible to amphotropic MLVs, we infected a sample of X3 cells with replication-competent amphotropic MLV(4070A) and used the rescued virus to infect naive NIH 3T3 cells. The latter cells also became highly susceptible to lacZ(X-MLV) infection, confirming that X3 cells contain a packagable retroviral vector that encodes the X-MLV receptor. To isolate the receptor cDNA, genomic DNA from clone X3 was subjected to PCR amplification by using primers specific for pBabe-X vector. PCR products of 4.5 and 1.3 kb were amplified, and these subsequently were cloned into pCR2.1TOPO vector and were sequenced. The 1.3-kb component encoded the human NADH dehydrogenase whereas the 4.5-kb component (termed X3) contained a region that appeared to be identical to an expressed sequence tag (EST 64781) derived from human JURKAT T leukemia cells.
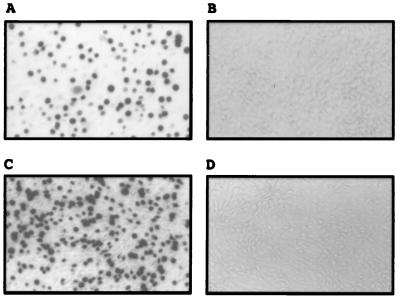
Figure 1.
Susceptibility of NIH 3T3 cells expressing the X3 cDNA to X-MLV pseudotype infection. Target cells were analyzed for their susceptibility to lacZ(X-MLV) pseudotype. (A) Infection of the puromycin-resistant NIH 3T3 cellular clone X3, previously transduced with the retroviral cDNA library vector and then with puro(X-MLV) pseudotype (see Materials and Methods). (B) Infection of NIH 3T3 cells. (C) Infection of NIH 3T3 cells transfected with pcDNA3.1-X3 (X3 cDNA cloned into the pCDNA3.1 mammalian expression vector). (D) Infection of NIH 3T3 cells transfected with only pCDNA3.1 vector.
The 4.5-kb X3 cDNA then was subcloned into the mammalian expression vector pcDNA3.1. NIH 3T3 fibroblasts transiently transfected with pcDNA3.1-X3 were highly susceptible to infection by lacZ(X-MLV) whereas cells transfected with vector alone were resistant (Fig. 1 C and D). Therefore, the 4.5-kb X3 cDNA encodes the X-receptor.
The X-Receptor Mediates Infections of both X-MLV and P-MLV.
We generated a murine NIH 3T3 cell clone (3T3X3A11) expressing the human X-receptor and found that it could be infected by lacZ(X-MLV) pseudotype virus (see Table 1). In contrast, expression of X-receptor in appropriate cells did not confer susceptibility to ecotropic, amphotropic, GALV, or FeLV-B (data not shown). Previous studies have indicated that X-MLVs and P-MLVs may use a common receptor in some cells (21, 22, 27, 28). To examine this, we transfected the X3 cDNA into CHO cells, which are naturally resistant to P-MLVs and only slightly sensitive to X-MLVs. As shown in Table 1, the resulting CX218 cell clone was highly sensitive to lacZ(X-MLV), weakly sensitive to lacZ(MCF13), and substantially more susceptible to lacZ(MCF247). The reduced susceptibility of these cells to MCF infections is in accordance with the relatively low titers of MCFs on human cells as reported (9) and shown for TE671 cells in Table 1. These results suggest that the human X-receptor has a preference for X-MLVs but also mediates infections of P-MLVs. Gene mapping evidence also implies that the mouse homologue of the X-receptor is closely related or identical to the P-MLV receptor gene Rmc1 (see below).
Table 1.
