Abstract
Several novel Legionella pneumophila virulence genes were previously discovered by use of signature-tagged mutagenesis (P. H. Edelstein, M. A. Edelstein, F. Higa, and S. Falkow, Proc. Natl. Acad. Sci. 96:8190-8195, 1999). One of these mutants appeared to be defective in multiplication in guinea pig lungs and spleens, yet it multiplies normally in guinea pig alveolar macrophages. Here we report further characterization of the mutated gene and its protein and the virulence role of the gene. The complete sequence of the gene, now called lvgA, is 627 bp long, and its protein product is approximately 27 kDa in size. lvgA was present in all 50 strains of L. pneumophila tested. No significant nucleic acid or protein homology was found in the GenBank database for the gene, nor were any distinctive motifs discovered in a search of other databases. The expression of both DotA and IcmX in the lvgA mutant was normal. Subcellular fractionation studies localized LvgA to the outer membrane fraction, and protease digestion studies suggested that at least some of the protein is surface expressed. No change in bacterial lipopolysaccharide composition or reactivity to serogroup-specific antisera was detected in the mutant. Growth competition studies with alveolar macrophages showed that the mutant was outcompeted by its parent 3-fold in 24 h and 24-fold in 48 h, in contrast to what was observed with the null phenotype in parallel testing with alveolar macrophages or with the A549 alveolar epithelial cell line. This macrophage defect of the mutant bacterium was due to slower growth, as the mutant invaded alveolar macrophages normally. Electron microscopy showed that the mutant bacterium resided in a ribosome-studded phagosome in alveolar macrophages, with no distinction from its parent. The lvgA mutant was outcompeted by its parent about sixfold in guinea pig lungs and spleens; prolonged observation of infected animals showed no late-onset virulence of the mutant. Transcomplementation of the mutant restored the parental phenotype in guinea pigs. The lvgA mutant was twofold more susceptible to killing by human β-defensin 2 but not to killing by other cationic peptides, serum complement, or polymorphonuclear neutrophils. lvgA is a novel virulence gene that is responsible for pleiotropic functions involving both extracellular and intracellular bacterial resistance mechanisms.
Legionella pneumophila, the most common cause of Legionnaires' disease, is a facultative gram-negative intracellular parasite. Legionnaires' disease is a type of pneumonia that affects mainly adults, especially those who have altered local lung defenses or who have cellular immune system-suppressing diseases. A multitude of L. pneumophila virulence factors have been described; the majority of these affect the ability of the bacterium to grow and survive within blood monocytes and alveolar macrophages or within free-living amoebae (5). To attempt to find L. pneumophila virulence factors responsible for growth or survival in vivo but not necessarily in macrophages, Edelstein et al. previously performed signature-tagged mutagenesis of L. pneumophila by using a guinea pig pneumonia model (12). Several different new and previously discovered putative virulence factors were found, and the transposon insertion mutations in all but one mutant were found to be responsible for significant defects in bacterial growth in guinea pig alveolar macrophages. This mutant, 47:1e, apparently grew as well in macrophages as did its parent, and partial sequencing of the interrupted gene revealed no homology with other known bacterial genes. In this study, we fully sequenced the 47:1e gene, which we now call lvgA, performed gene complementation studies, and characterized more completely the virulence characteristics of this gene. We demonstrate that lvgA is a novel guinea pig and macrophage virulence gene found in L. pneumophila, that it encodes an outer membrane protein, and that it is responsible for enhanced growth in macrophages detectable only by competition studies of the mutant and its parent.
MATERIALS AND METHODS
Bacteria, plasmids, and growth conditions.
L. pneumophila serogroup 1 strain F2310, also known as AA100jm, is a spontaneous streptomycin-resistant mutant of strain 130b (ATCC BAA-74), originally isolated from a patient with combined Legionnaires' disease and pneumococcal and meningococcal pneumonia (12, 27). AA100jm is virulent in guinea pigs, macrophages, and amoebae (12, 28). L. pneumophila strain F2341 contains a transposon insertion mutation in lvgA. F2341 was made by transposon mutagenesis of AA100jm with Tn903HT (Tn903 harboring a signature tag) (12). Complement-sensitive (K-12) and -resistant (K-29) Escherichia coli strains were obtained from Marcus Horwitz (20). E. coli strain K-29 and Pseudomonas aeruginosa ATCC 25619 were used as controls in neutrophil killing and defensin susceptibility assays, respectively. L. pneumophila strains having mutations in dotO-icmB, dotB, icmX, and dotF-icmG were made from strain F2310 by random transposon mutagenesis and used in studies of LvgA expression (12).
L. pneumophila bacteria were grown at 35 to 37°C in ambient air on 3-(N-morpholino)-propanesulfonic acid (MOPS)-buffered charcoal-yeast extract agar medium supplemented with α-ketoglutarate (BCYE-α agar) (14), in N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-buffered yeast extract broth supple mented with α-ketoglutarate (BYE-α broth), or in ACES-buffered charcoal-yeast extract broth supplemented with α-ketoglutaric acid (BCYE-α broth) (14). E. coli strain XL-1 blue (Stratagene) was grown at 37°C either on Luria-Bertani (LB) agar or in LB broth. Selective antimicrobial agents were added to the growth media when appropriate and included kanamycin at 30 μg/ml and chloramphenicol at 5 (L. pneumophila) or 30 (E. coli) μg/ml. A cosmid genomic library of L. pneumophila strain 130b (1) was a gift from Nicholas Cianciotto, as was plasmid pSU2719 (25). Plasmid pMMB207 (29) was a gift from Howard Shuman. Plasmid pUC18 was purchased from Life Technologies, Gaithersburg, Md.
Nucleic acid manipulations.
All nucleic acid manipulations were accomplished according to standard molecular biology techniques unless otherwise stated (2). PCR was performed by using Taq polymerase (Promega) unless otherwise stated.
Complete sequencing of the lvgA gene.
Genomic DNA from mutant clone 47:1e (AA100jm lvgA::Tn903HT) was digested with restriction enzymes known not to cut the transposon insertion upstream of the kanamycin resistance cassette and ligated into appropriately digested pUC18. The recombinant plasmids were electroporated into E. coli strain XL-1 blue, and the resulting Kmr transformants were characterized by restriction digest mapping. Plasmid DNAs from the desired transformant insertions were sequenced by using M13 primers and a primer walking technique. Because not all of the apparent open reading frame (ORF) sequence and its flanking sequence could be obtained by this approach, the remainder of the DNA sequence was obtained from a cosmid pool containing genomic 130b DNA. Approximately 800 cosmid clones were screened in pools of 16 individual clones each by using PCR with primers obtained from the partial sequence of lvgA. Two cosmid clones reacting with the lvgA-specific PCR primers were found; one was used to complete the full sequence of lvgA and its flanking sequences by using a primer walking technique. An ABI Big Dye Taq FS terminator sequencing kit (Applied Biosystems) was used to synthesize the dye-terminated DNA, which then was sequenced by using an ABI 377 automated sequencer (University of Pennsylvania Sequencing Facility). Whole sequence data were analyzed and aligned by using SeqMan II software, version 5.03 (DNASTAR Inc., Madison, Wis.). GenBank sequence database searching was performed with the BLASTX and BLASTN search algorithms. Deduced amino acid sequences were analyzed by using Motif (http://motif.stanford.edu/emotif/) and SOSUI (http://sosui.proteome.bio.tuat.ac.jp/sosuiframe0.html). Multiple sequence alignments were performed by using Megalign, version 5.03, with the Jotun Hein method (DNASTAR).
