Abstract
The outer membrane of gram-negative bacteria contains several proteins, and some of these proteins, the porins, have numerous biological functions in the interaction with the host; porins are involved in the activation of signal transduction pathways and, in particular, in the activation of the Raf/MEK1-MEK2/mitogen-activated protein kinase (MAPK) cascade. The P2 porin is the most abundant outer membrane protein of Haemophilus influenzae type b. A three-dimensional structural model for P2 was constructed based on the crystal structures of Klebsiella pneumoniae OmpK36 and Escherichia coli PhoE and OmpF. The protein was readily assembled into the β-barrel fold characteristic of porins, despite the low sequence identity with the template proteins. The model provides information on the structural features of P2 and insights relevant for prediction of domains corresponding to surface-exposed loops, which could be involved in the activation of signal transduction pathways. To identify the role of surface-exposed loops, a set of synthetic peptides were synthesized according to the proposed model and were assayed for MEK1-MEK2/MAPK pathway activation. Our results show that synthetic peptides corresponding to surface loops of protein P2 are able to activate the MEK1-MEK2/MAPK pathways like the entire protein, while peptides modeled on internal β strands are unable to induce significant phosphorylation of the MEK1-MEK2/MAPK pathways. In particular, the peptides corresponding to loops L5 (Lys206 to Gly219), L6B (Ser239 to Lys253), and L7 (Thr280 to Lys287) activate, as the whole protein, essentially JNK and p38.
Bacterial lipopolysaccharides (LPS) and associated outer membrane proteins (OMPs) are potent initiators of fever, coagulation disorders, multiple-organ failure, and shock in humans and experimental animals (4, 21, 46, 48-50, 66, 77). LPS induces transcription of several genes encoding proinflammatory mediators (30), and in the past few years the various immunobiological effects induced by the outer membrane pore-forming proteins have been compared to those induced by LPS (26, 32, 36).
Among the OMPs, porins are known to form diffusion channels, which allow small, polar molecules to diffuse across membranes. Porins are major components of the gram-negative bacterial outer membrane, and they also play an important role in bacterial pathogenesis and are involved in adherence, invasion, and serum resistance. Significant advances in our understanding of porin function have been facilitated by recent determinations of the crystal structures of several porins (13, 41, 47, 61, 70, 76). These structural analyses have revealed that bacterial porins exist as trimers in which the folding pattern of each monomer generally depends on 16 antiparallel β strands that cross the outer membrane and loops that connect the β strands on both sides of the membrane. The whole structure is an antiparallel β barrel with eight large loops of variable length on the external surface of the bacterial membrane and eight short periplasmic turns (13, 70). The barrel spans the entire outer membrane. In all porins, the cross section of the pore is restricted by one large loop (loop L3) that folds back into the channel and narrows it, determining the size of the pore and thus the molecular exclusion limit, as well as the physiological and conductivity properties of the pore (58).
Despite low sequence identity, members of the porin family whose structures have been determined superimpose surprisingly well across the β-barrel structure, but they differ in pore characteristics and more importantly in the surface-exposed loop. The importance of immunodominant surface-exposed loops of OMPs is the focus of recent studies, and the ability of gram-negative bacteria to cause recurrent infections is in part attributable to antigenic variability in all surface-exposed loops of OMPs (10, 33, 53, 74). Some of the surface-exposed loops are involved in the recognition of ligands, including small-molecule nutrients, agents such as bacteriophages or colicins, and probably eukaryotic target cells for bacterial pathogens. Moreover, it has been demonstrated that loops do not play an active structural role; thus, they are not involved in the assembly of integral β-structure membrane proteins (40).
All of the porins tested to date have been shown to bind fragment C1q, the first component of the classical pathway of the complement system, and they can activate the complement cascade (1, 19, 43). Recently, studies have focused on the interaction of loop L7 of porin OmpK36 from Klebsiella pneumoniae with C1q and comparison of this interaction with the C1q-LPS interaction (2). Most functional antibodies raised to nontypeable Haemophilus influenzae are directed to loop L5, which is thought to contain strain-specific and immunodominant epitopes (74). Furthermore, antibodies to loops L5 and L6 of strain NTHI showed complement-dependent bactericidal activity (53, 74). The loop regions are immunodominant, as determined by immunizing mice with whole bacterial cells (34, 73). However, whether only surface-exposed loops play an important role in the inflammatory and immunological response is still an open question.
Porin P2 of NTHI (74) is one of the best-characterized porins in terms of its functional characteristics. NTHI is a gram-negative bacterium which is a common cause of mucosal infections; its outer membrane contains six to eight major proteins (7, 52). P2 is the most abundant protein in NTHI and in H. influenzae type b outer membrane preparations; its molecular mass varies between 36 and 42 kDa, and it is present in all strains and functions as a porin (12). P2 contains 16 transmembrane regions and eight surface-exposed loops (3, 16, 64, 65). Sequence comparisons of P2 genes have indicated that the transmembrane regions are relatively conserved among strains, while there is considerable heterogeneity in the loop regions of the molecule (3, 8, 16, 64).
Activation of the inflammatory and immunological response is initiated by the interaction with the bacterium (57) or one of its components and involves phosphorylation of the main signal transduction networks (28, 29, 31). The LPS induces transcription and secretion of tumor necrosis factor alpha and activation of an Src gene family tyrosine kinase (Lyn), an extracellular signal-regulated kinase (ERK), and Rsk signal transduction proteins in mouse macrophages (31, 32). Also, porins from Salmonella enterica serovar Typhimurium have been implicated in the regulation of cell functions due to triggering of specific signaling pathways (29, 32). Activation of macrophages by porins results in the transduction of protein kinase C to the membrane, which is accompanied by nitric oxide release within the macrophages; nitric oxide plays a fundamental role in macrophage effector function (32).
It is still not known whether the surface-exposed loops also play an important role in signaling pathways. One of the best-known signal transduction systems is the mitogen-activated protein kinase (MAPK) cascade. MAPK plays a key role in the regulation of gene expression as well as cytoplasmic activities, and it is also involved in the regulation of cytokine responses (72). In mammalian systems, five different MAPK modules have been identified so far; single MAPK modules can signal independent of one another, and this specificity is manifest in different physiological responses (60). The five MAPK modules include the extracellular signal-regulated kinase 1 and 2 (ERK1-ERK2) cascade, which preferentially regulates cell growth and differentiation, as well as the c-Jun N-terminal kinase (JNK) and p38 MAPK cascades, which mainly regulate stress responses like inflammation and apoptosis. All MAPKs except extracellular signal-regulated kinase 3 (ERK3) are activated upon phosphorylation of both tyrosine and threonine residues by MAPK kinase (MEK). Many different MEKs have been described, and in vitro assays indicate that each of them has one or at most two specific targets in the MAPK pathways; MEK1 and MEK2 act on ERK1 and ERK2, respectively. The MAPK cascade activates transcription factors such as activating protein 1 (39) and nuclear factor κB (NF-κB) (63).
In this study, we developed a model of protein P2 from H. influenzae type b and demonstrated that peptides designed on the basis of the amino acid sequences of surface-exposed loops, but not peptides corresponding to the β barrel, are able to activate the MAPK cascade in U937 cells.
MATERIALS AND METHODS
Cell lines.
U937 monocytes (ATCC CRL-1593.2) were grown at 37°C in the presence of 5% CO2 in RPMI 1640 (Labtek, Eurobio) with HEPES supplemented with 10% heat-inactivated fetal calf serum, glutamine (2 mM), penicillin (100 U/ml), and streptomycin (100 U/ml) (Labtek, Eurobio) in 150-cm2 tissue culture flasks (Corning, New York, N.Y.). Before treatment of the cells, the serum concentration was reduced to 5% for 24 h at 37°C, and then serum-free media were used for at least 10 to 12 h. This should have prevented any interference from serum factors in the phosphorylation state of the proteins in the signaling cascade.
Bacteria and growth conditions.
H. influenzae type b subtype 1H strain ATCC 9795 was grown in CY medium (11) for 18 to 24 h at 37°C; cells were harvested at the end of the exponential growth phase.
Preparation of the porin.
The porin was isolated and purified from cells of H. influenzae type b strain ATTC 9795 by using the modified method described by Nurminen et al. (55, 56), as reported in a previous paper (26). The bacterial envelopes were treated with Triton X-100 buffer for 2 h at 37°C in a rotary shaker, dissolved in sodium dodecyl sulfate (SDS) buffer (4% [wt/vol] SDS in 0.1 M sodium phosphate, pH 7.2), and applied to an Ultragel ACA34 column equilibrated with 0.25% SDS-sodium azide buffer. The elution rate through the column was 8 ml h−1, and 2-ml fractions were collected. The fractions containing protein, identified by measuring the absorption at 280 nm, were extensively dialyzed and checked by SDS-polyacrylamide gel electrophoresis (PAGE) by using the method of Laemmli (42). The protein content of the porin preparation was determined by the method of Lowry et al. (44). All possible traces of LPS were revealed on SDS-PAGE gels stained with silver nitrate as described by Tsai and Frasch (67) and by the Limulus amoebocyte lysate assay (Limulus test) as described by Yin et al. (75). The pore-forming ability of our preparation was checked by a functional assay (liposome swelling assay) after incorporation into proteoliposomes as described by Nikaido and Rosenberg (54).