NIH3T3 and CHO cells transfected with the X-receptor cDNA expression vector are susceptible to xenotropic and polytropic MLVs infections
| Target cell | Titer of LacZ pseudotype, colony-forming units*
|
||
|---|---|---|---|
| X-MLV, NZB | MCF13 | MCF247 | |
| TE671 | 2.0 × 106 | 5 | 1.5 × 101 |
| NIH3T3 | 0 | 1.0 × 106 | 1.4 × 105 |
| 3T3X3A11† | 5.6 × 104 | 6.5 × 105 | 2.0 × 105 |
| CHOpcD | 2.2 × 102 | 0 | 0 |
| CX218‡ | 3.0 × 105 | 2.2 × 102 | 1.0 × 103 |
LacZ pseudotype infection of target cells after transfection of the X-receptor cDNA (X3). TE671 are human rhabdomyosarcoma cells; CHOpcD are CHO cells transfected with pcDNA3.1 mammalian expression vector only. Titers are average values of two infection studies.
3T3X3A11 is a G418-resistant NIH3T3 clone transfected with X3 cDNA.
CX218 is a G418-resistant CHO clone transfected with X3 cDNA.
Although we have produced CX218 derivatives that express large amounts of either xenotropic or polytropic envelope glycoproteins, the parental cultures had lost much of their susceptibility during the time required to obtain these derivatives. This instability of the cells has prevented us from precisely measuring the interferences caused by these viral glycoproteins. Despite this limitation, these studies have implied a significant degree of cross-interference between these viruses, in agreement with the conclusion that X-MLVs and P-MLVs both use the human X-receptor (results not shown).
The X-Receptor Protein.
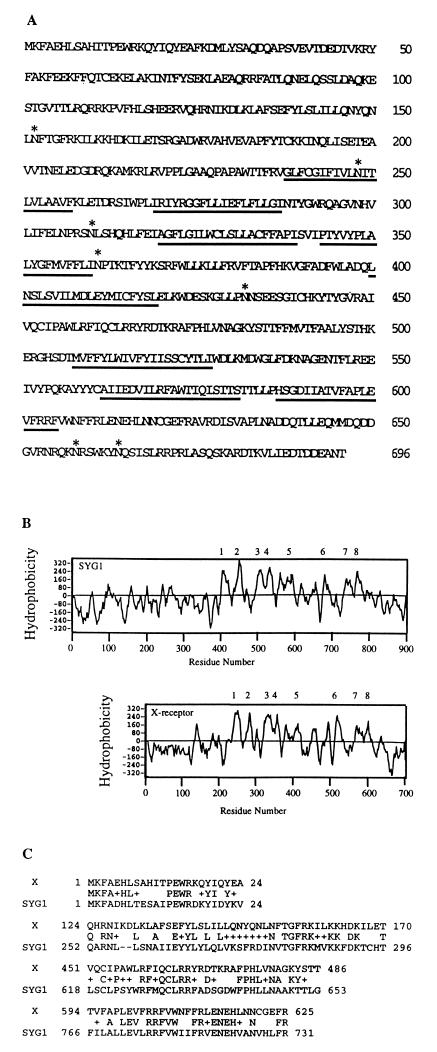
Fig. 2A shows the predicted amino acid sequence of the X-receptor. The ORF in the 5′ region of the 4.5-kb X3 cDNA encodes a protein of 696 amino acids. Although the X-receptor lacks an obvious signal sequence, suggesting that its amino terminus may occur in the cytosol, its Kyte-Doolittle hydrophobicity plot (35) implied the presence of eight or nine hydrophobic potential membrane-spanning sequences, consistent with occurrence in cellular membranes. blast [Basic Local Alignment Search Tool (http://www.ncbi.nlm.nih.gov/blast/)] comparisons with sequences in the databases suggested that the X-receptor is highly related to the yeast proteins SYG1 (P = 1 × 10−36) of unknown function and PHO81 (P = 7 × 10−7), which has been implicated indirectly in control of phosphate transport (36). SYG1 has a hydrophobicity plot that is very similar to that of the X-receptor (Fig. 2B), and it is also believed to be a membrane protein (37). Several regions of the X-receptor that are highly related in sequence to SYG1 are shown in Fig. 2C.
