Abstract
The periodontal pathogen Actinobacillus actinomycetemcomitans possesses myriad virulence factors, among them the ability to adhere to and invade epithelial cells. Recent advances in the molecular manipulation of this pathogen and the sequencing of strain HK 1651 (http://www.genome.ou.edu/act.html) have facilitated examination of the genetics of its interaction with epithelial cells. The related gram-negative organism, Haemophilus influenzae, possesses autotransporter adhesins. A search of the sequence database of strain HK 1651 revealed a homologue with similarity in the pore-forming domain to that of the H. influenzae autotransporter, Hap. A. actinomycetemcomitans mutants deficient in the homologue, Aae, showed reduced binding to epithelial cells. A method for making A. actinomycetemcomitans SUNY 465 transiently resistant to spectinomycin was used with conjugation to generate an isogenic aae mutant. An allelic replacement mutant was created in the naturally transformable A. actinomycetemcomitans strain ATCC 29523. Lactoferrin, an important part of the innate host defense system, protects against bacterial infection by bactericidal and antiadhesion mechanisms. Lactoferrin in human milk removes or cleaves Hap and another autotransporter, an immunoglobulin A1 protease, from the surface of H. influenzae, thereby reducing their binding to epithelial cells. Human milk whey had similar effects on Aae from A. actinomycetemcomitans ATCC 29523 and its binding to epithelial cells; however, there was little effect on the binding of SUNY 465. A difference in the genetic structure of aae in the two strains, apparently due to the copy number of a 135-base repeated sequence, may be the cause of the differential action of lactoferrin. aae is the first A. actinomycetemcomitans gene involved in adhesion to epithelial cells to be identified.
The gram-negative bacterium Actinobacillus actinomycetemcomitans is strongly implicated in the etiology of severe forms of juvenile and adult periodontitis. Colonization of the gingival sulcus and then the periodontal pocket by bacteria from dental plaque is the initial step in the development of periodontal disease. The ability of bacteria to adhere to surfaces in the oral cavity is essential for colonization. Earlier studies in our laboratory have shown that bacterial surface proteins and structures are important in the adhesion of A. actinomycetemcomitans to epithelial cells (33, 38). More recently, genes involved in the formation of long fibrils and bundled pili that are involved in the adherence of A. actinomycetemcomitans to solid surfaces have been discovered (9, 27, 44). The authors speculate that these genes may control the binding of A. actinomycetemcomitans to the tooth surface as a tenacious biofilm. This is possibly an early step of successful colonization of the oral cavity by A. actinomycetemcomitans.
Once established in the oral cavity, A. actinomycetemcomitans has been found inside gingival tissues (11, 49) and mucosal epithelium apart from the gingiva (47). The adhesive and invasive nature of A. actinomycetemcomitans has been examined with an in vitro model (35, 36, 52). No genes responsible for the attachment to soft tissue have been uncovered, whereas two genes related to invasion have been identified (29, 42, 48). One is homologous to apaH, a gene that encodes RGD, a sequence known to bind integrins (48). The apaH gene is a homolog of ialA, ygdP, and invA, genes associated with invasion by Bartonella bacilliformis (40), Escherichia coli K1 (8), and Rickettsia prowazekii (21), respectively. The proteins produced by these genes are members of the Nudix family of hydrolases which catalyze the dinucleoside polyphosphates, a class of signaling nucleotides (8, 12, 42). It has also been reported that A. actinomycetemcomitans invasion involves genes with sequence homology to spa genes, which are involved in protein export (29). The search for more adhesins and invasins has now been made easier with the advent of functional genomics and the whole-genome sequencing of A. actinomycetemcomitans and the closely related organism, Haemophilus influenzae.
One class of gram-negative adhesins that has been found in organisms of the family Pasteurellaceae (genera Haemophilus, Actinobacillus, and Pasteurella) is the autotransporter or type V secretion system (reviewed in reference 24). The family Pasteurellaceae contains several pathogens of the upper respiratory tract and oral cavity (31). The autotransporter proteins Hap (55) and Hia (54) of H. influenzae are implicated as adhesins in the adhesion of that organism to epithelial cells. The close relationship of A. actinomycetemcomitans to H. influenzae prompted a search for autotransporter adhesins in A. actinomycetemcomitans.
Proteins of the type V secretion system are termed “autotransporters” because they mediate their own transport from the periplasm to the exterior surface of the outer membrane (24). The extreme N terminus of an autotransporter is a signal sequence that targets the newly synthesized polypeptide to a component of the general (or type II) secretion pathway. The signal peptidase of the type II system cleaves the signal sequence and exports the remainder of the protein to the periplasm. The C-terminal region of the autotransporter then forms a β-barrel pore in the outer membrane through which the N-terminal “passenger domain” is threaded for presentation on the surface of the cell.
Comparison of autotransporter proteins from H. influenzae to the A. actinomycetemcomitans genome database revealed an open reading frame, termed aae, with characteristics of an autotransporter. We report here that mutation of this gene in two strains of A. actinomycetemcomitans resulted in a defect in adhesion to epithelial cells.
Lactoferrin is an iron-binding glycoprotein present in human milk and saliva and serves as part of the innate host defense system, possessing antibacterial (6) and antifungal (50) effects. Unsaturated lactoferrin (iron free and anion free) is able to kill A. actinomycetemcomitans (28), probably through damage to the cell envelope (18). Iron- containing lactoferrin interferes with the binding of A. actinomycetemcomitans to monolayers of fibroblasts and epithelial cells (2). Interestingly, lactoferrin cleaves two autotransporters from the surface of H. influenzae and thereby inhibits its adhesion to epithelial cell monolayers (45). Our studies for both of these phenomena showed similar effects of lactoferrin on A. actinomycetemcomitans strain ATCC 29523 cells and Aae protein but not on either SUNY 465 or its Aae.
MATERIALS AND METHODS
Bacterial strains, plasmids, and KB cells.
The bacterial strains used in this work are listed in Table 1. A. actinomycetemcomitans strains were grown using Trypticase soy broth plus yeast extract (TSB-YE; 30 g of Trypticase soy broth plus 6 g of yeast extract per liter) in a humidified 10% CO2 incubator at 37°C. E. coli strains were grown in Luria-Bertani broth (10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl per liter) at 37°C. For solid medium, liquid medium was augmented with 15 g of agar per liter.
TABLE 1.
Bacterial strains used in this study
| Strain | Characteristics | Source or reference |
|---|---|---|
| A. actinomycetemcomitans SUNY 465 | Clinical isolate, smooth phenotype, invasive; one copy of the aae repeat | 60 |
| A. actinomycetemcomitans VT 1006 | SUNY 465 carrying plasmid pPK1 | 51 |
| A. actinomycetemcomitans VT 1565 | aae mutant of SUNY 465 | This study |
| A. actinomycetemcomitans ATCC 29523 | Naturally transformable; four copies of the aae repeat | This study |
| A. actinomycetemcomitans VT 1568 | aae mutant of ATCC 29523 | This study |
| A. actinomycetemcomitans ATCC 29522 | Three copies of the aae repeat | American Type Culture Collection |
| A. actinomycetemcomitans SUNY 523 | Two copies of the aae repeat | 60 |
| E. coli JM109 | Cloning host for blue-white screen | Lab stock |
| E. coli DH5αλpir | Cloning host for mobilizable plasmids | Lab stock |
| E. coli SM10λpir | Conjugation host for mobilizable plasmids | Lab stock |
| E. coli VT 1561 | JM109 with 1.5-kb fragment in pGEM-T Easy | This study |
| E. coli VT1562 | DH5αλpir with Kmr in pGP704; mobilizable plasmid | This study |
| E. coli VT 1563 | DH5αλpir with 1.5-kb fragment in pVT1562 | This study |
| E. coli VT1564 | SM10λpir with 1.5-kb fragment in pVT1562 | This study |
| E. coli VT 1566 | JM109 with 3.1-kb fragment (whole aae gene) in pGEM-T Easy | This study |
| E. coli VT1567 | JM109 with Kmr inserted at the HindIII site in pVT1567 (disrupts aae gene) | This study |
KB, the epithelial cell line used, was derived from an oral epidermoid carcinoma and obtained from J. Moehring, University of Vermont. The cell culture medium was RPMI 1640 (Sigma, St. Louis, Mo.) plus 5% fetal bovine serum (Gibco-BRL, Grand Island, N.Y.). KB cells were cultured in a humidified 10% CO2 incubator at 37°C.