Macrophages and alveolar epithelial cells.
Guinea pig alveolar macrophages were prepared as previously described (12) and cultured in M199 (Life Technologies) supplemented with 10% fetal bovine serum (BioWhittaker, Walkersville, Md.). A549, a human alveolar epithelial cell line, was a gift from Michael Beers and was maintained in M199-10% fetal bovine serum. A549 cells were harvested at logarithmic growth phase by using a 10 mM EDTA-phosphate-buffered saline (PBS) solution and 5% trypsin-PBS. Alveolar epithelial cells (1.25 × 105/well) were cultured overnight in a 24-well culture tray in 5% CO2 in air at 37°C and then used for experiments.
Serum killing assays.
Normal serum was collected from healthy guinea pigs. The antibody titer of the serum against L. pneumophila serogroup 1 strain F889 was 1:32, as measured by indirect immunofluorescence as described previously but modified to detect guinea pig antibodies by use of fluorescein-labeled goat anti-guinea pig immunoglobulin G antibodies (Cappel Research Products, West Chester, Pa.) (10). Immune guinea pig serum was obtained from an animal immunized with Formalin-fixed L. pneumophila serogroup strain F889 and had an immunofluorescent-antibody titer of 1:256. Serum was heat inactivated at 56°C for 30 min when needed. Bacteria (108 CFU/ml) were incubated in phosphate buffer supplemented with Ca2+ and Mg2+ (pH 7.35) and with or without serum at 37°C for 1 h. The suspensions were diluted in decimal dilutions with Mueller-Hinton broth (MHB) (Difco Laboratories, Sparks, Md.), plated on BCYE-α agar, and incubated for 3 days. Surviving bacteria were enumerated by counting CFU on the plates.
Defensin killing assays.
To determine whether mutation of lvgA in L. pneumophila resulted in susceptibility to human defensins, assays of bacterial killing by the human defensins human β-defensin 2 (HβD-2) and human neutrophil peptide 1 (HNP-1) (both gifts from Tomas Ganz) were carried out by using a modification of a previously described assay (7). HβD-2 is found in human lungs, epidermis, and oral mucosa, and HNP-1 is found in human neutrophils (7). Plate-grown L. pneumophila bacteria or exponential-phase broth-grown P. aeruginosa bacteria were suspended in Sorenson's phosphate buffer (pH 7.4, 10 mM) (32). Sorenson's buffer alone had a greater killing effect on broth-grown than on plate-grown L. pneumophila F2310 and F2341, so that is why plate-grown bacteria were used. The assays were carried out with autoclaved siliconized microcentrifuge tubes (Fisherbrand siliconized low retention; Fisher Scientific).
Neutrophil killing assays.
Human peripheral polymorphonuclear leukocytes were purified by density gradient centrifugation and dextran sedimentation as described previously (4). Killing assays were performed with 20% human serum as previously described, with one exception (22). M199 rather than Hanks' balanced salt solution was used to suspend the bacteria and neutrophils, as the Legionella bacteria were killed by the salt solution.
Determination of flagellation.
Plate-grown Legionella bacteria were suspended in sterile distilled water and stained for the presence of flagella by using Ryu stain (Remel Laboratories, Lenexa, Kans.) as described previously (9, 24).
Intracellular growth assays.
Macrophages or alveolar epithelial cells were prepared as described above, infected with L. pneumophila (multiplicity of infection [MOI], 0.1), and incubated in 5% CO2 in air at 37°C. Culture supernatants were harvested at various times, diluted appropriately with MHB, and plated on BCYE-α agar. Bacterial competition studies took advantage of the kanamycin resistance of L. pneumophila strain F2341 (lvgA mutant) and the kanamycin susceptibility of its parent. In these competition studies, the concentration of the parent strain was determined by subtracting the total bacterial concentration determined by plating on nonselective BCYE-α agar from the bacterial concentration determined by plating on BCYE-α agar containing kanamycin. In some experiments, cultured cells were lysed in tissue culture wells by low-energy sonication as described previously (13); this process does not affect the viability of L. pneumophila.
Invasion assays.
Guinea pig alveolar macrophages (2.5 × 105 to 5 × 105 cells/well) were infected with bacteria (MOI, 100) in 24-well microplates, after which the plates were centrifuged at 800 × g for 8 min at room temperature. Five wells were inoculated for each bacterium. The infected macrophages were incubated at 37°C in 5% CO2 in air for 2 h; washed three times with warm M199; incubated with gentamicin (100 μg/ml) for 1 h; washed three times with warm M199, which was replaced with sterile distilled water; and lysed by low-energy sonication for 20 s. The lysate was plated quantitatively on BCYE-α agar. Previous experiments showed that there is no significant increase in intracellular L. pneumophila F2310 concentrations during the 1-h gentamicin incubation period (19), that the gentamicin incubation and subsequent washing steps kill and remove about 6 log10 extracellular bacteria, and that the residual intracellular bacterial concentration is higher than the extracellular concentration after gentamicin addition and washout (data not shown). The percentage of bacteria that were intracellular was calculated by dividing the bacterial concentration of the sonicate by that of the initial supernatant.
Electron microscopic observations.
To determine whether F2341 (lvgA mutant) resided in a ribosome-studded phagosome, electron microscopy of F2341-infected guinea pig alveolar macrophages was performed, and the results were compared with the electron microscopic findings for F2310-infected alveolar macrophages. Guinea pig alveolar macrophages were cultured on sterile plastic coverslips and then infected with L. pneumophila (MOI, 0.1). The infected cells were incubated for 2 or 3 days, fixed with 2% glutaraldehyde in PBS, and washed with ice-cold PBS. Fixed materials were processed by using standard techniques and examined by using an electron microscope, specifically seeking ribosome studding of the L. pneumophila-containing phagosome (University of Pennsylvania Biomedical Image Core Facility).
Transcomplementation of the L. pneumophila lvgA mutation.