Figure 2.
The amino acid sequence and hydrophobicity plot of X-receptor and sequence comparison to the yeast SYG1 protein. (A) The X-receptor is a large protein of 696 amino acids. Potential membrane-spanning domains (underlined) were identified by using the Kyte and Doolittle algorithm (35). Potential N-linked glycosylation sites (NXS/T) are shown by asterisks. (B) Hydrophobicity plots were generated by using the Kyte and Doolittle algorithm. X-receptor and SYG1 contained at least eight hydrophobic potential membrane spanning sequences, as indicated. The transmembrane region labeled 5 could possibly contain an additional membrane-spanning sequence. (C) Alignment of highly related regions between SYG1 and X-receptor (X) determined by blast comparisons. The consensus sequence is shown between the X-receptor and SYG1 sequence, where + indicates related amino acids.
Expression of X3 RNA in Human Tissues.
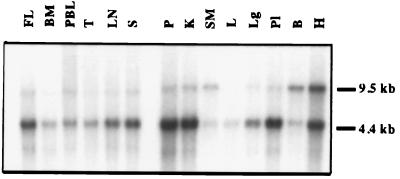
We analyzed the tissue distribution of X-receptor RNA by Northern blot analysis using X3 cDNA as a probe. A common X3 RNA with size slightly >4.4 kb was detected in all tissues tested (Fig. 3). This RNA was most abundant in the pancreas, kidney, placenta, and heart and most limited in skeletal muscle. Expression of this RNA was substantially greater in fetal liver, which is a site of active hematopoiesis, than in adult liver. Accordingly, expression of the 4.5-kb transcript was also substantial in other hematopoietic tissues, including spleen, lymph node, thymus, and peripheral blood lymphocytes. In addition, a 9.5-kb RNA was detected in all tissues except liver and bone marrow with relatively abundant expression in bone and heart.
Figure 3.
Northern blot analysis of Poly(A)+ RNA from various human tissues. The multiple-tissue Northern blots from human were probed with [32P]-labeled X3 cDNA. FL, fetal liver; BM, bone marrow; PBL, peripheral blood lymphocytes; T, thymus; LN, lymph node; S, spleen; P, pancreas; K, kidney; SM, skeletal muscle; L, liver; Lg, lung; Pl, placenta; B, brain; H, heart.
The Homologous X-Receptor Sequences of Mice Map at the Rmc1 Locus on Chromosome 1.
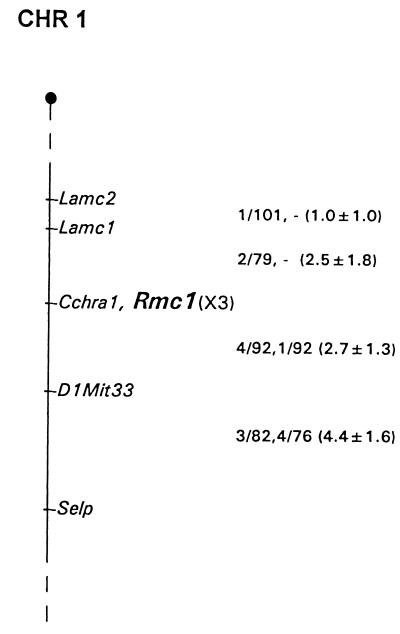
Southern hybridization was used to analyze DNAs from the parental mice of two sets of genetic crosses. ScaI digestion revealed X3-related fragments of 11.2 and 6.4 kb in NFS/N and 16.5 and 6.4 kb in M. Spretus. Digestion with HpaI produced fragments of 18.3 and 4.6 kb in NFS/N and 18.3 and 6.5 kb in M. musculus musculus. DNAs from the progeny of two sets of genetic crosses were typed for inheritance of these polymorphisms. The pattern of inheritance of the variant fragments was compared with inheritance of >1,200 markers previously typed in these crosses and was mapped to positions on all 19 autosomes and the X chromosome. Inheritance of the X3 fragments identified a single locus that mapped to distal Chr1 as shown in Fig. 4. Closest linkage was detected to Cchra1, for which no recombinants were identified in 162 mice, indicating that, at the upper limit of the 95% confidence interval, the X3-related sequences and Cchra1 are separated by no more than 1.83 centimorgans.