PCR amplification of aae.
Nucleotide sequences of PCR primers are indicated by dashed underlines in Fig. 1. Primers INT5 (AAG TTG CCC GAG TAA ATC G) and INT3 (CCG GGA CTT CTC ACG TTT AAC) were used to amplify an internal fragment of the aae gene with genomic DNA (obtained using PureGene [Gentra Systems, Minneapolis, Minn.]) as a template. After an initial denaturing period at 94°C (5 min), 40 cycles of denaturation at 94°C (15 s), annealing at 52.5°C (15 s), and elongation at 72°C (2 min) were performed in a Genius thermocycler (Techne, Princeton, N.J.). The 1.5-kb fragment was cloned into pGEM-T Easy (Promega, Madison, Wis.) to form plasmid pVT1561.
FIG.1.
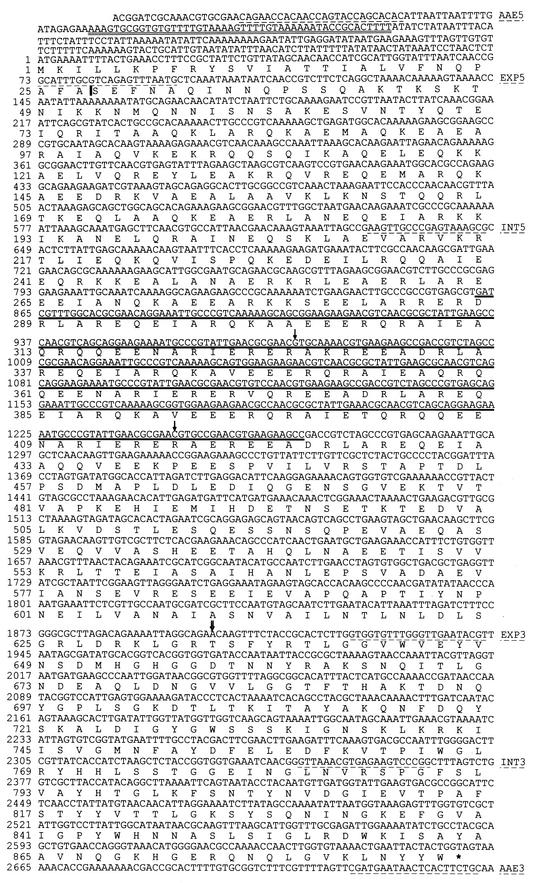
Nucleotide sequence of aae and the amino acid sequence of Aae. The PCR primers are indicated by dashed underlines, and the name of the primer is to the right. The doubly underlined sequence in the untranslated 5′ region is an inverted repeat presumed to be a terminator of the upstream gene. The vertical bar (|) in the amino acid sequence (after position 27) indicates the signal peptidase cleavage site. The solid underline beginning at position 862 indicates the repeat region. The single arrows (↓) at positions 977 and 1246 indicate the first and last bases, respectively, of the SUNY 465 deletion. The double arrow (⇓) between positions 1900 and 1901 indicates the insertion site for the sequence AATTAGACAGAA in SUNY 465.
To amplify the entire gene, primers Aae5 (CAG AAC CAC AAC CAG TAC CAG CAC AC) and Aae3 (GCA GAA GTG AGT TAT TCA TCG) were used with the same thermocycler conditions described above, except that the annealing temperature was 60°C and the elongation time was 4 min. The 3.1-kb fragment was cloned into pGEM-T Easy to form plasmid pVT1566.
Sequencing of aae.
A region that included the entire open reading frame was sequenced at the Vermont Cancer Center Sequencing Facility at the University of Vermont, using pVT1566 as template and, for sequencing primers, first the SP6 and T7 primers from pGEM-T Easy and then the primers depicted in Table 2.
TABLE 2.
Primers used for the sequencing of aae from strain SUNY 465
| Name | Direction | Sequence |
|---|---|---|
| AAE5 | Forward | CAG AAC CAC AAC CAG TAC CAG CAC AC |
| EXP5 | Forward | GCA TTT GCG TCA GAG TTT AAT G |
| INT5 | Forward | AAG TTG CCC GAG TAA AGC G |
| S5-2 | Forward | TCG CTC TAC TGC CCC TAC GGA TTT AC |
| S5-3 | Forward | GAA ATT CTG GTT GCC AAT GC |
| S5-4 | Forward | CCG GCA TTC TCA ACC TAT TAT G |
| AAE3 | Reverse | GCA GAA GTG AGT TAT TCA TCG |
| INT3 | Reverse | CCG GGA CTT CTC ACG TTT AAC |
| EXP3 | Reverse | ACG TAT TCA ACC CAA ACA CCA C |
| S3-2 | Reverse | GAG CTG CAA TTT CTT GCT CAC |
| S3-4 | Reverse | CCT CTG CCA CTT TAC GAT CTT C |
Plasmid-loss generation of isogenic mutant.
A mutagenesis system based on that described by Mintz and Fives-Taylor (39) was used to generate an aae mutant that is isogenic to SUNY 465. The A. actinomycetemcomitans-E. coli shuttle plasmid pPK1 (51) was used to make strain SUNY 465 transiently spectinomycin resistant by transformation (53). Plasmid pPK1 is a derivative of the shuttle plasmid pDL282, which was derived by the ligation of the cryptic A. actinomycetemcomitans plasmid pVT736-1 with a pUC19 derivative containing an Spr gene (see reference 51 for details). Whereas pPK1 maintains the ability to replicate in both E. coli and A. actinomycetemcomitans, it does not contain the plasmid maintenance genes of pVT736-1; therefore, it is lost if the strain is grown for 10 generations, about 16 h, in liquid medium not containing spectinomycin. Therefore, A. actinomycetemcomitans strains containing pPK1 can be easily cured of the plasmid by removing the selective pressure of spectinomycin. Construction of the mobilizable plasmid for site-directed mutagenesis of aae was as follows. The large fragment (containing the origin of replication and mobility element) from a PstI digest of pGP704 (37) was ligated to the aphA (kanamycin resistance [Kmr])-containing PstI fragment from p34S-Km3 (14). This plasmid, pVT1562, was transformed by electroporation into E. coli strain DH5αλpir. The internal fragment of aae was cut from pVT1561 with EcoRI and ligated into the EcoRI site in pVT1562 to generate pVT1563, also in the host DH5αλpir. For conjugation, plasmid pVT1563 was transformed by electroporation into the conjugation host, SM10αλpir, and the resulting strain was called VT 1564. The recipient strain, VT 1006, was a SUNY 465 derivative containing plasmid pPK1 (51).
To perform the mutagenesis, 1.0 ml of exponentially growing recipient cells (VT 1006) and 1.0 ml of exponentially growing donor cells (VT 1564) were pelleted in a centrifuge and each was resuspended in 50 μl of TSB-YE. Donor cells (10 μl) and recipient cells (50 μl) were mixed, poured onto a TSB-YE plate, and incubated in 10% CO2 at 37°C for 5 h. Thereafter, bacteria were scraped from the plate, resuspended in 1 ml of TSB-YE, diluted in TSB-YE, and plated on several TSB-YE plates containing 100 μg of kanamycin per ml and 100 μg of spectinomycin per ml. These plates were incubated in 10% CO2 at 37°C for 48 h.
Isolated colonies of putative transconjugants were grown separately overnight in 0.2 ml of TSB-YE with 100 μg of kanamycin per ml and replica plated on TSB-YE with 100 μg of kanamycin per ml and TSB-YE with 100 μg of spectinomycin per ml.