A DNA fragment containing the L. pneumophila A gene was amplified from L. pneumophila AA100jm genomic DNA by PCR with an upstream sense primer (mu1e U79; TACTGCAGTTTATCCGTCGATTTATTGTT) and a downstream antisense primer (mu1e L1003; CGGGATCCGGGGAGTTATGTGCTTTT); PstI and BamHI sites (underlined sequences) were incorporated into these primers, respectively. Amplification was performed by using Vent polymerase (New England Biolabs, Beverly, Mass.). The 958-bp PCR product of the lvgA gene was BamHI/PstI digested and then ligated into identically digested pSU2719. E. coli XL-1 blue was transformed with the ligated product by electroporation. Plasmid DNA of Cmr transformants was mapped by restriction digestion to confirm proper insertion of the desired DNA fragment into the plasmid. The cloned lvgA gene was sequenced by using M13 forward and reverse primers to verify that there was no erroneous incorporation of nucleotides during amplification. A single clone containing the whole lvgA gene ORF, 231 bp 5′ of the ORF, and 84 bp 3′ of the ORF was picked for further study, and the plasmid was designated pHT29c. The 731-bp DraI/SmlI Klenow-treated fragment from pHT29c, which contains the lvgA ORF and its predicted promoter, was then cloned into SmaI-digested pMMB207, creating pPE15. Complementing plasmid pPE15 contained the entire lvgA ORF as well as 77 bp 5′ and 31 bp 3′ of lvgA; it excluded the first 37 bp of the putative ORF 5′ of lvgA. The lvgA mutant of AA100jm, clone 47:1e, referred to in this report as F2341, was transformed with pPE15 by electroporation as previously described (6). Plasmid DNAs from several Cmr transformants were mapped by restriction digestion to confirm the presence of the desired plasmid. One of the transformants containing the desired plasmid was picked for further studies and was designated F2461. PCR testing of F2461 with lvgA-specific primers showed that it contained the full-length lvgA gene, in contrast to the noncomplemented mutant. In addition, immunoblot analysis of the transcomplemented mutant with LvgA antibodies demonstrated that the transcomplemented mutant made detectable LvgA of the same size as that found in the parent strain and that LvgA production was not detectable in the lvgA mutant. Appropriate negative controls for the mutant and parent strains were created by electroporating empty pMMB207 into F2341 and F2310, respectively, creating F2462 and F2463, respectively.
Detection of lvgA in other Legionella sp. strains.
The presence of lvgA in 50 different strains of L. pneumophila (15 serogroups) was detected by PCR with lvgA-specific primers U86 (TATAATTTTCTCTTGGGGATT) and L711 (TGTCAATTTTGGGCTAAT), which gave a 643-bp product. BCYE-α agar plate-grown bacteria were harvested into 5 mM EDTA to approximate the density of a no. 1 MacFarland barium sulfate standard. The bacterial suspensions were heated at 99 to 100°C for 20 min. PCR was carried out by using a 25-μl volume with 1 μl of the heated bacterial suspensions as a template.
Animal model.
The guinea pig model of L. pneumophila pneumonia was used as described previously (11). L. pneumophila was grown in BYE-α broth under the appropriate selective conditions and diluted in sterile water at a concentration of 3.3 × 106 CFU/ml. A total of 106 CFU was injected into the surgically exposed tracheas of Hartley strain male guinea pigs weighing ∼250 g. The animals were killed 2 days later. The right lower lung lobe and spleen were removed aseptically, weighed and ground in MHB, and diluted in decimal dilutions with the same type of broth. Diluted tissue homogenates were plated on BCYE-α agar with or without kanamycin. Another experiment extended the postinfection observation time to 7 days postinfection. Analysis of the amount of lung consolidation was performed by low-power microscopic analysis of blind-coded histologic sections of animal lungs that had been stained with hematoxylin and eosin as described previously (11). The inflammatory pattern was characterized by using a similar method but at a higher-power magnification.
Expression of LvgA and production of anti-LvgA polyclonal antibodies.
To produce LvgA antibodies for use in immunoblot studies, the protein was expressed in E. coli. The lvgA ORF was amplified from a plasmid containing the gene by PCR with primers (CGCGGATCCATGGCAGACGGCGATATC and CCGGAATTCTTTTGCATCAATGGACCC) having BamHI and EcoRI sites (underlined sequences) to allow directional cloning into pGEX-2T (Pharmacia). PfuTurbo DNA polymerase enzyme (Stratagene) was used in the PCR. The subcloned plasmid (pBH1) was transformed into E. coli BL-21. The sequence of the amplified gene was confirmed by sequencing in both directions with pGEX forward and reverse primers (Pharmacia). The expression of LvgA in BL-21 was accomplished by growth of the bacterium in ampicillin-containing LB broth at 25°C for 5 h after previous overnight growth at 37°C. Protein expression was then induced by the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and incubation at the same temperature for 1.5 h. The protein was purified from the sonicated bacterial supernatant by using glutathione S-transferase (GST) binding resin (Pharmacia). The ∼50-kDa expressed protein, including the GST tag, was used to immunize rabbits with a multi-injection protocol which included the use of both complete and incomplete Freund's adjuvants (Express Line Protocol; Lampire Biological Laboratories, Pipersville, Pa.). Rabbit serum collected approximately 50 days after the initiation of immunization was purified by using a protein A column (Sigma).
Immunoblot analysis.
To identify the expression of LvgA, DotA, and IcmX proteins in the lvgA mutant and its parent and the expression of LvgA in dot-icm mutants, immunoblotting was used (26, 34). Plate-grown colonies of bacteria were sonicated (for DotA and LvgA) or boiled (for IcmX), lysed in Laemmli sample buffer (2), and applied to sodium dodecyl sulfate-polyacrylamide gels (12.5 or 10% acrylamide Criterion precast gels; Bio-Rad). Separated proteins were transferred electrophoretically to Immobilon-P membranes (Millipore). The membranes were probed with polyclonal rabbit antibodies against LvgA (1:500), DotA (1:1,000; gift from Craig R. Roy), or IcmX (1:500; gift from Craig R. Roy) and alkaline-phosphatase conjugated anti-rabbit secondary antibodies (Boehringer Mannheim). The proteins were visualized by using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (DotA and IcmX) or by chemiluminescence (LvgA) with a Western Light Plus System (Applied Biosystems) according to the manufacturer's directions. For DotA and IcmX, blocking and antibody dilutions were performed with PBS containing 5% nonfat dry milk and 0.1% Tween 20. A proprietary blocking buffer (Applied Biosystems) was used for LvgA studies.
Subcellular localization of LvgA.
To determine whether LvgA was secreted into culture supernatants, culture supernatants were processed as previously described (40), except that antibiotic-free BYE-α broth was used as the culture medium. The subcellular location of LvgA was determined by using the method described by Roy and Isberg (35); the only exceptions to their protocol were the use of BYE-α broth and growth of the bacteria to log phase or stationary phase (optical density at 600 nm, 0.2 or 3.0, respectively). To determine whether LvgA was surface expressed and therefore subject to proteolytic cleavage, trypsin (bovine pancreatic trypsin type I; Sigma) digestion of whole and sonicated bacteria was performed as described previously (39) with BYE-α broth-grown log-phase bacteria. Lipopolysaccharide (LPS) preparations of L. pneumophila were made as described by Nolte and colleagues (30) with BYE-α broth-grown log-phase bacteria. Determination of the preservation of L. pneumophila serogroup 1 LPS antigen was performed as previously described with rabbit polyclonal anti-L. pneumophila serogroup 1 fluorescein-conjugated antibody (M Tech, Atlanta, Ga.) (9). Immunofluorescence staining of LvgA in whole bacterial cells, either unfixed or acetone treated, was attempted with rabbit polyclonal LvgA antibody as the primary antibody and goat anti-rabbit immunoglobulin G fluorescein-labeled antibody as the secondary antibody (U.S. Biochemicals).