N-terminal sequencing of H. influenzae type b porin.
A model 477A automatic protein sequencer (Applied Biosystems, Foster City, Calif.) was used to determine the N-terminal sequence of the H. influenzae type b porin. The phenylthiohydantoin (PTH) amino acids released during degradation were identified by using an Applied Biosystems 120A high-performance liquid chromatograph with a Brownlee C18 column. Separation was performed with an eluent gradient as suggested by Applied Biosystems. The column temperature was set at 54°C, and elution was monitored by measuring the absorbance at 269 nm. Data were analyzed with a Macintosh Ilsi computer (26).
Haemophilus strains and sequences from databases.
The P2 porin sequences of 11 strains were obtained from SwissProt (6). The SwissProt accession number for the P2 sequence of H. influenzae type b used to construct the model is P20149. Alignments and comparisons of the amino acid sequences in loop regions were performed by using the programs Homology (Insight II, version 98.0, molecular modeling system; Molecular Simulations, Inc., San Diego, Calif.) and Blast (45).
Generation of P2 structural model.
The initial model of the P2 monomer was generated by using MODELLER, a sophisticated software package that calculates protein three-dimensional structures based on sequence alignments, spatial restraints, and stereochemical considerations (59). The model for P2 was constructed by using the following three structures as templates: PhoE from Escherichia coli (pdb code 1PHO; 20% sequence identity) (13, 59), OmpF from E. coli (pdb code 2OMF; 19% sequence identity) (13, 14), and OmpK36 from K. pneumoniae (pdb code 1OSM; 17% sequence identity) (17). All parameters in MODELLER were set to their default values. In order to generate the full trimer by crystallographic symmetry, the model for the P2 monomer was inserted into the unit cell of PhoE (space group P321; a = b = 119.90 Å; c= 51.90 Å; α= β = 90.00°; γ = 120°); loops L1, L4, and L8 were removed from the model, and the model was subjected to 50 steps of conjugate gradient energy refinement, as implemented in CNS (9), to remove poor contacts. After refinement, coordinates for the full trimer were assembled with PDBSET, as implemented with the CCP4 suite of crystallographic programs (5). The quality of the model was assessed with Whatif (69).
Alignment of Haemophilus porin sequences from different strains was performed in order to check for mistakes in the model and to verify that the β strands were minimally disrupted.
Peptide synthesis.
Fluorenylmethoxycarbonyl (Fmoc)-protected amino acids were purchased from INBIOS (Pozzuoli, Italy), and 4-(2′,4′-dimetoxyphenyl-Fmoc-aminomethyl)-phenoxyacetamido-norleucyl-methoxybenzidrylamide(MBHA) resin was purchased from Nova Biochem (Darmstadt, Germany). Piperidine and pyridine, the reagents used for solid-phase peptide synthesis, were purchased from Fluka (Sigma-Aldrich, Milan, Italy), and trifluoroacetic acid (TFA) and acetic anhydride were obtained from Applied Biosystems. H2O, n,n-dimethylformamide (DMF) and CH3CN were supplied by LAB-SCAN (Dublin, Ireland).
Peptides corresponding to loop regions were prepared by standard 9-fluorenylmethoxycarbonyl polyamine solid-phase synthesis by using a PSSM8 multispecific peptide synthesizer (Biotechnology Instruments Department, Shimadzu Corporation, Kyoto Japan). The MBHA resin (substitution, 0.3 mmol/g) was used as the solid-phase support, and syntheses were performed on a scale of 15 μmol.
All amino acids (4 equivalents relative to resin loading) were coupled by the PyBop-HOBT-DIEA method, as follows: 1 equivalent of Fmoc-protected amino acid, 1 equivalent of benzotriazole-1-yl-oxy-trispyzolidino-phosphonium hexafluorophosphate (PyBop), 1 equivalent of n-hydroxybenzotriazole (HOBT) (0.5 mM HOBT in DMF), and 1.5 equivalent of diisopropyl ethyamine (DIEA) (1 mM DIEA in DMF). The Fmoc protecting group was removed with 30% (vol/vol) piperidine in DMF.
Peptides were fully deprotected and cleaved from the resin by treatment with a 90% TFA solution containing 5% thioanisole, 3% ethandithiol, and 2% anisole as scavengers.
The crude materials were purified to homogeneity by preparative reverse-phase high-performance liquid chromatography by using a Waters Delta Prep 3000 chromatographic system equipped with a model 481 UV Lambda Max detector. The samples were injected onto a Vydac (The Separation Group, Hesperia, Calif.) C18 column (22 mm by 25 cm; 5 μm) and eluted with a solvent mixture containing H2O-0.1% TFA (solution A) and CH3CN-0.1% TFA (solution B). A linear 5 to 95% solution B gradient over 50 min at a flow rate of 20 ml/min was employed. The collected fractions were lyophilized to dryness and analyzed by analytical reverse-phase high-performance liquid chromatography by using a Shimadzu class LC10 equipped with an SPD-M10AV diode array detector and a Phenomenex C18 analytical column (4.6 by 250 mm; 5 μm); a linear 5 to 95% solution B gradient over 50 min at a flow rate of 1 ml/min was used. The identities of purified peptides were confirmed by matrix-assisted laser desorption ionization spectrometry. High yields (70 to 80%) of all the purified peptides were obtained.
All the peptides were detoxified before being tested on cells. Detoxification was performed by using Detoxi Gel Affinity Pak columns supplied by Pierce (Rockford, Ill.) (38).
Cell stimulation with protein P2 and peptides and preparation of cell lysates.
U937 cells (3 × 106 cells/ml) were stimulated with different concentrations of stimuli for different times in 96-well polypropylene plates. After incubation, the cells were washed twice with ice-cold phosphate-buffered saline without Ca2+ and Mg2+ and resuspended in 250 μl of the appropriate lysis buffer (buffer A containing 150 mM NaCl, 50 mM Tris-HCl [pH 7.5], 5 mM EDTA, 1% Nonidet P-40, 1 mM Na2MoO4, 40 μg of phenylmethylsulfonyl fluoride per ml, 0.2 mM Na3VO4, 1 mM dithiothreitol, 10 μg of aprotinin per ml, 2 μg of leupeptin per ml, 0.7 μg of pepstatin per ml, 10 μg of soybean trypsin inhibitor per ml; or buffer B containing 137 mM NaCl, 20 mM Tris-HCl [pH 8.0], 5 mM EDTA, 1 mM Na6P2O7, 10% glycerol, 1% Triton X-100, 1 mM EGTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, 10 mM NaF, 10 mM β-glycerophosphate, 20 μg of aprotinin per ml, and 20 μg of leupeptin per ml); the sample was centrifuged for ECL Western blot analysis at 16,000 × g for 2 min at 4°C. The clarified cell lysates were used for ECL Western blot analysis.
Analysis of kinase phosphorylation by Western blotting.
Cell lysates were precleared with Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, Inc.) (20 μl) for 45 min. Immunoprecipitation was done with the appropriate antibodies and beads at 4°C overnight with gentle rotation. After incubation the beads were pelleted by centrifugation (6,000 × g), washed three times with 500 μl of specific lysis buffer, and boiled in 20 μl of Laemmli sample buffer with 5% β-mercaptoethanol for 5 min. The samples were pelleted, and the supernatant, containing cell phosphorylated proteins, was resolved by SDS-PAGE as described by Laemmli (42). Equal amounts of cell lysates (typically 50 μg) were separated on SDS-15% PAGE gels by using the buffer system described by Laemmli (42). Following electrophoresis, the separating gel was soaked in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol) for 5 min, and then the proteins were transferred to polyvinylidene difluoride membranes (0.45-μm-pore-size sheets) overnight at 30 V and 4°C. The blots were blocked for 1 h at room temperature in Tris-buffered saline (TBS) (150 mM NaCl, 20 mM Tris-HCl; pH 7.5) containing 1% bovine serum albumin (BSA) plus 1% blotting-grade blocker nonfat milk (Bio-Rad Laboratories), and subsequently the membranes were washed twice with TBS containing 0.05% Tween 20 (TTBS) before incubation with antiphosphorylated and nonphosphorylated kinase antibodies diluted 1:2,000 in TBS containing 1% BSA for 1 h at room temperature. After the polyvinylidene difluoride membranes were washed six times with TTBS for 3 min, they were incubated at room temperature for 2 h with anti-mouse or anti-rabbit immunoglobulin G (IgG) horseradish peroxidase secondary antibodies diluted 1:3,000. Then they were washed six times with TTBS and twice with PBS for 5 min. After this, proteins were visualized with enhanced chemiluminescence on Kodak film (Kodak X-OMAT LS). For Western blot analysis, the following phosphorylated antibodies were used: mouse monoclonal anti-phosphotyrosine (clone 4G10; Upstate Biotechnology, Inc., Lake Placid, N.Y.); phospho-p44/42 MAPK (Thr 202/Tyr 204) E10 monoclonal antibodies (isotype, mouse IgG1; anti-phospho-p44/42; New England Biolabs, Beverly, Mass.), which detects doubly phosphorylated threonine 202 and tyrosine 204 of p44 and p42 MAPKs (ERK1 and ERK2) and are produced by immunizing mice with a synthetic phospho-Thr202 and phospho-Tyr204 peptide corresponding to residues around Thr202 and Tyr204 of human p44 MAPK; rabbit polyclonal phospho-MEK1/2 antibody (anti-p-MEK1/2) (New England Biolabs), which detects MEK1/2 only when it is activated by phosphorylation at Ser217 and Ser221 and does not cross-react with other related family members; phospho-p38 antibody (anti-phospho-p38; New England Biolabs), which is a rabbit polyclonal antibody raised against a peptide mapping at the amino terminus of p38 of mouse origin identical to the corresponding human sequence and is directed against Thr180 and Tyr182-phosphorylated p38; and phospho-JNK antibody (anti-p-JNK) (Santa Cruz Biotechnology, Inc.), which is a mouse monoclonal IgG1 antibody raised against a peptide corresponding to a short amino acid sequence phosphorylated on Thr183 and Tyr185 of JNK of human origin. The mobility shift assay was performed with the following antibodies: anti-ERK1/2 (Upstate Biotechnology), which is a rabbit polyclonal antibody raised against a peptide corresponding to residues 333 to 367 of rat ERK1 recognizing ERK1 and ERK2 at 44 to 42 kDa; anti-MEK1 (Santa Cruz Biotechnology, Inc.), which is a mouse monoclonal IgG2b antibody raised against a recombinant protein corresponding to amino acids 1 to 393 representing full-length MEK1 of human origin; anti-MEK2 (Santa Cruz Biotechnology, Inc.), which is a rabbit polyclonal antibody raised against a peptide mapping at the amino terminus of MEK2 of human origin identical to the corresponding rat sequence; anti-p38 (Santa Cruz Biotechnology, Inc.), which is a rabbit polyclonal IgG1 antibody epitope corresponding to amino acids 213 to 360 mapping at the carboxy terminus of p38 of human origin; and anti-JNK (Santa Cruz Biotechnology, Inc.), which is a rabbit polyclonal IgG antibody produced by immunization with full-length (amino acids 1 to 384) human JNK produced in E. coli. Immunoreactivity was determined by using horseradish peroxidase-conjugated secondary antibodies and was visualized by ECL enhanced chemiluminescence (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom).