Natural transformation.
The entire aae gene amplified by PCR and cloned into pGEM-T Easy was restricted with HindIII, and the Kmr gene excised from p34S-Km3 with HindIII was inserted. The resulting plasmid, pVT1567, was restricted with DraI and run on a 0.5% agarose gel, and the large fragment containing the disrupted aae gene was extracted from the gel (Qiagen, Valencia, Calif.). The DNA was brought to 200 μl with TSB-YE. A 0.6-ml aliquot of overnight culture was centrifuged, and the bacterial pellet was resuspended in 25 μl of the DNA. This suspension was incubated at room temperature for 1 h. A 0.55-ml aliquot of warm (37°C), fresh TSB-YE was added, and the tubes were incubated at 37°C. After incubation for 2 h, 0.1 ml was plated on TSB-YE plus 100 μg of kanamycin per ml and the plates were incubated for 48 h at 37°C in 10% CO2. Several of the Kmr colonies were streaked onto a plate containing TSB-YE plus 100 μg of kanamycin per ml and incubated for 2 days. All of these putative aae mutants grew and were used to inoculate TSB-YE broth for the extraction of chromosomal DNA (as above) and subsequent analysis by Southern blotting. A verified clone was saved as VT 1568.
Adhesion assays.
Adhesion assays were performed as described previously (34). Briefly, bacteria at a multiplicity of infection of 100:1 were applied to confluent monolayers of KB cells and incubated for 2 h. The monolayers were washed twice with phosphate-buffered saline (PBS) plus MgCl2 and CaCl2, and the bacteria were released with 0.1% Triton X-100, which was subsequently diluted with PBS and plated on TSB-YE for quantification.
Expression of the passenger domain and antibody production.
Primers EXP5 (CCA TGG CTG CAT TTG CGT CAG AGT TTA ATG) and EXP3 (GGA TCC ACG TAT TCA ACC CAA ACA CCA C) were used to amplify the part of aae encoding the passenger domain of Aae (the underlined sequences are engineered NcoI and BamHI sites, respectively). The thermocycling parameters were an initial 5 min at 94°C, and then 40 cycles of denaturation at 94°C (15 s), annealing at 60°C (15 s), and elongation at 72°C (1.75 min). Recombinant passenger domain polypeptide (rAaePD) was purified using the HisBind kit (Novagen, Darmstadt, Germany) and used for antibody production (Covance, Princeton, N.J.). Crude antiserum was purified before use on a protein A column (Sigma) as specified by the manufacturer.
Protein gels and Western blots.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out with a Protean II gel apparatus (Bio-Rad, Philadelphia, Pa.), and proteins were transferred to nitrocellulose in a transfer apparatus (Hoeffer Scientific Instruments, San Francisco, Calif.). The membranes were blocked for 1 to 2 h in 5% nonfat dry milk in Tris-buffered saline (TBS) and then washed three times for 10 min each in TBS plus 0.1% Tween 20 (Sigma) (TBST). The membrane was incubated for 1 h with primary antibody (as indicated in the figure legends), diluted in TBST, and washed three times as above. A final 1-h incubation was performed with a 1:10,000 dilution of horseradish peroxidase-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, Pa.) in TBST and was followed by three washes as above. Detection was done with chemiluminescent ECL Western blotting reagents (Amersham Pharmacia, Piscataway, N.J.) as specified by the manufacturer.
Southern blots.
Site-directed mutagenesis was verified using Southern blot analysis. Genomic DNA was digested for 1 h with restriction enzymes as indicated in the figure legends and subjected to electrophoresis in 0.5% agarose gels. Depurination, denaturation, and neutralization of the DNA in the gel, transfer of the DNA to a Hybond N+ nylon membrane (Amersham Pharmacia), probe construction, and visualization were carried out using the ECL Direct nucleic acid-labeling kit (Amersham Pharmacia).
Bacterial ELISA.
A standard method for examining the surface expression of bacteria proteins is the bacterial enzyme-linked immunosorbent assay (ELISA). The assays were carried out as described previously (16). Briefly, bacteria were dried onto the ELISA plates by overnight incubation at 37°C and probed using anti-rAaePD as the primary antibody followed by peroxidase-conjugated anti-rabbit immunoglobulin G (IgG) as the secondary antibody. Hydrogen peroxide-containing buffer was used to generate the colored reaction, which was stopped with sulfuric acid and quantified in a plate reader (BioTek, Winooski, Vt.).
Immunofluorescence microscopy.
Surface expression of proteins was also tested by immunofluorescence microscopy. Bacteria were dried onto glass coverslips, fixed in 3.7% formaldehyde, and washed in PBS. Coverslips were incubated with the anti-rAaePD antibody for 20 min, washed with PBS, incubated for 20 min with fluorescein isothiocyanate-conjugated secondary antibody, and washed with PBS. Previously, antibodies to whole bacteria of strain SUNY 465 were created and purified (38). An aliquot of the purified antibody was conjugated to a blue fluorophore (Molecular Probes, Eugene, Oreg.). After incubation for 20 min with the blue fluorophore-conjugated antibody and a final wash in PBS, coverslips were inverted onto a drop of VectaShield (Vector Laboratories, Burlingame, Calif.) and sealed with nail polish. Digital micrographs were recorded with a charge-coupled device camera (Diagnostic Instruments, Sterling Heights, Mich.) attached to a fluorescence microscope (Nikon Instruments, Melville, N.Y.).
Adhesin capture.
The adhesin capture assay, a method of determining the interaction between bacterial proteins and KB cells, was essentially the same as that described elsewhere (10). KB cells were released from a monolayer by trypsin-EDTA treatment, centrifuged at 500 × g for 5 min, and resuspended in 200 μl of RPMI 1640. Extracts of A. actinomycetemcomitans were prepared by centrifugation of 1010 cells at 5,000 × g for 20 min, resuspension of the pellet in 200 μl of water, and incubation of the resuspended cells in a boiling-water bath for 10 min. After centrifugation, the supernatant was added to 106 KB cells that were resuspended in 200 μl of RPMI 1640 and incubated for 90 min at 37°C in 5% CO2. After the incubation, the assay milieu was centrifuged at 500 × g for 5 min to pellet KB cells. KB cells were washed twice (500 × g for 5 min) to remove loosely bound proteins, resuspended in SDS-PAGE loading buffer, and lysed by incubation in a boiling-water bath for 10 min. Western blot analyses were performed as described above.
Cleavage of Aae by human milk whey.
To determine if the similarity between Aae and the autotransporters of H. influenzae extends beyond sequence homology, the possible cleavage of Aae by human milk whey was examined. Milk whey, a gift from Andrew Plaut, New England Medical Center, Boston, Mass., was obtained by centrifugation of human milk twice to remove cells and lipids and diluted such that the final concentration of lactoferrin in the whey was 1.0 mg/ml. A. actinomycetemcomitans (approximately 107 cells) was mixed with the milk whey at a lactoferrin concentration of 0.5 mg/ml and diluted with RPMI 1640 to a volume of 100 μl. Mixtures were incubated at 37°C on a rotator for 1 h. Cells were centrifuged at 20,800 × g for 10 min, and supernatants were saved. Cell pellets were resuspended in 150 μl of loading buffer, and the supernatant concentrates were brought to 150 μl with loading buffer. Samples were incubated for 10 min in a boiling-water bath and examined by SDS-PAGE on a 7.5% gel. Western blot analyses with anti-rAaePD antibody were performed as above.
Effect of whey on adhesion.
Adhesion assays were performed as described above, except that A. actinomycetemcomitans was pretreated with whey as follows. Bacteria were incubated with 0.5 mg of whey per ml diluted with RPMI 1640 or with RPMI 1640 alone at a final volume of 100 μl for 90 min, as for the whey cleavage assay. After centrifugation at 20,800 × g for 10 min, supernatants were removed and cells were resuspended in RPMI 1640 and added to KB monolayers (108 bacteria/well).