Nucleotide sequence accession number.
The lvgA sequence has been deposited in the GenBank database at the National Center for Biotechnology Information under accession number AF181867.
RESULTS
Complete sequence and genetic structure of lvgA.
A 1,020-bp sequence surrounding the transposon insertion in clone 47:1e was sequenced in both directions. A 627-bp ORF was contained in the sequence; the Tn903 transposon insertion site was located 367 bp 3′ of the start of the large ORF. Both a predicted promoter site and a probable Shine-Dalgarno site were located 5′ of the start of the ORF. This 627-bp ORF was named lvgA, for Legionella virulence gene. Located 6 bp 5′ of lvgA was a 108-bp putative ORF in the same reading frame as lvgA but separated from lvgA by a termination codon in the same reading frame. No significant homology of lvgA with known genes was found in the National Center for Biotechnology Information GenBank database by use of either BLASTX (word size 2) or BLASTN (word size 7) search tools. Searches of a variety of databases with either nucleic acid or deduced protein input yielded no significant homologies; the negative searches included Blocks (http://blocks.fhcrc.org/blocks/), eMotif (http://motif.stanford.edu/emotif-search/), Interpro (http://www.ebi.ac.uk/interpro/scan.html), and ProfileScan (http://hits.isb-sib.ch/cgi-bin/PFSCAN). No signal sequence was identified. A comparison of the nucleic acid sequence of lvgA with that published for L. pneumophila strain Philadelphia 1 (http://genome3.cpmc.columbia.edu/∼legion/index.html) revealed 99.4% identity at the nucleic acid level and 96.2% identity at the deduced protein level. Of the 24 nucleic acid lvgA mismatches found between L. pneumophila strains F2310 and Philadelphia 1, 16 were silent. No recognized ORFs or predicted proteins were found within 500 bp of either end of lvgA in the Columbia University L. pneumophila genome database.
Presence of lvgA in other L. pneumophila strains.
To determine whether lvgA is confined to just L. pneumophila strains F2310 and Philadelphia 1 or can be found more globally within L. pneumophila strains, colony PCR with lvgA-specific primers was performed for 50 different L. pneumophila strains. All 50 L. pneumophila strains tested contained lvgA. Thus, lvgA is not unique to a particular L. pneumophila strain.
LvgA characteristics.
Immunoblot analysis of LvgA showed that it was detectable from sonicated, but not boiled, F2310 bacterial cells. LvgA had an apparent molecular size of 27.7 kDa and appeared as a single band under both reducing and nonreducing conditions. The apparent size of LvgA was the same for both the native L. pneumophila protein and the recombinant protein (after GST cleavage) in E. coli, indicating that the native protein is monomeric and is not extensively modified posttranslation. Strain F2341 (lvgA mutant) produced no detectable LvgA, as expected (Fig. 1, even-numbered lanes). Both F2310 and F2341 produced equivalent amounts of DotA and IcmX, as judged by the density of the specific stained bands when equivalent amounts of total bacterial protein were analyzed (data not shown). L. pneumophila strains having mutations in dotO-icmB, dotB, icmX, or dotF-icmG all produced normal amounts of LvgA, indicating that the dot-icm complex played no role in the production or transport of LvgA (data not shown).
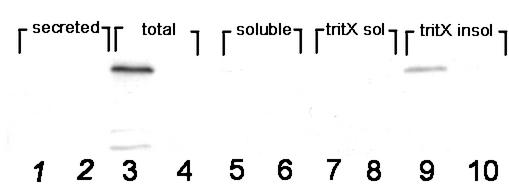
FIG. 1.
Immunoblot of various bacterial fractions with LvgA antibody. Odd-numbered lanes contain F2310 fractions (lvgA+); even-numbered lanes contain F2341 fractions (lvgA mutant). Lanes: 1 and 2, culture supernatants (secreted protein); 3 and 4, total bacterial sonicate; 5 and 6, soluble subfraction; 7 and 8, Triton X-100-soluble subfraction; 9 and 10, non-Triton X-100-soluble subfraction. Identical results were obtained in two additional independent experiments.
Subcellular localization of LvgA.
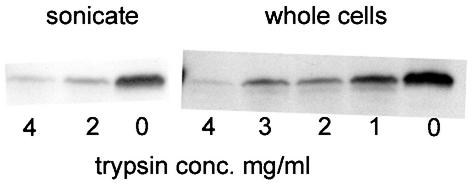
Bacterial cell fractionation studies showed that LvgA was located in the non-Triton X-100-soluble fraction (Fig. 1, lane 9), (fraction O, as defined by Roy and Isberg [35]). No secretion of LvgA into the culture medium could be detected. These findings were independent of growth phase, as both early-logarithmic- and late-stationary-phase cultures of F2310 (lvgA+) contained LvgA in the nonsecreted, non-detergent-soluble fraction. This result provided strong evidence that LvgA is an outer membrane protein. To determine whether LvgA is surface expressed, whole and sonicated bacterial cells were treated with trypsin. If LvgA is surface expressed, then trypsin exposure of both whole and sonicated bacterial cells will be expected to degrade the protein. If LvgA is not surface expressed, then trypsin will be expected to degrade the protein only in sonicated bacterial cells. LvgA was relatively trypsin resistant, but high trypsin concentrations degraded the protein in both intact and sonicated bacterial cells, indicating that it is at least partially surface expressed (Fig. 2). Coomassie blue-stained polyacrylamide gels of trypsin-treated whole bacterial cells showed that many bacterial proteins were not degraded by trypsin, excluding leaky or partially lysed bacterial cells as the explanation for the trypsin degradation of LvgA in whole bacterial cells. Since many outer membrane proteins are constituents of bacterial LPS, we determined whether there was any difference in LPS composition between F2310 (lvgA+) and F2341 (lvgA mutant). LPS gels stained by silver staining showed no difference in the 15 visualized bands between the two bacteria. Also, immunofluorescence staining of the bacterial strains with polyclonal antibody to the L. pneumophila serogroup 1 LPS antigen showed that both bacterial strains stained strongly with the antibody. Attempts to detect LvgA by immunofluorescence microscopy of F2310 with LvgA polyclonal antibody were unsuccessful, either in the presence or in the absence of acetone permeabilization of bacterial cells.
FIG. 2.
Immunoblot of F2310 (lvgA+) with LvgA antibody after trypsinization of sonicated bacterial cells or of whole bacterial cells with subsequent washing and sonication. Equivalent amounts of bacteria were added to each lane, as confirmed by Coomassie blue staining of gels before blot transfer. The trypsin concentrations (conc.) used are shown. The data are representative of findings of three independent experiments.