Figure 4.
Map location of the xenotropic/polytropic receptor gene on mouse Chr1. The map represents a 10-centimorgan segment of the distal end of Chr1. The entire Chr1 has a length of ≈107 centimorgans. To the right of the map are recombination fractions between adjacent loci, with the first fraction from the M. musculus musculus crosses and the second from M. spretus crosses. The numbers in parentheses represent recombinational distances ± standard errors. Marker loci were typed as previously described in this cross [C. A. Kozak and C. E. Buckler, Mouse Genome Database, Release 3.2, Mouse Genome Informatics, The Jackson Laboratory (http://www.informatics.jax.org/crossdata); ref. 38). The M. spretus crosses were not typed for Lamc1. Gene order was established by minimizing the number of recombinants. One single locus double recombinant involving Cchl2a was identified in the M. spretus crosses that is not included in the figure.
Previous studies had mapped the mouse gene responsible for susceptibility to polytropic and xenotropic viruses to distal Chr1 (21, 23, 38). The susceptibility gene for polytropic virus was assigned to this chromosomal region by using somatic cell hybrids and radiation hybrids that placed this gene distal to Lamc1 (21, 23). Genetic crosses mapped a gene responsible for weak susceptibility to a xenotropic virus to this same region and positioned a putative defective virus receptor in Mus castaneus near D1Mit33 (21, 38). The location of the X3 gene established in the present study closely corresponds to the expected location for the previously mapped phenotypes, and, therefore, we use the Rmc-1 designation for the locus identified by X3 (Fig. 4).
DISCUSSION
Functional Cloning of the Human Receptor for Xenotropic and Polytropic MLVs.
We report the functional cloning and initial characterization of a 4.5-kb human lymphocyte cDNA that encodes a cell surface receptor (the X-receptor) for both xenotropic and polytropic MLVs. Northern blot analyses suggest that the X-receptor is widely expressed in tissues including hematopoietic cells (see Fig. 3), consistent with its putative role in mediating disseminated systemic infections of mice that result in hematopoietic diseases (4, 5). In contrast, the receptor for amphotropic MLVs is expressed poorly in hematopoietic cells (17). This and other evidence (39) is consistent with the possibility that X-MLV retroviral vector pseudotypes may be useful for gene therapy targeting to specific hematopoietic cells.
Several lines of evidence in the present study strongly suggest that the X-receptor mediates infections of both X-MLVs and P-MLVs. First, expression of the human X-receptor in mouse NIH 3T3 cells (3T3X3A11) conferred susceptibility to infections by X-MLV (see Fig. 1 and Table 1). Although the 3T3X3A11 clone is likely to express high levels of the X-receptor, the infection titer of lacZ(X-MLV) on these cells was ≈40-fold lower than on human TE671. Similar differences in efficiencies of X-MLV infections also occur in different human cell lines (39). Second, in CHO cells that are naturally resistant to P-MLVs and only slightly susceptible to X-MLVs, expression of the human X-receptor in the CX218 cell clone and in other clones resulted in highly significant susceptibility to both X-MLVs and P-MLVs. These transfected cell clones were relatively more susceptible to X-MLVs than to P-MLV, consistent with the reduced susceptibility of human cells (e.g., TE671) to the P-MLV subgroup of viruses (see Table 1). It also has been observed previously that human cells are only weakly susceptible to P-MLVs (9). These results imply that the human X-receptor functions more strongly for X-MLVs than for P-MLVs. A third line of evidence derives from our mapping of the mouse X-receptor gene to the P-MLV receptor locus on the distal end of chromosome 1 (see Fig. 4). This analysis demonstrated that the human X-receptor gene hybridized to polymorphic restriction fragments of mouse DNA that mapped to a position on distal Chr1 consistent with the location of the P-MLV receptor gene Rmc-1 previously determined by susceptibility to infection (21, 23, 38). Fourth, the X-MLVs and P-MLVs are very closely related in their SU envelope sequences (40), consistent with utilization of a common receptor. Based on this evidence, we conclude that the human X-receptor functions strongly for X-MLVs and weakly for P-MLVs. Moreover, we infer from this and previous evidence (4, 21–26) that the homologous mouse protein may have essentially the opposite specificity. We recently have cloned the X-receptor cDNA from several strains of mice to directly test these interpretations.