LDH assay.
The lactate dehydrogenase (LDH) assay was carried out according to the manufacturer's instructions by using a cytotoxicity detection kit (Roche Diagnostic SpA, Milan, Italy). LDH is a stable cytoplasmic enzyme present in all cells and is rapidly released into cell culture supernatant when the plasma membrane is damaged. LDH activity was determined by a coupled enzymatic reaction in which the tetrazolium salt was reduced to formazan. An increase in the number of dead or damaged cells resulted in an increase in LDH activity in the culture supernatant.
Reproducibility.
Gels were scanned for densitometry by using Sigma Gel software, and the results shown below are averages of the values from three different experiments.
RESULTS
Purity of P2 porin preparations.
The purity of the porin preparation from H. influenzae type b subtype 1H was checked by SDS-PAGE as reported previously (26). SDS-PAGE revealed one band at a molecular mass of approximately 40 kDa, as previously reported by Coulton et al. (12). The isolated protein showed porin activity, as demonstrated by its ability to form transmembrane aqueous channels (data not shown). Moreover, the N-terminal sequence (AVVYNNEGTNV) of the isolated protein corresponds to the known N-terminal sequence of the protein reported previously (35, 51). By using the Limulus test, the LPS contamination in the porin preparation was estimated to be about 0.001% (wt/wt) compared with a standard H. influenzae type b LPS solution.
Alignments and homology modeling.
The sequences of P2 from 11 strains of H. influenzae aligned with the sequences of the three porins (PhoE, OmpF, and OmpK36) with known three-dimensional structures, which were used as templates, are shown in Fig. 1. Only these three porins were included in our study, since they were the only porins showing significant sequence identity with the P2 porin.
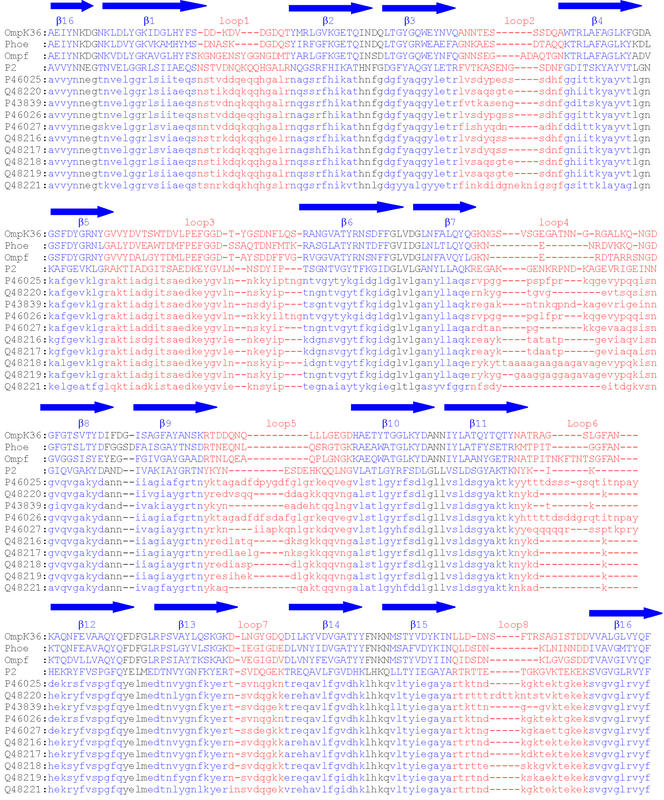
FIG. 1.
Multiple alignment of P2 porin from H. influenzae type b (SwissProt accession number P20149) with PhoE, OmpF, OmpK36, and P2 from strains included in this study. The dashes represent gaps in the alignment. The regions predicted to form β strands are labeled β1 to β16 and are blue. The putative external loops are marked L1 to L8 and are red. The SwissProt accession numbers of the P2 sequences are indicated on the left.
A structural alignment of PhoE, OmpF, and OmpK36 was constructed, and then the P2 porin sequence (SwissProt accession number P20149) was added to this initial alignment (Fig. 1). Thus, the alignment was edited manually and refined by taking into account additional structural information available for the variable regions. Correct alignment was critical for the generation of an accurate model, and each putative conserved residue was examined to verify that there was a sound structural basis for its conservation in the porins analyzed.
The sequences of Haemophilus P2 porins from 10 different strains were added only after construction of the structural homology model (Fig. 1). The other Haemophilus strain sequences were introduced into the alignment in order to verify that sequence variations were localized in loop regions and not within the β-barrel framework; in fact, most insertions and deletions are found in regions corresponding to loops facing the cell exterior.
The β-barrel framework of P2 was constructed remarkably well, with the well-defined main chain hydrogen-bonding pattern characteristic of the β-sheet motif, although no hydrogen bonding or backbone dihedral angle restraints were used in the calculation. The periplasmic turns were generally short and contained turn promoters (Asp, Asn, Glu, Gly, Pro, and Ser). The external loops were generally long and were associated with variable sequences. Larger loop regions were less well modeled by this procedure and showed a variety of conformations, depending on the starting alignment and other restraints. The P2 monomer model is shown in Fig. 2.
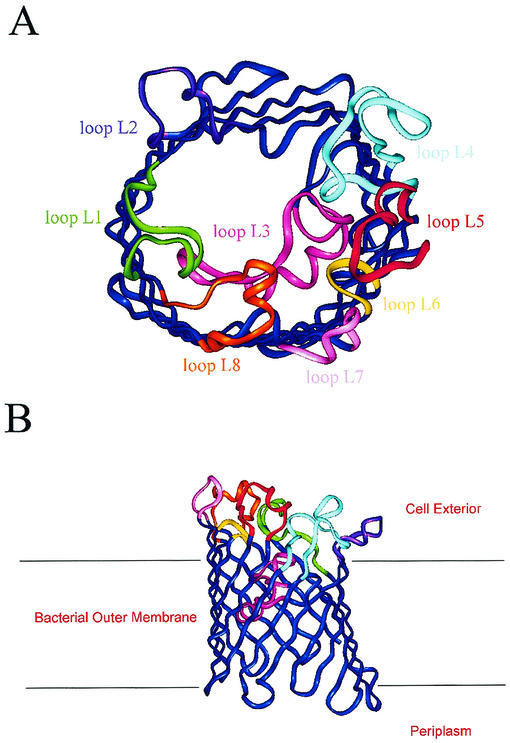
FIG. 2.
Three-dimensional model of the P2 monomer from H. influenzae type b based on the X-ray structures of K. pneumoniae OmpK36 and E. coli PhoE and OmpF. (A) Overview of the molecule. Surface loops are labeled L1 to L8 and are different colors. The extracellular space is located at the top, and the periplasmic space is at the bottom. (B) Horizontal slice through the barrel of the proposed model of P2 porin, highlighting the finding that loop L3 extends inside the barrel and constricts the pore. The position of the membrane bilayer is indicated by horizontal lines.