Nucleotide sequence accession number.
The GenBank accession number for the aae gene identified in this study is AY262734.
RESULTS
Identification of an H. influenzae autotransporter homologue in the A. actinomycetemcomitans genome database.
Searches of the A. actinomycetemcomitans HK 1651 genome database at the University of Oklahoma (46) using the BLAST (1) program with the autotransporter proteins Hap and Hia from H. influenzae as the query sequence revealed one nucleotide sequence with significant homology (Fig. 1). This search produced the sequence in A. actinomycetemcomitans most closely related to H. influenzae autotransporters, but whether that sequence was actually most closely related to autotransporters when compared to a larger database of proteins was unknown. To answer this question, a search of the GenBank database using the A. actinomycetemcomitans translated open reading frame (termed “Aae”) as query was performed. The results showed that this sequence was most homologous to IgA1 proteases and adhesion and penetration proteins of Haemophilus and Neisseria species. The significant homology to Neisseria autotransporters was not surprising since there appears to have been considerable horizontal transfer of DNA between Haemophilus and Neisseria species (13).
An interesting feature of Aae is that it does not appear to be a full-length version of the proteins to which it is homologous; the other proteins range from 1393 to 1764 amino acids, whereas Aae has 886 amino acids. The region of highest homology is the C terminus, the region in which autotransporters have a series of transmembrane domains that form a pore in the outer membrane. IgA1 protease, composed of 1552 amino acids, has some homology to the N-terminal region of Aae, but there is no homologue for its active site in the Aae sequence. Despite the homology in the C-terminal domain, there was no significant homology between the N-terminal region of Aae and other proteins in the database. Another interesting feature of the Aae sequence is that it has a region with three 45-amino-acid imperfect repeats (indicated by the solid underline in Fig. 1).
Since a signal sequence at the N terminus is an essential feature of autotransporters and the sequence of Aae appeared to be an N-terminal truncation of its homologues, the sequence was examined for a signal sequence by using the PSORT program (http://psort.nibb.ac.jp/form.html) (41) and the method of von Heijne (57). PSORT predicted the existence of a cleaved signal sequence ending at position 27 (Fig. 1). Although Aae appeared to be an autotransporter, it did not possess the proteolytic domain of the Haemophilus and Neisseria autotransporters to which it is most homologous. That it could be an adhesin was suggested by the fact that the H. influenzae autotransporter adhesin, Hap, when uncleaved (and still cell-associated) mediates adhesion to cultured epithelial cells (25, 55). Another H. influenzae autotransporter adhesin, Hia, has no known proteolytic activity and remains cell associated (54).
Cloning and sequencing of the aae gene in SUNY 465.
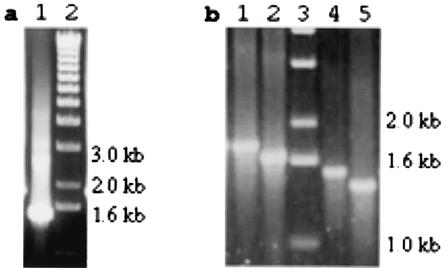
An internal fragment of aae was amplified by PCR from genomic DNA from strain SUNY 465, an invasive strain of A. actinomycetemcomitans (36), using primers INT5 and INT3. Interestingly, the apparent length of the fragment was about 300 bases shorter than predicted from the HK 1651 database (Fig. 2a). The sequencing of SUNY 465 aae revealed a 270-base deletion (two copies of the 135-bp sequence) in the repeat region. The deletion in the SUNY 465 aae gene (relative to the HK 1651 aae gene) is indicated in Fig. 1 by the thin arrows above the first and last bases in the aae sequence. A PCR screen of 30 strains of A. actinomycetemcomitans with the same primers showed at least two other alleles, presumably containing two and four copies of the repeat region (Fig. 2b). The use of PCR primers that more closely flank the repeat region showed that the length polymorphism was due to a different number of repeats in the various fragments (data not shown).
FIG. 2.
PCR of the INT5-INT3 fragment from several A. actinomycetemcomitans strains demonstrating four aae alleles. (a) SUNY 465 (lane 1); DNA markers (lane 2). (b) Strains 652, DB7A-173, SUNY 523, and SUNY 524 (lanes 1, 2, 4, and 5, respectively); DNA markers (lane 3).
The entire open reading frame was amplified using PCR with primers AAE5 and AAE3 and cloned into pGEM-T Easy. Sequencing of the cloned gene revealed that besides the expected deletion in the repeat region, there was a 12-base insertion of the sequence AATTAGACAGAA. The double arrow in Fig. 1 indicates the site of the insertion. This inserted sequence is close to an exact repeat (it differs in 1 base) of the 12 bases (AATTAGGCAGAA) immediately preceding it. Both the deletion and insertion are multiples of three bases, suggesting that the downstream region, most notably the autotransporter pore domain, is essential to the function of the protein.
Construction of an isogenic mutant of strain SUNY 465.
A total of 48 putative transconjugant colonies from the initial kanamycin- and spectinomycin-containing plate were picked for further study. All 48 strains were Sps, indicating loss of pPK1. Insertion of the plasmid was confirmed by Southern blot analysis (data not shown).
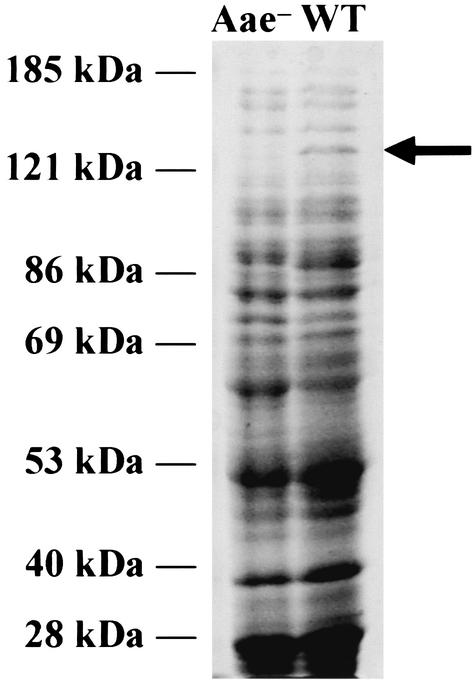
Analysis of SUNY 465 and its aae mutant by using a Coomassie blue-stained SDS-PAGE gel revealed a band above the 121-kDa marker in the wild type that is not present in the mutant (Fig. 3). The presence of a band at ∼130 kDa was unexpected, since the predicted molecular mass of Aae is 90 kDa (see Discussion).
FIG. 3.
SUNY 465 and Aae mutant extracts separated by SDS-PAGE (7.5% polyacrylamide) and stained with Coomassie blue. A high-molecular-mass band (arrow, ∼130 kDa) is absent in the Aae mutant (Aae-) but present in SUNY 465 wild type (WT).
Construction of an allelic replacement in strain ATCC 29523.
One method used to verify the function of a gene is to generate mutations in related strains and determine if the phenotypes change accordingly. An aae allelic replacement mutant was generated using linearized DNA in the naturally transformable A. actinomycetemcomitans strain ATCC 29523. The aae gene within the pGEM-T Easy plasmid was disrupted by insertion of a Kmr cassette at the HindIII site within the gene. The plasmid was cut with DraI, and the large fragment containing the disrupted gene was used in the natural transformation of ATCC 29523. Disruption of the gene was confirmed by Southern blot analysis using the INT5-INT3 internal fragment as the probe (data not shown).
Adhesion assays with the aae mutants of SUNY 465 and ATCC 29523.