Previously, it was proposed that some cells might contain two X-MLV receptors, with one being used also by P-MLVs (27). This hypothesis derived from the observation of nonreciprocal interference between X-MLVs and P-MLVs in M. dunni fibroblasts, with X-MLVs completely interfering with P-MLVs but P-MLVs causing only a 10-fold interference with X-MLVs (27). An alternative interpretation of this evidence is that nonreciprocal interference may be caused by differential competition of these viruses for a single receptor. If this were true, the degree of interference would be expected to depend on the specific viruses analyzed and on the quantities of their envelope glycoprotein expression. Consistent with this interpretation, the degree of interference between X-MLVs and P-MLVs in M. dunni fibroblasts depends on the specific virus isolates that are examined (28). Additional experiments will be needed to unambiguously investigate these alternative interpretations and to determine whether there may be additional receptors for X-MLVs and P-MLVs in mice or other species.
The X-Receptor Protein.
The primary structure of the X-receptor implies a protein of 696 amino acids with eight or nine hydrophobic potential membrane-spanning sequences (Fig. 2) and with seven NX(S/T) sites for potential N-linked glycosylation and seven dileucines that possibly could stimulate endocytosis via clathrin coated pits (see Fig. 2A) (41). The X-receptor is highly related to the yeast protein SYG1, both in its sequence (Fig. 2C) and in its hydrophobicity plot (Fig. 2B), and it is likely that these proteins have similar topologies in the membrane. Although the function of SYG1 is unknown, its amino terminal nonmembranous domain can replace the function of G α subunits in a yeast-signaling pathway (37). Based on this evidence for a cytoplasmic function and on the absence of obvious signal sequences in these proteins, we infer that their amino terminal domains occur in the cytosol. In addition, the X-receptor is widely expressed in tissues (Fig. 3) and has homology with the yeast protein PHO81 that has been implicated indirectly in control of phosphate transport (36). Recently, we have obtained evidence that the X-receptor is expressed on cell surfaces and that it functions as a transporter (N. Madani, C.S.T., D.K., and M. P. Kavanaugh, unpublished work).
General Implications.
Our results suggest that X-MLVs and P-MLVs comprise a single virus family that uses a common X-receptor encoded by a gene that maps on mouse Chr1. This gene is closely linked or identical to the polymorphic mouse Rmc1 gene that encodes the receptor for P-MLVs. A mutation that maps at this locus in M. castaneus causes complete resistance to P-MLV infections (38). Conversely, an allele of the Rmc1 gene in laboratory mice causes susceptibility to P-MLVs and complete resistance to X-MLVs whereas the allele in most wild mice confers susceptibility to both X-MLVs and P-MLVs (21–23). It seems likely that these polymorphisms have evolved in response to the highly pathogenic infections caused by this family of viruses (4–8) and that viral variants have coevolved to overcome these defenses. As a consequence, specific X-MLVs and P-MLVs can infect only certain populations of mice, and members of this virus family may interfere with each other only weakly or nonreciprocally. These considerations imply that retroviruses with distinct or even opposite host ranges can use a common polymorphic receptor. Availability of the X-receptor cDNA should facilitate further investigations of these issues, including identification of the polymorphisms that control infections by particular viruses of this family, of the adaptive changes in the viral envelope glycoproteins that overcome these controls, and of retroviruses in other species that also may use this or related receptors.