The model revealed several structural features that are common to porins with known three-dimensional structures. It presents a well-defined pore that is lined with hydrophilic and hydrophobic side chains having appropriate dimensions for the passage of low-molecular-weight solutes. In particular, the long loop L3 folds into the barrel, leaving a gap in the wall between strands β4 and β7. This loop constricts the size of the pore at about half the height of the barrel. L3 is shorter than the corresponding loops in the three reference proteins, thus determining a greater dimension of the pore.
The outer surface of the protein shows evidence of partitioning with respect to hydrophilicity. The stem of the porin presents a highly hydrophobic 25-Å band, which is compatible with estimates of the thickness of the nonpolar outer membrane. Moreover, the distribution of aromatic amino acid side chains within the model is similar to that established for other porins, with the majority of the Phe and Tyr chains pointing outward from the barrel and presumably playing an important role in interactions with phospholipids. In particular, tyrosines and phenylalanines cluster in two rings around the base and the top of the barrel, delimiting the position of the inner edge of the lipid bilayer.
The major limitation of the MODELLER package (59) is that the porin model is calculated as a monomer, while porins exist as trimers within the outer membrane. A more realistic model was produced with CNS by placing the monomers within the original crystallographic unit cell of the PhoE porin. Several cycles of conjugate gradient energy refinement provided the final model.
The three-dimensional homology model allowed definition of residues belonging to loop regions and of protein regions involved in the trimerization. The amino acid sequences of the loops differ significantly from the corresponding sequences of other porins.
The regions involved in the formation of the trimer are mainly hydrophobic; in particular, loop L2 is involved in monomer-monomer interactions within the porin trimer. Loops L1 and L4 are in close proximity between adjacent monomers, raising the possibility that these regions may interact to some extent across a monomer-monomer interface, and thus they seem to be involved in trimerization as well. Moreover, L4 is longer than the corresponding loops in the three reference proteins (Fig. 1).
Loop L3 is sequestered in the core of each monomer, influencing the pore dimension and function, as described above. Loops L6 and L7 are unusually short and essentially consist of β turns with only two or three connecting amino acid residues. Loop L8, which is the third longest loop, seems to fold back into the barrel interior and to contribute to the formation of the channel opening at the external side.
Analysis of the alignments of the sequences of the P2 proteins from 12 different strains allowed determination of loops showing the greatest sequence and length variability. In particular, loops L2, L4, and L5 showed the greatest sequence variability, while loops L4, L5 and L6 showed the greatest length variation (Fig. 1).
We found good agreement with previously proposed models only for the locations of loops L4 and L8 (3, 16, 65), while the the locations of the other loops differed significantly.
Peptides corresponding to the sequences of the predicted loops were synthesized. Table 1 shows the sequences of these peptides together with the sequences of control peptides.
TABLE 1.
Amino acid sequences of peptides
| Loop | Sequence | Protein fragment |
|---|---|---|
| L1 | NH2-NSTVDNQKQQHGALR-COOH | 25-39 |
| L2 | NH2-TKASENGSDNPGDITSK-COOH | 69-85 |
| L3 | NH2-RAKTIADGITSAEDKEYGVLNNSDYIPTSGN- COOH | 103-133 |
| L4 | NH2-REGAKGENKRPNDKAGEVRIGEIN-COOH | 157-180 |
| L5 | NH2-KYNESDEHKQQLNG-COOH | 206-219 |
| L6A | NH2-NYKIK-COOH | 246-250 |
| L6B | NH2-SGYAKTKNYKIKHEK-COOH | 239-253 |
| L7 | NH2-TSVDQGEK-COOH | 280-287 |
| L8 | NH2-RTRTTETGKGVKTEKEK-COOH | 315-331 |
| β1 | NH1-TNVELGGRLSIIAEQS-COOH | 9-24 |
| β3 | NH2-FYAQGYLETR-COOH | 57-66 |
| β8 | NH2-NGIQVGAKYD-COOH | 181-190 |
| β15 | NH2-LLTYIEGAYA-COOH | 305-314 |
| Scrambled | NH2-EQRHNEKKDTYSQYN-COOH | 206-219 |
| Aspy | NH2-RKIRKENRMK-COOH |
MAPK pathway signaling by isolated P2 porin in U937 cells.
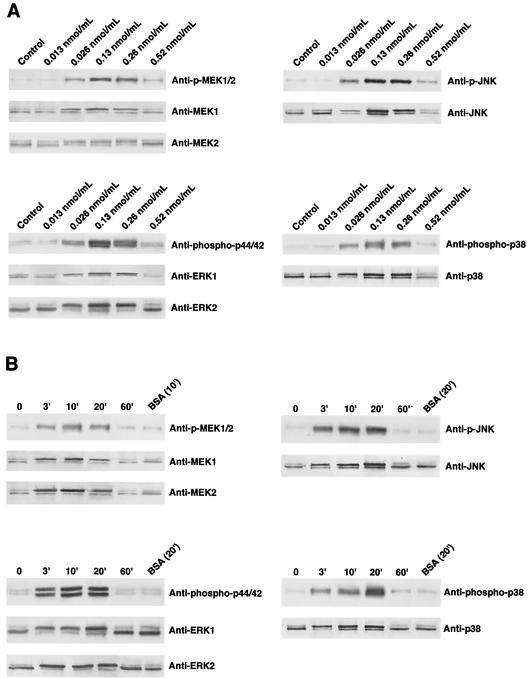
The P2 porin isolated from H. influenzae type b strain ATCC 9795 was able to induce tyrosine phosphorylation in U937 cells, as demonstrated previously for THP-1 cells (28). In particular, the activation was dose dependent; porin concentrations as low as 0.013 nmol/ml (500 ng/ml) still induced activation, while a concentration of 0.52 nmol/ml (20 μg/ml) did not further increase the phosphorylation (Fig. 3A). Thus, in our experiments we used a protein concentration of 0.13 nmol/ml.
FIG. 3.
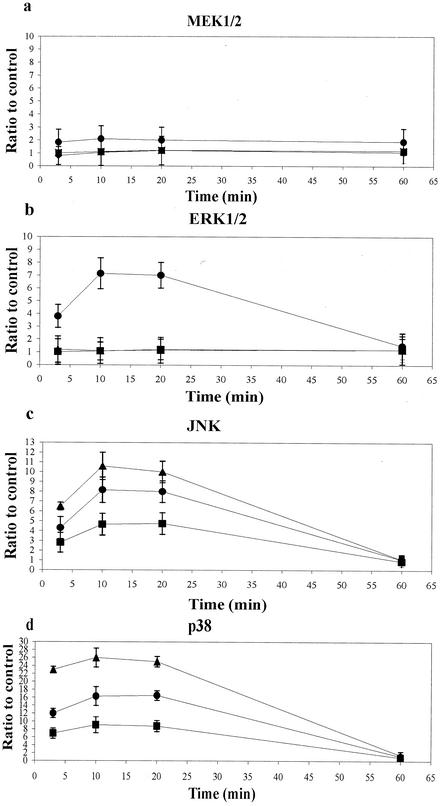
MEK1/2, ERK1/2, JNK, and p38 activation in U937 cells in response to various doses of P2 porin (0.013, 0.026, 0.13, 0.26, and 0.52 nmol/ml) after 20 min of stimulation (A) and after different stimulation times (3, 10, 20, and 60 min) with a dose of 0.13 nmol/ml (B). Total cellular proteins (3 × 106 cells/ml) were analyzed by SDS-PAGE and were immunoblotted with MEK1/2-, ERK1/2-, JNK- or p38-phosphospecific antibody. Shifts in band mobility on SDS-PAGE gels due to phosphorylation were obtained with anti-MEK1/2, anti-ERK1/2, anti-JNK, and anti-p38. BSA was used as a nonspecific stimulus control.
We next examined activation of the MEK1-MEK2/MAPK pathway in untreated cells, as well as P2 porin-stimulated U937 cells. To verify that P2 porin stimulates the MEK1-MEK2/MAPK signal transduction pathway, U937 cells (3 × 106 cells/ml) were treated with 0.13 nmol (5 μg) of porin per ml. Cell lysates were prepared at different times after the stimulation. The lysates obtained were immunoprecipitated with antibodies that specifically recognized the phosphorylated and nonphosphorylated forms of each enzyme. We had previously detected related proteins in untreated U937 cells using anti-MEK1, anti-MEK2, anti-ERK1/2, anti-JNK, and anti-p38 antibodies. A small difference was detected between cells that were not incubated with serum and cells that were treated with serum (data not shown). Using anti-p-MEK1/2 antibodies, we observed immunoreactive bands of phosphorylated enzymes. MEK1/2 phosphorylation reached its peak by 10 min and went back to a standard value by 60 min (Fig. 3B). The shift in the electrophoretic mobility of the bands at 10 min obtained with the anti-MEK1 and -MEK2 antibodies confirmed the results obtained with anti-p-MEK1/2 antibodies. Using anti-phospho-p44/42 antibodies, we observed that porins increased the appearance of the phosphorylated form of this enzyme as well. A time course of ERK1/2 activation following porin treatment is shown in Fig. 3. When anti-phospho-p44/42 antibodies were used, bands of phosphorylation appeared by 3 min, reached the highest level of expression by 20 min, and returned to standard levels by 60 min. The shift in the electrophoretic mobility of the bands at 20 min obtained with anti-ERK1/ERK2 antibodies confirmed the results obtained with anti-phospho-p44/42 antibodies. Using an anti-phospho-p38 antibody, we observed immunoreactive bands in U937 cell lysates treated with porin. The shift in the electrophoretic mobility of the bands at 20 min obtained with anti-p38 antibody confirmed the results obtained with anti-phospho-p38 antibody (Fig. 3B). Using the anti-p-JNK monoclonal antibody, we also observed by Western blot analysis phosphorylation obtained after stimulation with porin. The shift in the electrophoretic mobility of the bands at 20 min obtained with anti-JNK antibody confirmed the results obtained with anti-p-JNK monoclonal antibody (Fig. 3B).