Growth and flagellar expression of the lvgA mutant.
L. pneumophila strain F2341 (lvgA mutant) grew as well as its parent in BYE-α broth, as determined by monitoring of its optical density with time (data not shown). The mutant possessed single or double monopolar or bipolar flagella indistinguishable from those of its parent.
Susceptibility to extracellular killing.
Initial studies with guinea pigs and their alveolar macrophages showed that L. pneumophila strain F2341 (lvgA mutant) apparently grew as well as its parent in the macrophages but that the mutant bacterium was less virulent than its parent in the guinea pig pneumonia model. To determine whether this phenotype could be attributed to the susceptibility of the mutant bacterium to non-macrophage-related host defenses, the resistance of the bacterium to killing by serum complement, neutrophils, and defensins was determined. The mutant bacterium was as resistant as its parent to killing by serum complement and human neutrophils. After 1 h of incubation in 20% fresh nonimmune guinea pig serum or heated 20% L. pneumophila immune guinea pig serum, there was no change in the bacterial concentration; incubation in combined 20% fresh nonimmune serum and heated immune serum resulted in a decrease in the bacterial concentration of log10 3.8 CFU/ml. The serum-sensitive E. coli control strain showed a decrease in concentration of log10 4.2 CFU/ml after incubation in 20% fresh serum but showed no change in concentration after incubation in heated 20% L. pneumophila immune serum. Similarly, incubation of the L. pneumophila A mutant strain with human neutrophils for 1 h resulted in a decrease in bacterial concentration of log10 0.55 CFU/ml, whereas incubation of the same strain in all reaction components except for neutrophils resulted in a decrease of log10 0.45 CFU/ml, an insignificant difference. The neutrophil-sensitive E. coli control strain was killed by neutrophils under the same conditions, with a change in concentration of log10 −2.85 CFU/ml; under the same conditions without neutrophils, there was an increase in bacterial concentration of log10 0.08 CFU/ml.
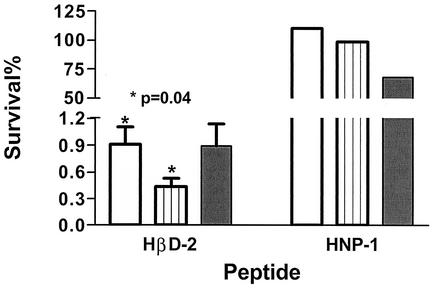
L. pneumophila strain F2341 (lvgA mutant) was significantly more sensitive to HβD-2 (5 μg/ml) than its parent, strain F2310 (Fig. 3). After 1 h of incubation with the peptide, the F2341 concentration was 0.43% (95% confidence interval [CI], 0.01 to 0.85%; n = 3) of its concentration under the same conditions in the absence of the peptide. In contrast, F2310 survival was about twice that of the mutant (0.91%; 95% CI, 0.09 to 1.7%); the difference between the survival concentrations of the two bacterial strains was significant (P value, 0.04, as determined by a paired t test; 95% CI for the difference, 0.05 to 0.90%; three independent experiments). Lower concentrations of HβD-2 had minimal killing effects on the L. pneumophila bacteria. The limited amount of material precluded extensive studies with HNP-1 (5 μg/ml), but one experiment showed that the F2310 concentration after 1 h of incubation with the peptide was not significantly different from that found for F2341 under the same conditions (Fig. 3). Both F2310 and F2341 grew equally well on BCYE-α medium containing polymyxin B (80 U/ml, or ≈8 μg/ml).
FIG. 3.
Mean percent survival of L. pneumophila strains F2310 (lvgA+) and F2341 (lvgA mutant) and P. aeruginosa after 1 h of incubation with HβD-2 or HNP-1 (both at 5 μg/ml) in comparison to that of the same bacteria incubated in buffer without the defensins. Symbols: open bar, F2310; hatched bar, F2341; closed bar, P. aeruginosa. Error bars, shown only for HβD-2, represent the standard error of the mean. Three independent experiments were performed in duplicate for HβD-2, and one experiment was performed in duplicate for HNP-1. Note the two different axis scales.
Intracellular growth and invasion.
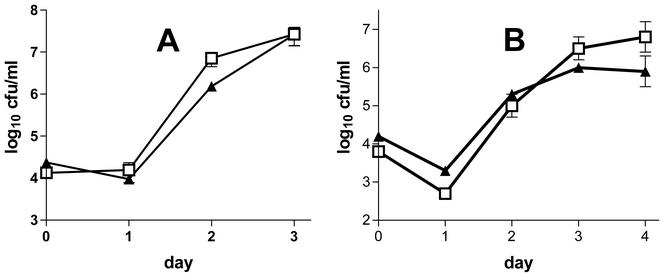
Multiple studies of L. pneumophila strain F2341 (lvgA mutant) showed that it grew in guinea pig alveolar macrophages and A549 cells apparently as well as its parent (Fig. 4). There was a consistent difference in bacterial concentrations of ≈0.5 log10 unit on the second day of macrophage infection, a difference that we originally dismissed as being both biologically and statistically insignificant. However, when the mutant and parent bacteria were combined in the same tissue culture wells, the parent strain outcompeted the mutant strain by 4- to 24-fold (Fig. 5). Microscopic observation of F2341-infected alveolar macrophages at later stages of infection showed almost complete lysis of all macrophages after 3 to 4 days postinfection, a pattern indistinguishable from that seen with F2310-infected macrophages. F2341 was internalized as effectively as its parent, as shown by gentamicin protection studies; 0.06% of extracellular F2341 bacteria were internalized after 1 h of incubation with alveolar macrophages, as opposed to 0.07% internalization of F2310 bacteria (P value, 0.75, as determined by an unpaired t test; one experiment performed in quadruplicate). A second gentamicin protection experiment confirmed these results.
FIG. 4.
Growth of L. pneumophila strains F2310 (lvgA+) and F2341 (lvgA mutant) in explanted guinea pig alveolar macrophages (A) or A549 cells (B). The L. pneumophila concentration (log10 CFU per milliliter) in tissue culture supernatants was plotted against the day of infection. Cells were infected on day 0. Several other experiments yielded similar results. Day 0 bacterial concentrations are based on counts of the bacterial inocula. Means and standard errors of the means for triplicate wells are shown. Note the different scales in the two panels. Symbols: □, F2310; ▴, F2341. The data are representative of more than two independent experiments.
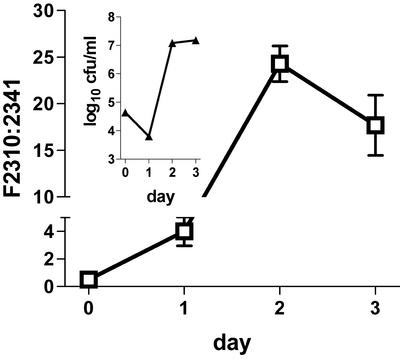
FIG. 5.