Acknowledgments
We are extremely grateful to Richard Sutton for supplying the retroviral cDNA library, Jean-Michel Heard for providing the xenotropic (NZB) envelope gene, and Yasuhiro Takeuchi and Garry Nolan for providing the packaging cells. We are grateful to Sandra Ruscetti, Leonard Evans, and Christine Holland for donating important materials. We are also indebted to our coworkers Susan Kozak, Emily Platt, Navid Madani, and Shawn Kuhmann for their encouragement and helpful suggestions and to Susan Kozak for help with the Northern blot analyses. This research was supported by National Institutes of Health Grants CA25810 and CA54149 and by The Wellcome Trust. C.S.T. is a Wellcome Trust International Prize Fellow.
ABBREVIATIONS
- MLV
murine leukemia virus
- X-MLV
xenotropic murine leukemia virus
- P-MLV
polytropic murine leukemia virus
- MCF
mink cell focus-inducing P-MLV
- X-receptor
human X-MLV receptor
- Chr1
chromosome 1
- CHO
Chinese hamster ovary
- kb
kilobase
Footnotes
References
- 1.Hunter E. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 71–120. [Google Scholar]
- 2.Levy J A. Curr Top Microbiol Immunol. 1978;79:111–213. doi: 10.1007/978-3-642-66853-1_4. [DOI] [PubMed] [Google Scholar]
- 3.Rein A, Schultz A. Virology. 1984;136:144–152. doi: 10.1016/0042-6822(84)90255-1. [DOI] [PubMed] [Google Scholar]
- 4.Fischinger P J, Nomura S, Bolognesi D. Proc Natl Acad Sci USA. 1975;72:5150–5155. doi: 10.1073/pnas.72.12.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartley J W, Wolford N K, Old L J, Rowe W P. Proc Natl Acad Sci USA. 1977;74:789–792. doi: 10.1073/pnas.74.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cloyd M W, Hartley J W, Rowe W P. J Exp Med. 1980;151:542–552. doi: 10.1084/jem.151.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenberg N, Jolicoeur P. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 475–585. [PubMed] [Google Scholar]
- 8.Kabat D. Curr Top Microbiol Immunol. 1989;148:1–42. doi: 10.1007/978-3-642-74700-7_1. [DOI] [PubMed] [Google Scholar]
- 9.Cloyd M W, Thompson M M, Hartley J W. Virology. 1985;140:239–48. doi: 10.1016/0042-6822(85)90362-9. [DOI] [PubMed] [Google Scholar]
- 10.Albritton L M, Tseng L, Scadden D, Cunningham J M. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 11.O’Hara B, Johann S V, Klinger H P, Blair D G, Rubinson H, Dunn K J, Sass P, Vitek S M, Robbins T. Cell Growth Differ. 1990;1:119–127. [PubMed] [Google Scholar]
- 12.Miller D G, Edwards R H, Miller A D. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeijl M V, Johann S V, Cross E, Cunningham J, Eddy R, Shows T B, O’Hara B. Proc Natl Acad Sci USA. 1994;91:1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Kavanaugh M P, North R A, Kabat D. Nature (London) 1991;352:729–731. doi: 10.1038/352729a0. [DOI] [PubMed] [Google Scholar]
- 15.Kim J W, Closs E I, Albritton L M, Cunningham J M. Nature (London) 1991;352:725–728. doi: 10.1038/352725a0. [DOI] [PubMed] [Google Scholar]
- 16.Olah Z, Lehel C, Anderson W B, Eiden M V, Wilson C A. J Biol Chem. 1994;269:25426–25431. [PubMed] [Google Scholar]