The duration of the treatment with porins at the concentrations used was not toxic for the cells; in fact, the treatment did not induce any significant release of LDH in the cell supernatants (data not shown). The observed effect was specific because when we used a different protein, such as BSA, as a stimulus, we were not able to observe any phosphorylation of the enzymes investigated in this study. The biological activity in inducing a mechanism of signal transduction in our experiments was exclusively due to porins and not to eventual traces of LPS that could have contaminated the porin preparation. The minimum amount of LPS able to produce phosphorylation in our in vitro model was 100 ng/ml, which was much more than the amount of LPS present in our preparations (about 10 pg/μg of porin).
MAPK pathway signaling activation by peptides designed by using the model of P2 porin.
Once it was established that porin P2 from H. influenzae type b was able to activate cell phosphorylation and in particular the MEK/MAPK pathway in U937 cells, we evaluated whether the peptides corresponding to variable loop regions were alone able to activate phosphorylation. Synthesized peptides were individually tested with U937 cells.
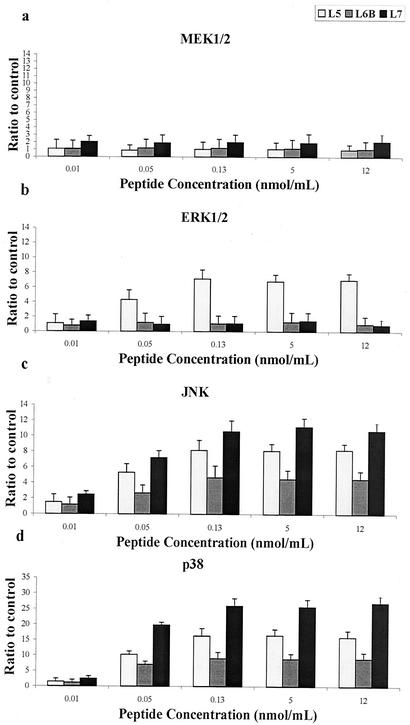
Peptide concentrations of 0.01, 0.05, 0.13, 5.0, 12, and 26 nmol/ml were assayed in our experiments. The results obtained for active peptides are shown in Fig. 4; the results obtained for inactive peptides are not shown. A concentration of 26 nmol/ml was toxic for cells, as verified by LDH release (data not shown). As observed for the entire protein, signals from an active peptide were visible by 3 min after treatment; there was an activity peak at 10 min, which persisted for at least 20 min, and the amount returned to the standard amount by 60 min. The results obtained for active peptides are shown in Fig. 5; data for inactive peptides are not shown. Thus, for our experiments we used a concentration of 0.13 nmol/ml and an activation time of 10 min.
FIG. 4.
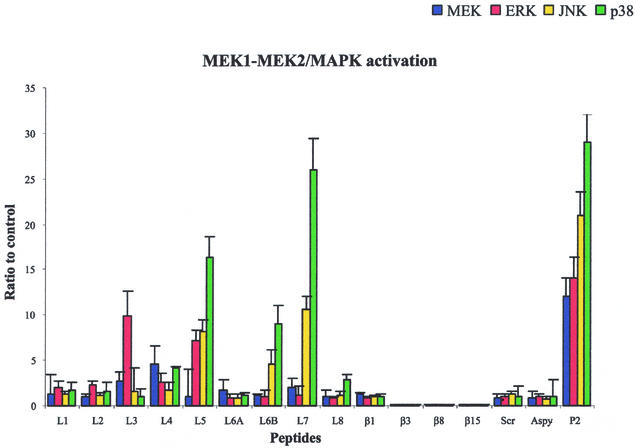
MEK1-MEK2/MAPK activation in response to various doses of active peptides. U937 cells (3 × 106 cells/ml) were stimulated with various peptide concentrations, as indicated on the horizontal axis. Total cellular protein was harvested after 10 min, analyzed by SDS-PAGE, and immunoblotted with MEK1/2-, ERK1/2-, JNK-, or p38-phosphospecific antibody. Gels were scanned for densitometry analysis with the Sigma Gel software, and the ratio of the value for each peptide dose to the value for an unstimulated control is shown. The data are averages from three different experiments, and the error bars indicate the standard errors of the means.
FIG. 5.
Time course of MEK1-MEK2/MAPK activation in response to active peptides. Total cellular protein (from 3 × 106 U937 cells/ml) was harvested at the times indicated on the horizontal axis, analyzed by SDS-PAGE, and immunoblotted with MEK1/2-, ERK1/2-, JNK-, or p38-phosphospecific antibody. Gels were scanned for densitometry analysis with the Sigma Gel software, and the ratio of the value for each stimulation time to the value for an unstimulated control is shown. The data are averages from three different experiments, and the error bars indicate the standard errors of the means. Symbols: •, loop L5; ▪, loop L6B; ▴, loop L7.
A 15-amino-acid peptide corresponding to the protein sequence from Asn25 to Arg39 (loop L1) induced only a slight increase in the appearance of the phosphorylated enzymes MEK1/2, ERK1/2, JNK, and p38, comparable to the increase induced by the control (Fig. 6). A 17-amino-acid peptide corresponding to the protein fragment from Thr69 to Lys85 (loop L2) was also unable to induce phosphorylation of MEK1/2, ERK1/2, JNK, and p38. A long 31-amino-acid peptide corresponding to the internal loop L3 (protein fragment from Arg103 to Asn133) was able to activate MEK1/2 and ERK1/2 but was unable to induce significant activation of JNK and p38. The 24-amino-acid peptide corresponding to the protein sequence from Arg157 to Asn180 (loop L4) was able to induce slight activation of MEK1/2, ERK1/2, JNK, and p38. A 14-amino-acid peptide corresponding to the protein fragment from Lys206 to Gly219 (loop L5) strongly induced an increase in the immunoreactive bands of ERK1/2, JNK, and p38 and less effectively activated the phosphorylation of MEK1/2 (Fig. 6). A 15-amino-acid peptide containing the sequence of loop L6 (protein fragment from Ser239 to Lys253; L6B) induced an increase in the immunoreactive bands of phosphorylated JNK and p38, while it only slightly activated MEK1/2 and ERK1/2. A shorter 5-amino-acid peptide corresponding to the exact sequence of loop L6 (protein fragment from Asn246 to Lys250; L6A), as obtained from the model, was unable to induce any activation of MEK1/2, ERK1/2, JNK, and p38. The 8-amino-acid peptide corresponding to loop L7 (protein sequence from Thr280 to Lys287) was the most active peptide in the phosphorylation of JNK and p38 and was less effective in the activation of MEK1/2 and ERK1/2. The increase in the immunoreactive bands of phosphorylated JNK and p38 was comparable to the activation induced by the entire protein at concentrations of 0.13, 5, and 12 nmol/ml. The 17-amino-acid peptide corresponding to loop L8 (protein fragment from Arg315 to Lys331) was able to induce slight activation only of p38.
FIG. 6.
MEK1-MEK2/MAPK activation in response to optimal doses and times for each peptide and for the P2 porin. Total cellular protein (from 3 × 106 U937 cells/ml) was harvested, analyzed by SDS-PAGE, and immunoblotted with MEK1/2-, ERK1/2-, JNK-, or p38-phosphospecific antibody. Gels were scanned for densitometry analysis with the Sigma Gel software, and the ratio of the value for each peptide to the value for an unstimulated control is shown. The data are averages from three different experiments, and the error bars indicate the standard errors of the means.
Control peptides were synthesized and tested in order to verify the reliability of our experiments and to verify that the induction of signal transduction pathways is exclusively due to amino acid sequences corresponding to loops. In particular, (i) four peptides corresponding to β strands β1 (protein sequence from Thr9 to Ser24), β3 (Phe57 to Arg66), β8 (Asn181 to Asp190), and β15 (Leu305 to Ala314) of the P2 porin, which are considered to be located within the transmembrane region, were tested in order to verify that conserved regions located in the interior of the membrane were not able to induce signal transduction; (ii) an 11-amino-acid peptide, designated Aspy, which corresponded to a highly hydrophilic sequence, was tested in order to demonstrate that even a highly hydrophilic sequence was not able to induce any response; and (iii) a scrambled peptide corresponding to the sequence of loop L5, in which the same amino acid residues were present but in a different sequence, was tested in order to verify that the results obtained with L5 were associated with the particular sequence. The four peptides corresponding to β strands β1, β3, β8, and β15 were unable to induce significant phosphorylation with the concentrations and activation times used for the other peptides. Furthermore, the scrambled peptide and Aspy did not induce any phosphorylation of the MEK1-MEK2/MAPK pathways as well (Fig. 6).