Ratio of concentrations of L. pneumophila strains F2310 (lvgA+) and F2341 (lvgA mutant) in explanted guinea pig alveolar macrophages. Equal concentrations of F2310 and F2341 were added to the same tissue culture wells, and the ratio of F2310 to F2341 was determined by observing the differential growth on media with and without kanamycin. The inset shows the total (F2341 plus F2310) bacterial concentrations. Cells were infected on day 0. Means and standard errors of the means for triplicate wells are shown. Note the two different axis scales. The data are representative of two independent experiments.
Electron microscopy.
To determine whether the lvgA mutation affected the ribosomal studding of phagosomes ordinarily observed in L. pneumophila infection of macrophages (21), electron microscopy of guinea pig alveolar macrophages infected with either the mutant or its parent was performed. This examination showed that both bacterial strains were located within qualitatively identical ribosome-studded phagosomes (data not shown).
Guinea pig infection studies.
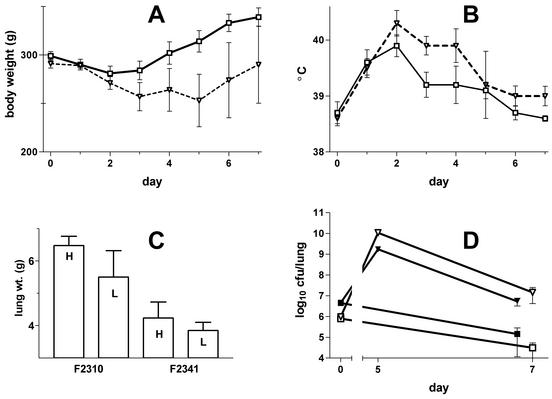
F2341 (lvgA mutant) was significantly less virulent for guinea pigs than F2310 (lvgA+) in lung infection studies; in all of these studies, the bacteria were inoculated into the trachea under direct observation. Four guinea pigs were inoculated with a mixture of F2310 and F2341 (F2310/F2341 inoculum ratio, 2.5). All animals developed fever and weight loss and were killed 50 h postinfection; F2341 was present in the lungs and spleens at 15% (95% CI, 8 to 23%) and 18% (95% CI, 14 to 20%) F2310 levels, respectively. In three other parallel (noncompetition) infection studies, in which guinea pigs received either F2310 or F2341 in nearly equivalent inocula (F2310/F2341 inoculum ratios, 0.9, 1.2, and 1.3), the animals infected with F2341 had mean L. pneumophila lung concentrations 18% of the concentrations in those infected with the parent strain (pooled data; n = 10 for each bacterium; 95% CI, 8 to 28%). Guinea pigs infected with F2341 had higher weights and lower temperatures than animals infected with F2310 (data not shown). To determine whether animals infected with F2341 would become as ill as those infected with F2310, but at a later time period, infected animals were studied for 7 days rather than for the 2 days used in the lung clearance studies (Fig. 6). The inoculum ratios were almost identical for the two bacteria (F2310/F2341 inoculum ratios, 1.0 and 1.2 for the high- and low-inoculum groups, respectively). This study showed that animals infected with F2341 had significantly lower temperatures (P value on days 3 and 4 postinfection, <0.04, as determined by a t test; pooled data) and higher body weights than animals infected with F2310 (Fig. 6B and A, respectively). Two of seven animals infected with F2310 died of pneumonia, both on day 5 postinfection, whereas all eight F2341-infected animals survived. As was found in the shorter-observation-period animal studies, the clearance of F2341 from the lungs of 7-day survivors was significantly greater than that seen in animals infected with F2310 (P value, <0.01, as determined by a t test; pooled data) (Fig. 6D). Lung weights were also significantly higher in the F2310-infected 7-day survivors (P value, <0.01, as determined by a t test; pooled data) (Fig. 6C). Animals infected with F2341 did become ill, as measured by the presence of fever and weight loss. Thus, F2341 was significantly less virulent in guinea pigs than its parent, but the mutant bacterium was not completely avirulent.
FIG. 6.
Results of guinea pig pneumonia study. Guinea pigs were infected by the intratracheal route on day 0 with L. pneumophila strain F2310 (lvgA+) or F2341 (lvgA mutant). (A) Body weights. (B) Body temperatures. Only data for the lowest inoculum sizes used (log10 6.0 and 5.9 CFU for F2310 and F2341, respectively) are shown for graphic clarity, as results obtained with higher inoculum sizes were very similar; there were four animals in each group. (C) Lung weights (wt.) for all survivors infected with either high (H) (log10 6.7 and 6.6 CFU for F2310 and F2341, respectively) or low (L) (see above) inoculum sizes; there were four animals in both F2341 groups, three in the low-inoculum F2310 group, and two in the high-inoculum F2310 group. (D) Lung concentrations of L. pneumophila for each of the four experimental groups. Day 0 bacterial concentrations are based on the inoculum size delivered into the trachea, day 5 values are for animals with fatal pneumonia (one animal in each F2310 inoculum group), and day 7 values are for survivors. For all panels, means and standard errors of the means are shown. Symbols for panels A, B, and D: ▿, F2310 low inoculum; ▾, F2310 high inoculum; ▪, F2341 high inoculum; □, F2341 low inoculum.
Infection of guinea pigs with either F2310 or F2341 resulted in lung inflammation and consolidation, although the histologic patterns differed qualitatively for the different infections. Infection with the parent strain resulted in more neutrophilic lung infiltration than infection with the lvgA mutant strain. An average of 45% (two independent experiments; n = 7; 95% CI, 19 to 70%) of the total lung area was consolidated in lungs taken from animals infected with the lvgA mutant strain for 2 days; this value was greater than the average 23% consolidation observed in animals infected with the parent strain (two independent experiments; n = 14; 95% CI, 14 to 32%), but this difference was not significant (P value, 0.09, as determined by an unpaired t test with Welch's correction). The degrees of lung consolidation were almost exactly the same for animals infected with these bacteria at 7 days after infection (one experiment; n = 7 in each group; means of 64 and 66% for mutant and parent, respectively; P value, 0.8, as determined by an unpaired t test). In striking contrast, neutrophilic infiltration was significantly greater in the lungs of animals infected with the parent strain than in those of animals infected with the mutant strain at both 2 and 7 days after infection. Macrophages represented the major type of inflammatory cells observed in the lungs of animals infected with either bacterium. Neutrophils represented an average of 40% (n = 14; 95% CI, 32 to 47%) of the total inflammatory cells present in the lungs of animals infected with the parent at 2 days after infection, versus 24% (n = 7; 95% CI, 17 to 31%) for infection with the mutant (P value, 0.009, as determined by an unpaired t test). At 7 days after infection, neutrophils represented an average of 43% (n = 7; 95% CI, 25 to 60%) of lung inflammatory cells for parent infection, versus 2% (n = 7; 95% CI, 0 to 4%) for mutant infection (P value, 0.001, as determined by an unpaired t test with Welch's correction).