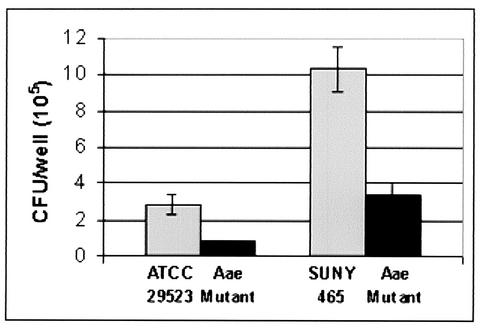
Adhesion assays were performed to determine if the putative autotransporter, Aae, is involved in the adhesion of A. actinomycetemcomitans to epithelial cells. Figure 4 shows that there was close to a 70% reduction in adhesion of the aae mutant compared with that of the wild type for each strain. These data, together with the Aae sequence similarity to autotransporter adhesins, suggested that Aae is an adhesin.
FIG. 4.
Adhesion of wild-type A. actinomycetemcomitans and Aae mutant strains to KB monolayers. SUNY 465 and ATCC 29523 strains were used. Experiments were performed with quadruplicate wells for each strain. Results shown are from a typical experiment; bars represent the standard deviation of the replicates.
Expression of the passenger domain and antibody production.
To better characterize Aae, the passenger domain was expressed using the pET28a vector and the E. coli BL21(DE3) host (Novagen). Primers EXP5 and EXP3 were constructed such that EXP5 contained an NcoI site and EXP3 contained a BamHI site in order to make use of the His6 C-terminal tag for purification. The His6 tag was placed at the C terminus because the native protein is predicted to be attached to the cell at the C terminus, with the N terminus being free. If the extreme N terminus is important to adhesion, the presence of a His6 tag in that position could interfere with that adhesion process.
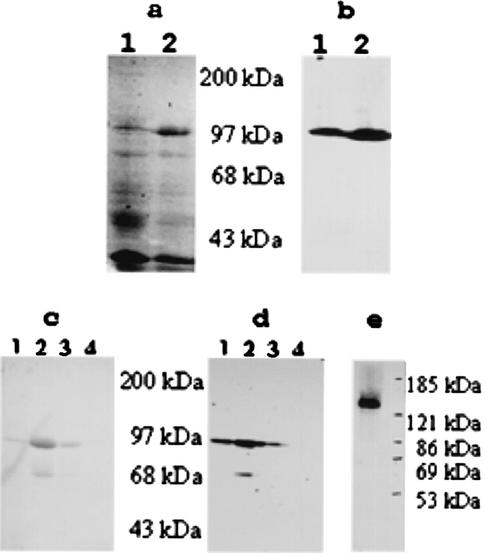
A Coomassie blue-stained SDS-PAGE gel (Fig. 5a) and the corresponding Western blot (Fig. 5b) generated using anti-His6 antibodies showed that the promoter in the BL21(DE3) host is leaky; however, ample protein was being produced. Both the gel and the blot indicated that the apparent molecular mass of Aae is ∼90 kDa, not the expected 63 kDa. A Coomassie blue-stained gel with the first four fractions from the purification of rAaePD revealed, in addition to the ∼90-kDa band, a band at ∼65 kDa (Fig. 5c). To determine if the lower band (especially evident in lane 2) was a contaminant or a degradation product, a Western blot analysis was performed (Fig. 5d). The 65- kDa band reacted with the anti-6-HIS antibody, indicating that it resulted from breakdown of the rAaePD peptide. The specificity of the anti-rAaePD antibody is shown in Fig. 5e by the presence of a single band with a molecular mass of about 130 kDa, the same size as the band representing Aae on the gel in Fig. 3.
FIG. 5.
Expression and purification of the passenger domain. (a) Extracts of the host strain uninduced (lane 1) and induced by 1 mM isopropyl-β-d-thiogalactopyranoside (lane 2), run on a 7.5% polyacrylamide gel. (b) Western blot of the same gel probed with a 1:1,000 dilution of anti-His6 antibody (Novagen), showing the existence of the His6 tag. (c) A Coomassie blue gel of the first four fractions from the purification of rAaePD. (d) Corresponding Western blot with conditions as for panel b. (e) Western blot of a SUNY 465 extract, with gel and probe conditions as for panel b. Note that the band is about ∼130 kDa, as seen in Fig. 3.
Bacterial ELISA and immunofluorescence microscopy.
The location of Aae is predicted to be on the bacterial surface; thus a bacterial ELISA, which detects surface components, was performed. Cells from both wild-type and aae mutant strains of SUNY 465 were dried onto the ELISA plate and probed with the anti-rAaePD antibody. The surface-associated antibody reaction was three times greater with the wild-type cells than with the aae mutant cells (data not shown).
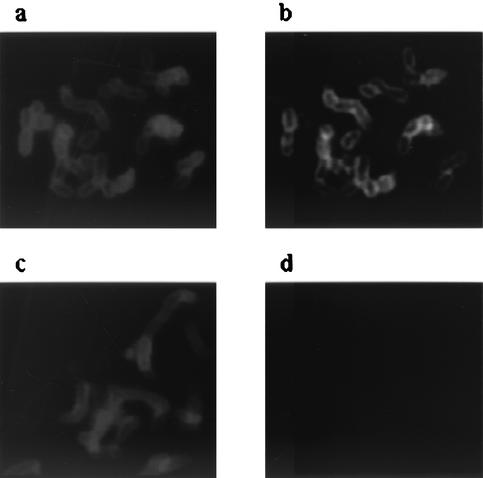
A confirmatory test of the presentation of Aae on the bacterial surface was carried out using SUNY 465 wild-type and Aae mutant and both anti-rAaePD and anti-SUNY 465 whole-cell antibody in immunofluorescence microscopy. Figure 6 shows that the Aae mutant reacted with only the anti-SUNY 465 antibody (Fig. 6c); no reactivity occurred with anti- rAaePD (Fig. 6d). By contrast, the wild type reacted with both anti-rAaePD (Fig. 6b) and SUNY 465 whole-cell antibody (Fig. 6a). Taken together, these data showed that the Aae protein is on the surface of A. actinomycetemcomitans.
FIG. 6.
Immunofluorescence microscopy of SUNY 465 and the Aae mutant. (a and b) Wild-type cells treated with anti-SUNY 465 antibody (a) and anti-rAaePD antibody (b). (c and d) Aae mutant treated with anti-SUNY 465 antibody (c) and anti-rAaePD antibody (d). Primary antibodies were used at a 1:10,000 dilution, secondary antibodies were used at a 1:100 dilution, and blue fluorophore-conjugated antibody was used at a 1:100 dilution.
Adhesin capture assay.
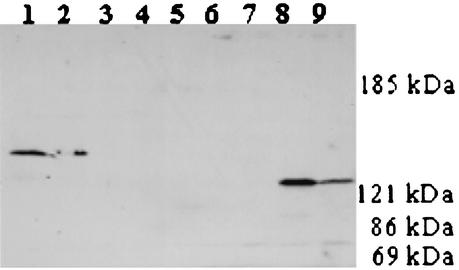
Aae was shown to be present on the bacterial surface; it could therefore interact directly with epithelial cells. To investigate this possibility, we used the adhesin capture method. KB cells were incubated with extracts of wild- type and aae mutant bacteria, washed, and analyzed by SDS-PAGE followed by Western blotting with anti-rAaePD antibody as the probe. As shown in Fig. 7, strong Aae bands were generated by both wild-type strains (lanes 1 and 2 and lanes 8 and 9), but neither of the Aae mutants (lanes 3 to 8) nor KB cells alone (lane 5) generated Aae bands. Whereas the bands representing Aae from the two wild types were different and indicate molecular masses of 140 and 130 kDa for SUNY 465 and strain ATCC Aae, respectively, the sizes were those expected for each strain based upon experimental data. Strain ATCC 29523 was determined by PCR analysis to have the same large allele as strain HK 1651 (data not shown). These data indicated that Aae is in fact an adhesin that interacts directly with KB cells.
FIG. 7.
Adhesin capture. Lanes: 1 and 2 ATCC 29523 wild type; 3 and 4, Aae mutant of ATCC 29523; 5, KB not exposed to bacterial extracts; 6 and 7, Aae mutant of SUNY 465; 8 and 9 SUNY 465 wild-type. This was a 5% gel; the anti-rAaePD antibody was used at a 1:20,000 dilution.