DISCUSSION
Molecules spanning the outer membrane of gram-negative bacteria are involved in host-cell interactions. Thus, it is important to define the bacterial molecules that react with complementary structures on eukaryotic cells, since this probably represents an important step in the host response to bacterial infection. It has been demonstrated that porins play a fundamental role in the pathogenesis of gram-negative infections (18, 62). Moreover, they stimulate an immunological response, inducing the release of several cytokines (20, 22, 24, 36, 37), and they stimulate endothelial cells, inducing the expression of adhesin molecules (15) and leukocyte transmigration through endothelial cells (25). Porins induce tyrosine phosphorylation in THP-1 cells and C3H/HeJ macrophages (28). In a previous study (29), porins purified from S. enterica serovar Typhimurium were shown to activate activating protein 1 and NF-κB through the MAPK cascade. Activation of activating protein 1 and NF-κB was regulated mainly by the p38 and JNK pathways.
In this study, we verified the signaling activity of porin P2 from H. influenzae type b and investigated in detail which domains of the protein are involved in signaling pathways. As the three-dimensional structure has not yet been determined, in this study we used molecular modeling techniques to generate a three-dimensional homology model of the porin using the X-ray structures of PhoE and OmpF from E. coli and OmpK36 from K. pneumoniae. The homology model enabled us to identify more precisely the structural as well as functional roles of individual residues and of loop regions. Despite low sequence identity, structurally known proteins of the porin family superimpose surprisingly well. Thus, homology modeling appears to be a useful method for predicting the structure of any member of the family whose three-dimensional structure remains unresolved. A highly accurate structure-based family alignment can be constructed and used as a framework for alignment of new members.
Thus, we studied the sequence homology that P2 exhibits with porins whose high-resolution structures have been determined, and we constructed a theoretical model for P2. Although the level of sequence homology of P2 was quite low, the alignment of its sequence with the sequences of PhoE, OmpF, and OmpK36 was generally unambiguous. The β strands are connected by short turns on the periplasmic side and by long loops on the extracellular side. For the β scaffold and most of the short turns, construction of the model was straightforward. The locations and orientations of amino acid side chains could be approximated, but determination of exact conformations was premature. Loop regions were less well modeled, and more reliable calculation of their conformation requires further experimental data. Thus, our model is not meant to represent the detailed conformation of the loops but rather to indicate the lengths and positions of loops within the overall β-barrel framework, and it provides a preliminary structural model for determining the structure-function relationship of this surface protein. The quality of the model can be appreciated in terms of the conservation of the hydrophobicity of residues pointing toward the outside of the barrel and also the rings of aromatic residues at the proposed lipid-water interface of the molecule. All insertions and deletions are found in extracellular loops apart from one periplasmic turn, and the loops show the greatest sequence and length variability.
Our model is improved compared to earlier models (3, 16, 65) of H. influenzae type b porin. A highly accurate selection of a set of three proteins with known three-dimensional structures that belong to the same porin family allowed construction of an highly accurate structure-based family alignment. The recent availability of numerous different H. influenzae type b porin sequences increased the reliability of the alignment, which was previously constructed by using less data. The overall structure is comparable to that proposed by Srikumar et al. (65) previously.
Comparisons of the pairing of β strands and the positions of the loop regions present in the three-dimensional model with those predicted by Srikumar et al. (65) showed good agreement only for the locations of loops L4 and L8, while the remaining loops differ significantly. The differences in the models account for the fact that the results of homology modeling depend critically on the sequence alignment used. Different sequence alignments produce shifts in the positions of β strands and loops which may accumulate along the sequence and produce great differences toward the C terminus of the protein. Our model may be improved compared to earlier models, and it was used in our study to select peptide sequences to be tested in our experiments. Our results, together with further experiments on active peptides, may provide useful information on the validity of the model, which should be definitively resolved once high-resolution structural information on H. influenzae type b is available.
The results obtained in this study showed that porins from H. influenzae type b are able to induce activation of signaling pathways in U937 cells, and we also identified one of the phosphorylation pathways that was activated through the MEK1-MEK2/MAPK cascade. Porin P2, like porins from S. enterica serovar Typhimurium (29), activates mainly but not exclusively the JNK and p38 pathways.
We tested peptides that correspond to the amino acid sequences of variable loop regions facing the cell exterior and thus are probably involved in the initial interaction with the host cell. The results obtained show that peptides corresponding to the sequences of surface-exposed loops are able to activate the MEK1-MEK2/MAPK cascade; moreover, the peptides that exhibit the greatest diversity in terms of both length and sequence are involved mainly in signaling pathways. In particular, they are able to activate the MAPK pathway like the entire porin P2, thus confirming the validity of the proposed model.
Loop L1, which in the trimer is located in close proximity to adjacent monomers and interacts to some extent across the monomer-monomer interface, does not induce significant activation of MEK1/2, ERK1/2, JNK, and p38. The peptide corresponding to loop L2 is important for monomer-monomer interactions within the porin trimer, and it is unable to induce any phosphorylation. The peptides corresponding to the internal loop L3 are active in the phosphorylation of MEK1/2 and ERK1/2, while they are unable to activate the phosphorylation of JNK and p38. L4, although it is located like loop L1 at the monomer-monomer interface, induces slight phosphorylation of MEK1/2, ERK1/2, JNK, and p38. Loop L4 is the second longest loop after internal loop L3; thus, it may be that the length of the peptide allows partial exposure of this loop at the surface, which makes it also partially active. The peptide corresponding to loop L5 is very active in the phosphorylation of ERK1/2, JNK, and p38. The peptide containing the sequence of loop L6 (L6B) activates the phosphorylation of JNK and p38; the shorter peptide corresponding to the exact sequence of loop L6 (L6A) is unable to induce any activation of MEK1/2, ERK1/2, JNK, and p38. Our results show that a 5-amino-acid peptide, although corresponding to a surface loop, is too short to induce any activation. The peptide corresponding to loop L7, although it is only 8 amino acid long, is the most efficient peptide for activation of the phosphorylation of JNK and p38. Loop L8, which is the third longest loop and folds back into the barrel interior, contributing to the formation of the channel opening at the external side, is unable to activate these pathways significantly. All the control peptides were unable to activate the MEK1-MEK2/MAPK signal transduction pathway.
Our experiments were sufficient to determine which regions of the protein are involved in signal transduction and thus which regions should be subjected to further analyses, which will require determination of the minimal and maximal lengths of each active peptide and design of cyclic peptides to reduce the conformational freedom, together with a detailed mutational analysis of the selected improved peptides. Moreover, our results are further supported by the analysis of the structural model. In fact, we showed that peptides corresponding to loop variable regions are able to stimulate the MEK1-MEK2/MAPK signal transduction pathway. In particular, surface peptides corresponding to loops L5, L6, and L7 showed prominent JNK and p38 activation, comparable to the activation induced by the entire protein. The peptides corresponding to loops L1, L2, L3, L4, and L8, which are involved in formation of the trimer or in determination of the pore dimension and functionality, are unable to activate significantly the same pathways; these peptides show slight but variable activation of the signal transduction pathway, which may be related to the degree of exposure to the surface.
Cell activation takes place through signal transduction networks both in vivo (32) and in vitro (29). Our results are further supported by data reported previously; in fact, loops L5 and L6 of NTHI were reported to be the target for bactericidal serum activity against several strains (53, 74), and loop L7 of K. pneumoniae was reported to be involved in the interaction with C1q (2). Thus, our results show that other biological activities of porins are linked to epitopes located on variable loop and that the high degree of surface accessibility of the loop regions is probably an important factor in the interaction with target cells. Accessibility of epitopes on the bacterial surface is essential for antibodies to be protective and is likely to play a critical role in specific binding to cellular receptors. It is still not known which cellular receptors are involved in porin stimulation (23, 27), while it is known which receptors are involved in LPS signal transduction (68, 71).
In conclusion, in this study we demonstrated that the peptides corresponding to loops are the domains of porins mainly responsible for the activation of signaling pathways; these results may prove to be important in the development of therapies to fight conditions such as septic shock.