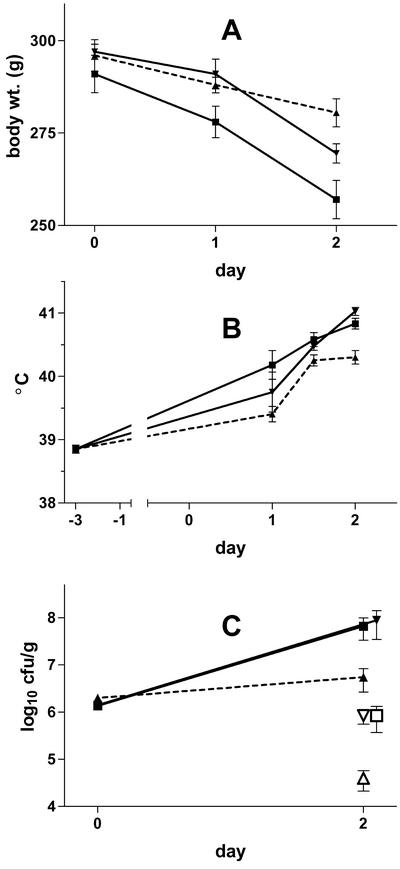
Guinea pig virulence of the transcomplemented lvgA mutant.
Transcomplementation of lvgA in F2341 (lvgA mutant) restored the virulence of the bacterium to the levels observed for the parent strain (Fig. 7). Animals infected with the transcomplemented mutant had body temperatures and spleen and lung clearances of L. pneumophila that were indistinguishable from those observed for animals infected with the parent strain containing an empty vector. Animals infected with the transcomplemented mutant showed slightly more weight loss than animals infected with the parent strain, although this difference was not significant.
FIG. 7.
Infection of guinea pigs with the transcomplemented lvgA mutant. Guinea pigs were infected on day 0 with equal amounts of F2341 (lvgA mutant) transcomplemented with lvgA on pMMB207, F2310 (lvgA+) with empty vector, or F2341 with empty vector. All animals were killed 50 h later. (A) Body weights (wt.). (B) Body temperatures. (C) Lung (filled symbols) and spleen (open symbols) concentrations (day 0 concentrations represent inoculum sizes). Data represent means and standard errors of the means. Symbols: ▪, transcomplemented mutant; ▾, parent; ▴, mutant.
DISCUSSION
These results show that lvgA is a novel L. pneumophila virulence factor which may act both through altering intracellular growth and through resistance to one or more pulmonary defensins. The discovery of lvgA as a virulence factor was possible only by testing bacterial mutants in guinea pigs, as the macrophage growth defect of the lvgA mutant is extremely subtle and would have been missed by the usual cell culture screening methods. These factors suggest that more such undiscovered L. pneumophila virulence factors may exist.
Since the initial macrophage testing of L. pneumophila F2341 (lvgA mutant) showed no significant defect in macrophage growth, we explored several possible reasons for the attenuated virulence of the mutant in guinea pigs, focusing on extra-alveolar macrophage lung host defense mechanisms. We speculated that either during initial deposition in the lungs or during the release of the bacterium from infected macrophages, the non-macrophage-dependent lung defenses could affect the survival or lung clearance of the mutant bacterium. Our studies demonstrated that the mutation did not confer serum complement sensitivity or neutrophil killing susceptibility, two major mechanisms for host defenses against pathogens in general. However, we did show that one lung antimicrobial peptide, HβD-2, was significantly more active against the mutant than against its parent. This defensin susceptibility did not appear to be the result of global susceptibility to defensins or cationic peptides, in view of the resistance of the bacterium to polymyxin B and probably its resistance to HNP-1. However, sufficient studies were not performed with HNP-1 to confirm that the mutant bacterium was truly resistant to this defensin.
The twofold increase in the sensitivity of the mutant bacterium to HβD-2 may be biologically significant. HβD-2 concentrations in normal human lungs are not known with certainty but are probably in the range of 1 μg/ml (15). Localized concentrations of HβD-2 in inflamed lungs may be considerably higher, such that resistance to extracellular or intracellular HβD-2 activity could be an important L. pneumophila virulence mechanism (15). HβD-2 is an inducible bactericidal cationic peptide produced by skin and respiratory epithelial cells, monocytes, and alveolar macrophages (8, 36, 37). A number of host inflammatory mediators increase HβD-2 production; these include gamma interferon and tumor necrosis factor α, the levels of both of which are increased in L. pneumophila infection (3, 8, 18, 36, 37). Intracellular HβD-2 concentrations and the subcellular localization are unknown, but it is possible that this defensin plays a role in killing intraphagocytic bacteria (23). HβD-2 is thought to be most important in host lung mucosal immunity against infection and in aiding in the clearance of inhaled bacteria, and it could play this role in Legionnaires' disease (36, 37). In both normal and inflamed lungs, even a small effect on bacterial survival could tip the balance in favor of the host, especially early in the course of infection. Proof of the role of HβD-2 in host defenses against L. pneumophila will require studies with L. pneumophila-susceptible animals that are defective in HβD-2 production, an animal model that does not yet exist, to our knowledge.
Cationic peptide resistance is important for the virulence of many pathogenic bacteria. Some of the known gram-negative bacterial cationic peptide virulence factors include pagP and the sap family for Salmonella enterica serovar Typhimurium, htrB for Haemophilus influenzae, and rcp for L. pneumophila (16, 17, 33, 38). S. enterica serovar Typhimurium pagP controls LPS acylation, and the sap family virulence factors act as peptide and ion transporters (16, 17, 31). Starner and colleagues described the enhanced susceptibility of nontypeable H. influenzae htrB mutants to HβD-2 but not HβD-3 and attributed the specific HβD-2 resistance to a defect in lipo-oligosaccharide acylation (38). Robey and colleagues reported that an L. pneumophila rcp mutant had reduced sensitivity to cationic peptides, including polymyxin B and C18G; neither the susceptibility of this mutant to HβD-2 nor studies of its LPS were reported, but an abnormality of LPS acylation could be expected because of the homology of rcp to pagP (33). The rcp mutant, in contrast to the lvgA mutant, showed substantially reduced growth in several different cell types. Unlike the H. influenzae htrB mutant and probably the L. pneumophila rcp mutant, the L. pneumophila lvgA mutant had no discernible abnormality in LPS structure, although it is possible that such an LPS abnormality existed but was not detected by our methods. The degree of increased sensitivity of the L. pneumophila rcp and H. influenzae htrB mutants to the peptides relative to what was observed for their parents was of the same order of magnitude as that which we observed for the increased sensitivity of F2341 relative to its parent.
We were able to demonstrate that the lvgA mutant grew less well in macrophages than its parent, but only by using bacterial competition studies and alveolar macrophages. The lvgA mutant was outcompeted by its parent starting 1 day after infection. There appeared to be a plateau in the parent/mutant ratio by day 2. Whether the mutant would eventually “catch up” to the parent with prolonged observed is impossible to know because of the entire loss of the cell monolayer by infection day 3 as a result of bacterial infection and cytotoxicity.