- 17.Kavanaugh M P, Miller D G, Zhang W, Law W, Kozak S L, Kabat D, Miller A D. Proc Natl Acad Sci USA. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller D G, Miller A D. J Virol. 1994;68:8270–8276. doi: 10.1128/jvi.68.12.8270-8276.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson C A, Farrell K B, Eiden M V. J Virol. 1994;68:7697–7703. doi: 10.1128/jvi.68.12.7697-7703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeuchi Y, Vile R G, Simpson G, O’Hara B, Collins M K L, Weiss R A. J Virol. 1992;66:1219–1222. doi: 10.1128/jvi.66.2.1219-1222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozak C A. J Virol. 1983;48:300–303. doi: 10.1128/jvi.48.1.300-303.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak C A. J Virol. 1985;55:690–695. doi: 10.1128/jvi.55.3.690-695.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunter K, Housman D, Hopkins N. Somatic Cell Mol Genet. 1991;17:169–183. doi: 10.1007/BF01232974. [DOI] [PubMed] [Google Scholar]
- 24.Ruscetti S, Davis L, Feild J, Oliff A. J Exp Med. 1981;154:907–920. doi: 10.1084/jem.154.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruscetti S, Matthai R, Potter M. J Exp Med. 1985;162:1579–1587. doi: 10.1084/jem.162.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bassin R H, Ruscetti S, Ali I, Haapala D K, Rein A. Virology. 1982;123:139–151. doi: 10.1016/0042-6822(82)90301-4. [DOI] [PubMed] [Google Scholar]
- 27.Miller A D, Wolgamot G. J Virol. 1997;71:4531–4535. doi: 10.1128/jvi.71.6.4531-4535.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chesebro B, Wehrly K. Virology. 1985;141:119–129. doi: 10.1016/0042-6822(85)90188-6. [DOI] [PubMed] [Google Scholar]
- 29.Deng H, Unutmaz D, KewalRamani V N, Littman D R. Nature (London) 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 30.Whitehead I, Kirk H, Kay R. Mol Cell Biol. 1995;15:704–710. doi: 10.1128/mcb.15.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitamura T, Onishi M, Kinoshita S, Shibuya A, Miyajima A, Nolan G P. Proc Natl Acad Sci USA. 1995;92:9146–9150. doi: 10.1073/pnas.92.20.9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cosset F L, Takeuchi Y, Battini J L, Weiss R A, Collins M K L. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tailor C S, Kabat D. J Virol. 1997;71:9383–9391. doi: 10.1128/jvi.71.12.9383-9391.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green . Genetics and Probability in Animal Breeding Experiment. New York: Oxford Univ. Press; 1981. [Google Scholar]
- 35.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida K, Ogawa N, Oshima Y. Mol Gen Genet. 1989;217:40–46. doi: 10.1007/BF00330940. [DOI] [PubMed] [Google Scholar]
- 37.Spain B H, Koo D, Ramakrishnan M, Dzudzor B, Colicelli J. J Biol Chem. 1995;270:25435–25444. doi: 10.1074/jbc.270.43.25435. [DOI] [PubMed] [Google Scholar]
- 38.Lyu M S, Kozak C A. J Virol. 1996;70:830–833. doi: 10.1128/jvi.70.2.830-833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porter C D, Collins M K L, Tailor C S, Parkar M H, Cosset F L, Weiss R A, Takeuchi Y. Hum Gene Ther. 1996;7:913–919. doi: 10.1089/hum.1996.7.8-913. [DOI] [PubMed] [Google Scholar]
- 40.Battini J L, Heard J M, Danos O. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamer I, Haft C R, Paccaud J P, Maeder C, Taylor S, Carpentier J L. J Biol Chem. 1997;272:21685–21691. doi: 10.1074/jbc.272.35.21685. [DOI] [PubMed] [Google Scholar]