Editor: J. N. Weiser
REFERENCES
- 1.Alberti, S., G. Marques, S. Camprubi, S. Merino, J. M. Tomas, F. Vivanco, and V. J. Benedi. 1993. C1q binding and activation of the complement classical pathway by Klebsiella pneumoniae outer membrane proteins. Infect. Immun. 61:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberti, S., F. Rodriquez-Quinones, T. Schirmer, G. Rummel, J. M. Tomas, J. P. Rosenbusch, and V. J. Benedi. 1995. A porin from Klebsiella pneumoniae: sequence homology, three-dimensional model, and complement binding. Infect. Immun. 63:903-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arbing, M. A., J. W. Hanrahan, and J. W. Coulton. 2001. Mutagenesis identifies amino acid residues in extracellular loops and within the barrel lumen that determine voltage gating of porin from Haemophilus influenzae type b. Biochemistry 40:14621-14628. [DOI] [PubMed] [Google Scholar]
- 4.Arditi, M., J. Zhou, R. Dorio, G. W. Rong, S. M. Goyert, and K. S. Kim. 1993. Endotoxin-mediated endothelial cell injury and activation: role of soluble CD14. Infect. Immun. 61:3149-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey, S. 1994. Collaborative Computational Project number 4. Acta Crystallogr. Sect. D 50:760-763.15299374 [Google Scholar]
- 6.Bairoch, A., and R. Apweiler. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28:45-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barenkamp, S. J., Jr., R. S. Munson, and D. M. Granoff. 1982. Outer membrane protein and biotype analysis of pathogenic nontypable Haemophilus influenzae. Infect. Immun. 36:535-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell, J., S. Grass, D. Jeanteur, and R. S. Munson, Jr. 1994. Diversity of the P2 protein among nontypeable Haemophilus influenzae isolates. Infect. Immun. 62:2639-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kusrewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 10.Chong, P., Y. P. Yang, R. Fahim, P. McVerry, C. Sia, and M. Klein. 1993. Immunogenicity of overlapping synthetic peptides covering the entire sequence of Haemophilus influenzae type b outer membrane protein P2. Infect. Immun. 61:2653-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coulton, J. W., and I. F. Wan. 1983. The outer membrane of Haemophilus influenzae type b: cell envelope associations of major proteins. Can. J. Microbiol. 29:280-287. [DOI] [PubMed] [Google Scholar]
- 12.Coulton, J. W., A. C. Chin, and V. Vachon. 1992. Recombinant porin of Haemophilus influenzae type b. J. Infect. Dis. 165(Suppl. 1):S188-S191. [DOI] [PubMed]
- 13.Cowan, S. W., T. Schirmer, G. Rummel, M. Steiert, R. Ghosh, R. A. Pauptit, J. N. Jansonius, and J. P. Rosenbusch. 1992. Crystal structures explain functional properties of two E. coli porins. Nature 358:727-733. [DOI] [PubMed] [Google Scholar]
- 14.Cowan, S. W., R. M. Garavito, J. N. Jansonius, J. A. Jenkins, R. Karlsson, N. Köning, E. F. Pai, R. A. Pauptit, P. J. Rizkallh, J. P. Rosenbusch, G. Rummel, and T. Schirmer. 1995. The structure of OmpF porin in a tetragonal crystal form. Evaluation of comparative protein modeling by MODELLER. Structure 3:1041-1050. [DOI] [PubMed] [Google Scholar]
- 15.Donnarumma, G., F. Brancaccio, G. Cipollaro de l'Ero, A. Folgore, A. Marcatili, and M. Galdiero. 1996. Release of GM-CSF, sELAM and sICAM-1 by human vascular endothelium stimulated with Gram-negative and Gram-positive components. Endothelium 4:11-22. [Google Scholar]
- 16.Druim, B., J. Dankert, H. M. Jansen, and L. van Alphen. 1993. Genetic analysis of the diversity in outer membrane protein P2 of non-encapsulated Haemophilus influenzae. Microb. Pathog. 14:451-462. [DOI] [PubMed] [Google Scholar]
- 17.Dutzler, R., G. Rummel, S. Alberti, S. Hernandez-Alles, P. Phale, J. Rosenbusch, V. Benedi, and T. Schirmer. 1999. Crystal structure and functional characterization of OmpK36, the osmoporin of Klebsiella pneumoniae. Structure 7:425-434. [DOI] [PubMed] [Google Scholar]
- 18.Evans, T. J., E. Strivens, A. Carpenter, and J. Cohen. 1993. Differences in cytokine response and induction of nitric oxide synthase in endotoxin-resistant and endotoxin-sensitive mice after intravenous gram-negative infection. J. Immunol. 150:5033-5040. [PubMed] [Google Scholar]
- 19.Galdiero, F., M. A. Tufano, L. Sommese, A. Folgore, and F. Tedesco. 1984. Activation of complement system by porins extracted from Salmonella typhimurium. Infect. Immun. 46:559-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galdiero, F., G. Cipollaro de l'Ero, N. Benedetto, M. Galdiero, and M. A. Tufano. 1993. Release of cytokines induced by Salmonella typhimurium porins. Infect. Immun. 61:155-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galdiero, F., L. Sommese, P. Scarfogliero, and M. Galdiero. 1994. Biological activities, lethality, Shwartzman reaction and pyrogenicity of Salmonella typhimurium porins. Microb. Pathog. 16:111-119. [DOI] [PubMed] [Google Scholar]
- 22.Galdiero, M., G. Cipollaro de l'Ero, G. Donnarumma, A. Marcatili, and F. Galdiero. 1995. Interleukin-1 and interleukin-6 gene expression in human monocytes stimulated with Salmonella typhimurium porins. Immunology 86:612-619. [PMC free article] [PubMed] [Google Scholar]
- 23.Galdiero, M., F. Brancaccio, C. Nazzaro, and L. De Martino. 1998. Salmonella typhimurium porin internalization by leukocytes. Res. Microbiol. 149:625-630. [DOI] [PubMed] [Google Scholar]
- 24.Galdiero, M., L. De Martino, A. Marcatili, I. Nuzzo, M. Vitiello, and G. Cipollaro de l'Ero. 1998. Th1 and Th2 cell involvement in immune response to Salmonella typhimurium porins. Immunology 94:5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galdiero, M., A. Folgore, M. Molitierno, and R. Greco. 1999. Porins and lipopolysaccharide (LPS) from Salmonella typhimurium induce leucocyte transmigration through human endothelial cells in vitro. Clin. Exp. Immunol. 116:453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galdiero, M., M. D'Amico, F. Gorga, C. Di Filippo, M. D'Isanto, M. Vitiello, A. Longanella, and A. Tortora. 2001. Haemophilus influenzae porin contributes to signaling of the inflammatory cascade in rat brain. Infect. Immun. 69:221-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galdiero, M., M. D'Isanto, M. Vitiello, E. Finamore, L. Peluso, and M. Galdiero. 2001. Porins from Salmonella enterica serovar Typhimurium induce TNF-alpha, IL-6 and IL-8 release by CD14-independent and CD11a/CD18-dependent mechanisms. Microbiology 147:2697-2704. [DOI] [PubMed] [Google Scholar]
- 28.Galdiero, M., M. Vitiello, M. D'Isanto, L. Peluso, and M. Galdiero. 2001. Induction of tyrosine phosphorylated proteins in THP-1 cells by Salmonella typhimurium, Pasteurella haemolytica and Haemophilus influenzae porins. FEMS Immunol. Med. Microbiol. 13:121-130. [DOI] [PubMed] [Google Scholar]
- 29.Galdiero, M., M. Vitiello, E. Sanzari, M. D'Isanto, A. Tortora, A. Longanella, and S. Galdiero. 2002. Porins from Salmonella enterica serovar Typhimurium activate the transcription factors activating protein 1 and NF-kappaB through the Raf-1-mitogen-activated protein kinase cascade. Infect. Immun. 70:558-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldfeld, A. E., C. Doyle, and T. Maniatis. 1990. Human tumor necrosis factor alpha gene regulation by virus and lipopolysaccharide. Proc. Natl. Acad. Sci. USA 87:9769-9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta, D., Y. P. Jin, and R. Dziarski. 1995. Peptidoglycan induces transcription and secretion of TNF-alpha and activation of lyn, extracellular signal-regulated kinase, and rsk signal transduction proteins in mouse macrophages. J. Immunol. 155:2620-2630. [PubMed] [Google Scholar]
- 32.Gupta, S., D. Kumar, H. Vohra, and N. K. Ganguly. 1999. Involvement of signal transduction pathways in Salmonella typhimurium porin activated gut macrophages. Mol. Cell. Biochem. 194:235-243. [DOI] [PubMed] [Google Scholar]
- 33.Haase, E. M., A. A. Campagnari, J. Sarwar, M. Shero, M. Wirth, C. U. Cumming, and T. F. Murphy. 1991. Strain-specific and immunodominant surface epitopes of the P2 porin protein of nontypeable Haemophilus influenzae. Infect. Immun. 59:1278-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haase, E. M., K. Yi, G. D. Morse, and T. F. Murphy. 1994. Mapping of bactericidal epitopes on the P2 porin protein of nontypeable Haemophilus influenzae. Infect. Immun. 62:3712-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen, E. J., C. Hasemann, A. Clausell, J. D. Capra, K. Orth, C. R. Moomaw, C. A. Slaughter, J. L. Latimer, and E. Miller. 1989. Primary structure of the porin protein of Haemophilus influenzae type b determined by nucleotide sequence analysis. Infect. Immun. 57:1100-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henderson, B., S. Poole, and M. Wilson. 1996. Bacterial modulins: a novel class of virulence factors which cause host tissue pathology by inducing cytokine synthesis. Microbiol. Rev. 60:316-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iovane, G., P. Pagnini, M. Galdiero, G. Cipollaro de l'Ero, M. Vitiello, M. D'Isanto, and A. Marcatili. 1998. Role of Pasteurella multocida porin on cytokine expression and release by murine splenocytes. Vet. Immunol. Immunopathol. 66:391-404. [DOI] [PubMed] [Google Scholar]