Effects of human milk whey on Aae.
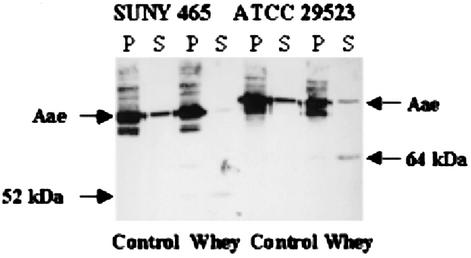
Another feature of autotransporter adhesins of H. influenzae is that they are cleaved by the lactoferrin in human milk (45). Using samples kindly provided by A. G. Plaut, we incubated wild-type A. actinomycetemcomitans with human milk whey, fractionated treated and untreated cells, and carried out PAGE and Western blot analysis on the fractions. Aae cleavage by components of the whey would be indicated by the presence of a “small” band in supernatants of whey-treated cells that was not present in supernatants of untreated cells. Since the cleavage site specificity of lactoferrin is not yet known, the expected size of the cleavage product could only be estimated to be less than ∼90 kDa (the apparent molecular mass of the recombinant passenger domain). Figure 8 shows bands at ∼50 and 64 kDa in the whey-treated supernatants of SUNY 465 and ATCC 29523, respectively, indicating cleavage of the Aae passenger domain. In lanes representing untreated cells and pellet fractions, no similar low-molecular-weight bands were evident. No other lower-molecular-weight bands were seen on Coomassie blue-stained gels or Western blots, suggesting that the shift in molecular mass is not due to nonspecific degradation.
FIG. 8.
Cleavage of Aae by human milk whey. Pellets and supernatants from bacteria incubated with RPMI medium (Control) or milk whey diluted to 0.5 mg of lactoferrin per ml with RPMI medium (Whey) are shown. The anti-rAaePD antibody was used at a 1:40,000 dilution. P, pellet; S, supernatant.
Adhesion assays with length polymorphism strains.
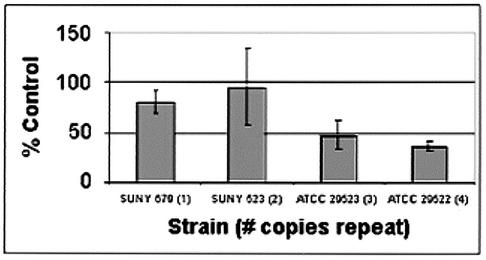
A comparison of the effects of whey on adhesion by strains containing one to four copies of the Aae repeat revealed a substantial decrease in adhesion by strains with three and four copies compared with that of SUNY 465, a strain with only one copy (Fig. 9). SUNY 523, a strain with two copies (i.e. only one additional copy), adhered at essentially the same level as SUNY 465, suggesting that a single extra copy of the repeat could not effectively reduce adhesion.
FIG. 9.
Effect of whey on adhesion of length polymorphism strains. Adhesion to monolayers of KB cells is shown as a percentage of untreated SUNY 465. The number in parentheses next to the strain name indicates the number of copies of the repeat in the strain. Each strain was tested in quadruplicate. Results shown are from a typical experiment; bars represent the standard deviation of the replicates.
DISCUSSION
Most research into bacterial pathogens, especially gram-negative organisms, has concentrated on those in the gastrointestinal tract (19) and has led to considerable knowledge of the pathogenic molecules and mechanisms used by these organisms. There has also been an effort to elucidate the mechanisms of oral bacterial pathogenesis (32, 35), but, by comparison, such knowledge for these organisms is limited.
The advent of genomics has enabled researchers to look for homologues of known virulence factors in other organisms, such as oral organisms. While there may be dead ends to this sort of comparative genomics, there are also many big rewards. We have used this approach to find an autotransporter in A. actinomycetemcomitans by using the closely related H. influenzae as the comparison organism. This is probably the key: the use of a reasonably close relative for the comparison organism.
An open reading frame, aae, with homology to autotransporter genes of H. influenzae and Neisseria species was discovered in the A. actinomycetemcomitans database. Although this open reading frame at first appeared to encode an N-terminal truncation with respect to its homologues, the Aae protein did carry an N-terminal signal sequence. Antibodies made against the putative passenger domain reacted with epitopes on the surface of wild-type strains but not with aae mutant strains, indicating that the passenger domain is presented on the surface of the bacteria.
One difference between Aae and its homologues in Neisseria and Haemophilus is its anomalous apparent molecular mass on denaturing polyacrylamide gels. This is not an uncommon phenomenon (15, 58). One possible explanation for the anomalous apparent molecular mass of rAaePD is the net charge of the residues in the passenger domain. There are 106 negatively charged and 87 positively charged residues in the expressed protein, resulting in a net charge of −19. Replacement of acidic residues with basic residues in the human papillomavirus type 16 E7 protein generates a protein with the predicted mobility rather than the anomalous high mobility of the native protein (5).
Mutants with mutations in aae derived from two different strains of A. actinomycetemcomitans showed a marked decrease in adhesion to KB cells. Upstream of the start codon of aae is an inverted repeat (Fig. 1, double underline) that is probably a terminator for the upstream gene (an open reading frame homologous to bipA, a transcription factor). The open reading frame immediately downstream from aae (homologous to bcp, a gene encoding a protein that comigrates with bacterioferritin) is transcribed in the opposite direction. These facts suggest that aae is transcribed alone. If so, this would imply that the mutation in aae is solely responsible for the adhesion defect.
Data presented here support the premise that adhesion to epithelial cells can occur in the absence of fimbriae; in Porphyromonas gingivalis, the adhesion is due to gingipains (10). There appears to be a connection between the expression of fimbrillin (a fimbria-associated protein) and gingipains, making it difficult to separate the roles of each in adhesion. Rough strains of A. actinomycetemcomitans that form transparent colonies tend to be more fimbriated than are smooth strains (26). In A. actinomycetemcomitans, smooth strains show a wide variation in adhesion to epithelial cells (34), possibly due to extracellular vesicles and amorphous material (33). This further complicates dissection of the role of nonfimbrial adhesins in A. actinomycetemcomitans. Do the vesicles and amorphous material, which are likely to be composed of outer membrane material, contain large amounts of Aae or other nonfimbrial adhesins? These earlier studies showed that the amorphous material and vesicles that could be washed off the more adhesive strains could increase adhesion of the other strains (33), suggesting that nonfimbrial adhesins are contained in that extracellular material. In either event, the role of extracellular material in the adhesion of A. actinomycetemcomitans to epithelial cells needs to be investigated further.
It is also interesting that both the gingipains of P. gingivalis and Aae of A. actinomycetemcomitans appear to be heat stable, given the method of lysis used (10). The oral cavity is subject to mechanical stress from several sources: the tongue, hot and cold food and drink, and in some cases even oral hygienic techniques. Bacteria that cause periodontal disease are able to withstand this assault and remain attached to tissues in the oral cavity. It is perhaps not surprising, then, that adhesins of periodontal pathogens are very stable.
The secreted gingipains that mediate the adhesion of P. gingivalis possess both adhesin and proteinase activities (43). The Hap autotransporter of H. influenzae also has these traits (25). Is it possible that Aae is also a proteinase, given the similar niche and adhesion strategies of these two pathogens? The sequence of Aae rules out an IgA1-like active site, such as Hap, but Aae does possess a potential zinc finger domain, HEETAH, similar to that of a dipeptidyl peptidase III enzyme (20). Mutational analysis of the HELLGH zinc-binding domain of dipeptidyl peptidase III shows a requirement for a third amino acid between the glutamic acid and second histidine residues, rather than the more common HEXXH zinc finger domain. More studies are required to determine if Aae has proteolytic activity. It may simply be that Aae is more similar in a functional way to the cell-associated H. influenzae autotransporter adhesin, Hia (54).
Lactoferrin is an important part of the host's innate defenses (18, 30). The binding of iron by lactoferrin is very strong and suggests that one of its antimicrobial activities is the sequestering of iron away from the microorganisms. The N-terminal domain of lactoferrin is a cationic peptide called lactoferricin, which can cause damage to the outer membrane of gram-negative bacteria (7, 17, 59). Lactoferrin also has the ability to bind to A. actinomycetemcomitans in a nonbactericidal manner (2). Low concentrations of lactoferrin in the oral cavity may be a risk factor in A. actinomycetemcomitans-associated periodontal disease (22).
Earlier reports by others showed that lactoferrin decreased A. actinomycetemcomitans adhesion to fibroblasts and the basement membrane (3, 4). Consistent with that is our results here, which showed that lactoferrin decreased the adhesion of A. actinomycetemcomitans strains ATCC 29522 and ATCC 29523 to epithelial cells. Our finding that lactoferrin did not affect the adhesion of strains SUNY 465 and SUNY 523 is a discrepancy that we believe may be explained by the presence of fewer copies of the Aae repeat in these strains. One notable feature of lactoferrin is the N-terminal peptide called lactoferricin, which is highly charged. The 15-residue part of lactoferricin that has been shown to interact with bacteria has a net charge of +6 (23). Each copy of the repeat at the protein level contains 55% charged residues, with a net charge of −6. The shorter form of Aae in SUNY 465 may effectively reduce its ability to interact with lactoferricin. This may be the first hint of the significance of the length polymorphisms of the aae gene.
Identification of an autotransporter adhesin of A. actinomycetemcomitans is just the first step in gaining an understanding of the genetic mechanisms involved in the interactions of this organism with epithelial cells. With the anti-rAaePD antibody and expressed recombinant passenger domain, it should be possible to identify the ligand on the surface of KB cells to which Aae binds by using the same method used to find the receptor for the Yersinia invasin (56). Gram-negative bacteria that have autotransporters often have several different ones, indicating that further searching of the HK 1651 database is warranted.
Acknowledgments
We thank Joan E. Lippmann for producing the blue fluorophore- conjugated anti-SUNY 465 antibody. We also thank Richard Ellen and Gary Ward for their critical analysis of the manuscript.
This work was supported by Public Health Service grant RO1DE09760.
Editor: J. T. Barbieri
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Alugupalli, K. R., and S. Kalfas. 1995. Inhibitory effect of lactoferrin on the adhesion of Actinobacillus actinomycetemcomitans and Prevotella intermedia to fibroblasts and epithelial cells. APMIS 103:154-160. [PubMed] [Google Scholar]
- 3.Alugupalli, K. R., and S. Kalfas. 1997. Characterization of the lactoferrin- dependent inhibition of the adhesion of Actinobacillus actinomycetemcomitans, Prevotella intermedia and Prevotella nigrescens to fibroblasts and to a reconstituted basement membrane. APMIS 105:680-688. [DOI] [PubMed] [Google Scholar]
- 4.Alugupalli, K. R., S. Kalfas, S. Edwardsson, and A. S. Naidu. 1995. Lactoferrin interaction with Actinobacillus actinomycetemcomitans. Oral Microbiol. Immunol. 10:35-41. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong, D. J., and A. Roman. 1993. The anomalous electrophoretic behavior of the human papillomavirus type 16 E7 protein is due to the high content of acidic amino acid residues. Biochem. Biophys. Res. Commun. 192:1380-1387. [DOI] [PubMed] [Google Scholar]
- 6.Arnold, R. R., M. Brewer, and J. J. Gauthier. 1980. Bactericidal activity of human lactoferrin: sensitivity of a variety of microorganisms. Infect. Immun. 28:893-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellamy, W., M. Takase, K. Yamauchi, H. Wakabayashi, K. Kawase, and M. Tomita. 1992. Identification of the bactericidal domain of lactoferrin. Biochim. Biophys. Acta 1121:130-136. [DOI] [PubMed] [Google Scholar]
- 8.Bessman, M. J., J. D. Walsh, C. A. Dunn, J. Swaminathan, J. E. Weldon, and J. Shen. 2001. The gene ygdP, associated with the invasiveness of Escherichia coli K1, designates a nudix hydrolase, Orf176, active on adenosine (5′)-pentaphospho-(5′)- adenosine (Ap5A). J. Biol. Chem. 276:37834-37838. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharjee, M. K., S. C. Kachlany, D. H. Fine, and D. H. Figurski. 2001. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J. Bacteriol. 183:5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, T., K. Nakayama, L. Belliveau, and M. J. Duncan. 2001. Porphyromonas gingivalis gingipains and adhesion to epithelial cells. Infect. Immun. 69:3048-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christersson, L. A., B. Albini, J. J. Zambon, U. M. Wikesjo, and R. J. Genco. 1987. Tissue localization of Actinobacillus actinomycetemcomitans in human periodontitis. I. Light, immunofluorescence and electron microscopic studies. J. Periodontol. 58:529-539. [DOI] [PubMed] [Google Scholar]
- 12.Conyers, G. B., and M. J. Bessman. 1999. The gene, ialA, associated with invasion of human erythrocytes by Bartonella bacilliformis, designates a nudix hydrolase active on dinucleoside 5′-polyphosphate. J. Biol. Chem. 274:1203-1206. [DOI] [PubMed] [Google Scholar]
- 13.Davis, J., A. L. Smith, W. R. Hughes, and M. Golomb. 2001. Evolution of an autotransporter: domain shuffling and lateral transfer from pathogenic Haemophilus to Neisseria. J. Bacteriol. 183:4626-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dennis, J. J., and G. J. Zylstra. 1998. Improved antibiotic-resistance cassettes through restriction site elimination using Pfu DNA polymerase PCR. BioTechniques 25:772-776. [DOI] [PubMed] [Google Scholar]
- 15.Dunker, A. K., and R. R. Rueckert. 1969. Observations on molecular weight determinations on polyacrylamide gel. J. Biol. Chem. 244:5074-5080. [PubMed] [Google Scholar]
- 16.Elder, B. L., D. K. Boraker, and P. M. Fives-Taylor. 1982. Whole-bacterial cell enzyme-linked immunosorbent assay for Streptococcus sanguis fimbrial antigens. J. Clin. Microbiol. 16:141-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellison, R. T., III, T. J. Giehl, and F. M. LaForce. 1988. Damage of the outer membrane of enteric gram-negative bacteria by lactoferrin and transferrin. Infect. Immun. 56:2774-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellison, R. T., III. 1994. The effects of lactoferrin on gram-negative bacteria, p. 71-90. In T. W. Hutchins, S. V. Rumball, and B. Lonnerdal (ed.), Lactoferrin: structure and function. Plenum Press, New York, N.Y. [DOI] [PubMed]
- 19.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukasawa, K., K. M. Fukasawa, H. Iwamoto, J. Hirose, and H. Harada. 1999. The HELLGH motif of rat liver dipeptidyl peptidase III is involved in zinc coordination and the catalytic activity of the enzyme. Biochemistry 38:8299-8303. [DOI] [PubMed] [Google Scholar]
- 21.Gaywee, J., W. Xu, S. Radulovic, M. J. Bessman, and A. F. Azad. 2002. The Rickettsia prowazekii invasion gene homolog (invA) encodes a nudix hydrolase active on adenosine (5′)-pentaphospho-(5′)-adenosine. Mol. Cell Proteomics 1:179-183. [DOI] [PubMed] [Google Scholar]
- 22.Groenink, J., E. Walgreen-Weterings, K. Nazmi, J. G. Bolscher, E. C. Veerman, A. J. van Winkelhoff, and A. V. Nieuw Amerongen. 1999. Salivary lactoferrin and low-Mr mucin MG2 in Actinobacillus actinomycetemcomitans-associated periodontitis. J. Clin. Periodontol. 26:269-275. [DOI] [PubMed] [Google Scholar]
- 23.Haug, B. E., and J. S. Svendsen. 2001. The role of tryptophan in the antibacterial activity of a 15-residue bovine lactoferricin peptide. J. Pept. Sci. 7:190-196. [DOI] [PubMed] [Google Scholar]
- 24.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendrixson, D. R., and J. W. St Geme III. 1998. The Haemophilus influenzae Hap serine protease promotes adherence and microcolony formation, potentiated by a soluble host protein. Mol. Cell 2:841-850. [DOI] [PubMed] [Google Scholar]
- 26.Inouye, T., H. Ohta, S. Kokeguchi, K. Fukui, and K. Kato. 1990. Colonial variation and fimbriation of Actinobacillus actinomycetemcomitans. FEMS Microbiol. Lett. 57:13-17. [DOI] [PubMed] [Google Scholar]
- 27.Kachlany, S. C., P. J. Planet, M. K. Bhattacharjee, E. Kollia, R. DeSalle, D. H. Fine, and D. H. Figurski. 2000. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J. Bacteriol. 182:6169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalmar, J. R., and R. R. Arnold. 1988. Killing of Actinobacillus actinomycetemcomitans by human lactoferrin. Infect. Immun. 56:2552-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laing Gibbard, L. P., G. Lepine, and R. P. Ellen. 1998. DNA fragments of Actinobacillus actinomycetemcomitans involved in invasion of KB cells. J. Dent. Res. 77(SI-B):770. [Google Scholar]
- 30.Levay, P. F., and M. Viljoen. 1995. Lactoferrin: a general review. Haematologica 80:252-267. [PubMed] [Google Scholar]
- 31.Mannheim, W. 1984. Family III. Pasteurellaceae, p. 550-575. In N. R. Kreig (ed.), Bergey's manual of systematic bacteriology, The Williams & Wilkins Co., Baltimore, Md.