- 38.Issekutz, A. C. 1983. Removal of gram-negative endotoxin from solutions by affinity chromatography. J. Immunol. Methods 61:275-281. [DOI] [PubMed] [Google Scholar]
- 39.Karin, M., Z. Liu, and E. Zandi. 1997. AP-1 function and regulation. Curr. Opin. Cell Biol. 9:240-246. [DOI] [PubMed] [Google Scholar]
- 40.Koebnik, R. 1999. Structural and functional roles of the surface-exposed loops of the beta-barrel membrane protein OmpA from Escherichia coli. J. Bacteriol. 181:3688-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kreusch, A., A. Neubuser, E. Schiltz, J. Weckesser, and G. E. Schulz. 1994. Structure of the membrane channel porin from Rhodopseudomonas blastica at 2.0 A resolution. Protein Sci. 3:58-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 277:680-685. [DOI] [PubMed] [Google Scholar]
- 43.Loos, M., and F. Clas. 1987. Antibody-independent killing of gram-negative bacteria via the classical pathway of complement. Immunol. Lett. 14:203-208. [DOI] [PubMed] [Google Scholar]
- 44.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 45.Madden, T. L., R. L. Tatusov, and J. Zhang. 1996. Applications of network BLAST server. Methods Enzymol. 266:131-141. [DOI] [PubMed] [Google Scholar]
- 46.Mathison, J. C., E. Wolfson, and R. J. Ulevitch. 1988. Participation of tumor necrosis factor in the mediation of gram negative bacterial lipopolysaccharide-induced injury in rabbits. J. Clin. Investig. 81:1925-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer, J. E., M. Hofnung, and G. E. Schulz. 1997. Structure of maltoporin from Salmonella typhimurium ligated with a nitrophenyl-maltotrioside. J. Mol. Biol. 266:761-775. [DOI] [PubMed] [Google Scholar]
- 48.Michie, H. R., K. R. Manogue, D. R. Spriggs, A. Revhaug, S. O'Dwyer, C. A. Dinarello, A. Cerami, S. M. Wolff, and D. W. Wilmore. 1988. Detection of circulating tumor necrosis factor after endotoxin administration. N. Engl. J. Med. 318:1481-1486. [DOI] [PubMed] [Google Scholar]
- 49.Morrison, D. C., and R. J. Ulevitch. 1978. The effects of bacterial endotoxins on host mediation systems. Am. J. Pathol. 93:526-617. [PMC free article] [PubMed] [Google Scholar]
- 50.Morrison, D. C., and J. L. Ryan. 1987. Endotoxins and disease mechanisms. Annu. Rev. Med. 38:417-432. [DOI] [PubMed] [Google Scholar]
- 51.Munson, R. S., Jr., and R. W. Tolan, Jr. 1989. Molecular cloning, expression, and primary sequence of outer membrane protein P2 of Haemophilus influenzae type b. Infect. Immun. 57:88-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murphy, T. F., K. C. Dudas, J. M. Mylotte, and M. A. Apicella. 1983. A subtyping system for nontypable Haemophilus influenzae based on outer-membrane proteins. J. Infect. Dis. 147:838-846. [DOI] [PubMed] [Google Scholar]
- 53.Neary, J. M., K. Yi, R. J. Karalus, and T. F. Murphy. 2001. Antibodies to loop 6 of the P2 porin protein of nontypeable Haemophilus influenzae are bactericidal against multiple strains. Infect. Immun. 69:773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nikaido, H., and E. Y. Rosenberg. 1983. Porin channels in Escherichia coli: studies with liposomes reconstituted from purified proteins. J. Bacteriol. 153:241-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nurminen, M., K. Lounatmaa, M. Sarvas, P. H. Makela, and T. Nakae. 1976. Bacteriophage-resistant mutants of Salmonella typhimurium deficient in two major outer membrane proteins. J. Bacteriol. 127:941-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nurminen, M. 1985. Enterobacterial surface antigens: methods for molecular characterization. FEMS Symp. 25:294-299. [Google Scholar]
- 57.Rosenshine, I., S. Ruschkowski, V. Foubister, and B. B. Finlay. 1994. Salmonella typhimurium invasion of epithelial cells: role of induced host cell tyrosine protein phosphorylation. Infect. Immun. 62:4969-4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saint, N., K. L. Lou, C. Widmer, M. Luckey, T. Schirmer, and J. P. Rosenbusch. 1996. Structural and functional characterization of OmpF porin mutants selected for larger pore size. II. Functional characterization. J. Biol. Chem. 271:20676-20680. [PubMed] [Google Scholar]
- 59.Sali, A., L. Potterton, F. Yuan, H. van Vlijmen, and M. Karplus. 1995. Evaluation of comparative protein modeling by MODELLER. Proteins 23:318-326. [DOI] [PubMed] [Google Scholar]
- 60.Schaeffer, H. J., and M. J. Weber. 1999. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol. Cell. Biol. 19:2435-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schirmer, T., T. A. Keller, Y. F. Wang, and J. P. Rosenbusch. 1995. Structural basis for sugar translocation through maltoporin channels at 3.1 A resolution. Science 267:512-514. [DOI] [PubMed] [Google Scholar]
- 62.Shenep, J. L., P. M. Flynn, E. F. Barrett, G. L. Stidhman, and D. F. Westen-Kirchner. 1988. Serial quantitation of endotoxemia and bacteremia during therapy for gram-negative bacterial sepsis. J. Infect. Dis. 157:237-244. [DOI] [PubMed] [Google Scholar]
- 63.Siebenlist, U., G. Franzoso, and K. Brown. 1994. Structure, regulation and function of NF-kappa B. Annu. Rev. Cell Biol. 10:405-455. [DOI] [PubMed] [Google Scholar]
- 64.Sikkema, D. J., and T. F. Murphy. 1992. Molecular analysis of the P2 porin protein of nontypeable Haemophilus influenzae. Infect. Immun. 60:5204-5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Srikumar, R., D. Dahan, M. F. Gras, M. J. Ratcliffe, L. van Alphen, and J. W. Coulton. 1992. Antigenic sites on porin of Haemophilus influenzae type b: mapping with synthetic peptides and evaluation of structure predictions. J. Bacteriol. 174:4007-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tracey, K. J., Y. Fong, D. G. Hesse, K. R. Manogue, A. T. Lee, G. C. Kuo, S. F. Lowry, and A. Cerami. 1987. Anti-cachectin/TNF monoclonal antibodies prevent septic shock during lethal bacteraemia. Nature 330:662-664. [DOI] [PubMed] [Google Scholar]
- 67.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 68.Van der Bruggen, T., S. Nijenhuis, E. van Raaij, J. Verhoef, and B. S. van Asbeck. 1999. Lipopolysaccharide-induced tumor necrosis factor alpha production by human monocytes involves the raf-1/MEK1-MEK2/ERK1-ERK2 pathway. Infect. Immun. 67:3824-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vriend, G. 1990. WHAT IF: a molecular modeling and drug design program. J. Mol. Graphics 8:52-56. [DOI] [PubMed] [Google Scholar]
- 70.Weiss, M. S., and G. E. Schulz. 1992. Structure of porin refined at 1.8 A resolution. J. Mol. Biol. 227:493-509. [DOI] [PubMed] [Google Scholar]
- 71.Willis, S. A., and P. D. Nisen. 1996. Differential induction of the mitogen-activated protein kinase pathway by bacterial lipopolysaccharide in cultured monocytes and astrocytes. Biochem. J. 313:519-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yagisawa, M., K. Saeki, E. Okuma, T. Kitamura, S. Kitagawa, H. Hirai, Y. Yazaki, F. Takaku, and A. You. 1999. Signal transduction pathways in normal human monocytes stimulated by cytokines and mediators: comparative study with normal human neutrophils or transformed cells and the putative roles in functionality and cell biology. Exp. Hematol. 27:1063-1076. [DOI] [PubMed] [Google Scholar]
- 73.Yi, K., and T. F. Murphy. 1994. Mapping of a strain-specific bactericidal epitope to the surface-exposed loop 5 on the P2 porin protein of non-typeable Haemophilus influenzae. Microb. Pathog. 17:277-282. [DOI] [PubMed] [Google Scholar]
- 74.Yi, K., and T. F. Murphy. 1997. Importance of an immunodominant surface-exposed loop on outer membrane protein P2 of nontypeable Haemophilus influenzae. Infect. Immun. 65:150-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yin, E. T., C. Galanos, S. Kinsky, R. A. Bradshaw, S. Wessler, O. Luderity, and M. F. Sarmiento. 1972. Picogram-sensitive assay for endotoxin: gelation of Limulus polyphemus blood cell lysate induced by purified lipopolysaccharides and lipid A from Gram-negative bacteria. Biochim. Biophys. Acta 261:284-289. [DOI] [PubMed] [Google Scholar]
- 76.Zeth, K., K. Diederichs, W. Welte, and H. Engelhardt. 2000. Crystal structure of Omp32, the anion-selective porin from Comamonas acidovorans, in complex with a periplasmic peptide at 2.1 A resolution. Structure Fold Des. 8:981-992. [DOI] [PubMed] [Google Scholar]
- 77.Ziegler, E. J. 1988. Protective antibody to endotoxin core: the emperor's new clothes? J. Infect. Dis. 158:286-290. [DOI] [PubMed] [Google Scholar]