- 32.Meyer, D. H., and P. M. Fives-Taylor. 1993. Models of invasion of enteric and periodontal pathogens into epithelial cells: a comparative analysis. Crit. Rev. Oral Biol. Med. 8:389-409. [DOI] [PubMed] [Google Scholar]
- 33.Meyer, D. H., and P. M. Fives-Taylor. 1993. Evidence that extracellular components function in adherence of Actinobacillus actinomycetemcomitans to epithelial cells. Infect. Immun. 61:4933-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer, D. H., and P. M. Fives-Taylor. 1994. Characteristics of adherence of Actinobacillus actinomycetemcomitans to epithelial cells. Infect. Immun. 62:928-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer, D. H., J. E. Lippmann, and P. M. Fives-Taylor. 1996. Invasion of epithelial cells by Actinobacillus actinomycetemcomitans: a dynamic, multistep process. Infect. Immun. 64:2988-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer, D. H., P. K. Sreenivasan, and P. M. Fives-Taylor. 1991. Evidence for invasion of a human oral cell line by Actinobacillus actinomycetemcomitans. Infect. Immun. 59:2719-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mintz, K. P., and P. M. Fives-Taylor. 1994. Adhesion of Actinobacillus actinomycetemcomitans to a human oral cell line. Infect. Immun. 62:3672-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mintz, K. P., and P. M. Fives-Taylor. 2000. impA, a gene coding for an inner membrane protein, influences colonial morphology of Actinobacillus actinomycetemcomitans. Infect. Immun. 68:6580-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell, S. J., and M. F. Minnick. 1995. Characterization of a two-gene locus from Bartonella bacilliformis associated with the ability to invade human erythrocytes. Infect. Immun. 63:1552-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakai, K., and M. Kanehisa. 1991. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins 11:95-110. [DOI] [PubMed] [Google Scholar]
- 42.Paju, S., M. Saarela, S. Alaluusua, P. Fives-Taylor, and S. Asikainen. 1998. Characterization of serologically nontypeable Actinobacillus actinomycetemcomitans isolates. J. Clin. Microbiol. 36:2019-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pike, R. N., J. Potempa, W. McGraw, T. H. Coetzer, and J. Travis. 1996. Characterization of the binding activities of proteinase-adhesin complexes from Porphyromonas gingivalis. J. Bacteriol. 178:2876-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Planet, P. J., S. C. Kachlany, R. DeSalle, and D. H. Figurski. 2001. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. USA 98:2503-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu, J., D. R. Hendrixson, E. N. Baker, T. F. Murphy, J. W. St Geme III, and A. G. Plaut. 1998. Human milk lactoferrin inactivates two putative colonization factors expressed by Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 95:12641-12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roe, B. A., F. Z. Najar, S. Clifton, T. Ducey, L. Lewis, and D. W. Dyer. 1997. Actinobacillus Genome Sequencing Project. [Online.] University of Oklahoma, Norman, Okla. http://www.genome.ou.edu.
- 47.Rudney, J. D., R. Chen, and G. J. Sedgewick. 2001. Intracellular Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in buccal epithelial cells collected from human subjects. Infect. Immun. 69:2700-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saarela, M., J. E. Lippmann, D. H. Meyer, and P. M. Fives-Taylor. 1999. The Actinobacillus actinomycetemcomitans apaH gene is implicated in invasion of epithelial cells. J. Dent. Res. 78(Spec. issue):1225. [Google Scholar]
- 49.Saglie, F. R., F. A. Carranza, Jr., M. G. Newman, L. Cheng, and K. J. Lewin. 1982. Identification of tissue-invading bacteria in human periodontal disease. J. Periodont. Res. 17:452-455. [DOI] [PubMed] [Google Scholar]
- 50.Samaranayake, Y. H., L. P. Samaranayake, E. H. Pow, V. T. Beena, and K. W. Yeung. 2001. Antifungal effects of lysozyme and lactoferrin against genetically similar, sequential Candida albicans isolates from a human immunodeficiency virus- infected southern Chinese cohort. J. Clin. Microbiol. 39:3296-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sreenivasan, P. K., and P. M. Fives-Taylor. 1994. Isolation and characterization of deletion derivatives of pDL282, an Actinobacillus actinomycetemcomitans/Escherichia coli shuttle plasmid. Plasmid 31:207-214. [DOI] [PubMed] [Google Scholar]
- 52.Sreenivasan, P. K., D. H. Meyer, and P. M. Fives-Taylor. 1993. Requirements for invasion of epithelial cells by Actinobacillus actinomycetemcomitans. Infect. Immun. 61:1239-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sreenivasan, P. K., D. J. LeBlanc, L. N. Lee, and P. M. Fives-Taylor. 1991. Transformation of Actinobacillus actinomycetemcomitans by electroporation, utilizing constructed shuttle plasmids. Infect. Immun. 59:4621-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.St Geme, J. W., III, and D. Cutter. 2000. The Haemophilus influenzae Hia adhesin is an autotransporter protein that remains uncleaved at the C terminus and fully cell associated. J. Bacteriol. 182:6005-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.St Geme, J. W., III, M. L. de la Morena, and S. Falkow. 1994. A Haemophilus influenzae IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol. Microbiol. 14:217-233. [DOI] [PubMed] [Google Scholar]
- 56.Van Nhieu, G. T., and R. R. Isberg. 1994. Isolation and identification of eukaryotic receptors promoting bacterial internalization. Methods Enzymol. 236:307-318. [DOI] [PubMed] [Google Scholar]
- 57.von Heijne, G. 1986. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14:4683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ward, G. E., G. W. Moy, and V. D. Vacquier. 1986. Dephosphorylation of sea urchin sperm guanylate cyclase during fertilization. Adv. Exp. Med. Biol. 207:359-382. [DOI] [PubMed] [Google Scholar]
- 59.Yamauchi, K., M. Tomita, T. J. Giehl, and R. T. Ellison III. Antibacterial activity of lactoferrin and a pepsin-derived lactoferrin peptide fragment. Infect. Immun. 61:719-728. [DOI] [PMC free article] [PubMed]
- 60.Zambon, J. J. 19865. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 12:1-20. [DOI] [PubMed]