Virtually all studies of L. pneumophila putative virulence genes have used intracellular growth or cytotoxicity studies to determine whether a gene is a virulence factor. This study demonstrates that this cellular infection method may not detect all virulence genes, even ones that cause significant defects in macrophage multiplication, as determined by competition studies. Bacterial competition studies may be required to exclude attenuated intracellular virulence of an L. pneumophila gene mutation, despite negative cell infection or cytotoxicity studies.
Several guinea pig studies demonstrated that lvgA is an L. pneumophila virulence factor in this animal model. A competition study in which both the parent and the mutant were given to the same guinea pigs in equivalent amounts demonstrated that the mutant was outcompeted by the parent by about five- to sevenfold after 2 days. Several parallel noncompetition studies confirmed the greater lung clearance of the mutant than of the parent and that animals infected with the mutant appeared less ill. A clinical study with a longer observation period confirmed that infection with the mutant attenuated the clinical illness, compared to that observed with the parent, and that the lvgA mutant did not cause late-onset illness. The lvgA mutant bacterium is not avirulent in guinea pigs, however, as infection with this bacterium causes animal illness and lung inflammation. Thus, lvgA is only one of multiple L. pneumophila virulence genes responsible for causing pneumonia.
Guinea pig infection with the lvgA mutant bacterium resulted in considerable lung inflammation, as has been observed for other L. pneumophila bacteria with mutations in other virulence genes (28; unpublished observations). In addition, partial suppression of the acute neutrophil response in the lungs of animals infected with the lvgA mutant was seen, as was almost complete elimination of the neutrophil response to infection with the mutant bacterium in the recovery phase of lung inflammation. These results may indicate that LvgA promotes a neutrophilic inflammatory response, either directly or indirectly, or that the more efficient lung clearance of the mutant bacterium results in less of a proneutrophilic stimulus, perhaps one elicited by macrophages.
Whether the lvgA mutant is attenuated in guinea pig virulence solely because of its slower growth in alveolar macrophages is unanswered by this study. The enhanced susceptibility of the lvgA mutant to HβD-2 suggests that both HβD-2-dependent extracellular and HβD-2-independent intracellular factors play a role in the pathogenesis of Legionnaires' disease although, as discussed above, it is possible that intracellular HβD-2 plays a role in host defenses. It is possible that HβD-2 and other extracellular factors play roles during the initial entry of the bacterium into the lungs, before phagocytosis of the bacterium by lung cells, or that an extracellular factor plays a role in host defenses during the period between the release of the bacterium from one phagocyte and its entry into a new host cell. Why lvgA would have a pleiotropic virulence effect is unanswered.
The mechanism by which lvgA exerts its phenotype is unknown. We have strong evidence that LvgA is an outer membrane protein that is surface expressed and not secreted. Based on this probable location, LvgA could function in a variety of ways, such as acting as part of a bacterial sensing system or as a trigger for the host cell or immune system response. LvgA is unlikely to function as a macrophage or other immune cell binding ligand, as the lvgA mutant bacterium was phagocytosed normally by alveolar macrophages and because the bacterium was killed normally by neutrophils. A variety of L. pneumophila surface-expressed virulence factors have been described; these include lipid A, flagellae, pili, a porin, MIP, a 25-kDa outer membrane protein, and a heat shock protein (Hsp60) (5). LvgA is not clearly related to any of these previously described virulence factors on genetic or structural grounds. The lvgA mutant has no apparent effect on LPS composition or production of flagella, and the lvgA gene has no significant DNA homology with any other described bacterial gene. LvgA also has no apparent structural motifs or significant protein homology with previously bacterial proteins.
In conclusion, L. pneumophila lvgA is a novel virulence factor. This virulence factor is necessary for full virulence of the bacterium in guinea pigs and presumably humans. It is unique in that an lvgA mutant shows subtly decreased growth in alveolar macrophages, yet infection of the whole animal with the mutant results in attenuated disease seemingly out of proportion to the macrophage growth defect. The susceptibility of the mutant to a pulmonary defensin and the relative to complete lack of a neutrophilic response to infection suggest that lvgA has a pleiotropic effect on macrophage growth, local host defenses, and the global host inflammatory response to infection. It is very likely that many more such L. pneumophila virulence factors exist that have subtle or nonexistent cell infection phenotypes but are still capable of causing disease because of synergistic pleiotropic functions.
Acknowledgments
John F. Kennedy and Edward Schecter provided excellent technical assistance. Tom Ganz provided several defensins and advice on their use. Raphael Valdivia suggested performing macrophage competition studies. John Lambris provided useful suggestions for the characterization of LvgA. Donald Nierlich provided help in gene nomenclature, and Nina Salama critically reviewed the manuscript.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Arroyo, J., M. C. Hurley, M. Wolf, M. S. McClain, B. I. Eisenstein, and N. C. Engleberg. 1994. Shuttle mutagenesis of Legionella pneumophila: identification of a gene associated with host cell cytopathicity. Infect. Immun. 62:4075-4080. [DOI] [PMC free article] [PubMed]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Blanchard, D. K., J. Y. Djeu, T. W. Klein, H. Friedman, and W. E. Stewart. 1987. Induction of tumor necrosis factor by Legionella pneumophila. Infect. Immun. 55:433-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyum, A. 1968. Isolation of mononuclear cells and granulocytes from human blood. Isolation of monuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand. J. Clin. Lab. Investig. Suppl. 97:77-89. [PubMed] [Google Scholar]
- 5.Cianciotto, N. P. 2001. Pathogenicity of Legionella pneumophila. Int. J. Med. Microbiol. 291:331-343. [DOI] [PubMed] [Google Scholar]
- 6.Cianciotto, N. P., and B. S. Fields. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. USA 89:5188-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, A. M., and T. Ganz. 2000. Human antimicrobial peptides: analysis and application. BioTechniques 29:822-831. [DOI] [PubMed] [Google Scholar]
- 8.Duits, L. A., B. Ravensbergen, M. Rademaker, P. S. Hiemstra, and P. H. Nibbering. 2002. Expression of β-defensin 1 and 2 mRNA by human monocytes, macrophages and dendritic cells. Immunology 106:517-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edelstein, P. H. 1985. Legionnaires' disease laboratory manual. National Technical Information Service, Springfield, Va.
- 10.Edelstein, P. H. 2002. Detection of antibodies to Legionella spp., p. 468-476. In N. R. Rose, R. G. Hamilton, and B. Detrick (ed.), Manual of clinical laboratory immunology. American Society for Microbiology, Washington, D.C.
- 11.Edelstein, P. H., K. Calarco, and V. K. Yasui. 1984. Antimicrobial therapy of experimentally induced Legionnaires' disease in guinea pigs. Am. Rev. Respir. Dis. 130:849-856. [DOI] [PubMed] [Google Scholar]
- 12.Edelstein, P. H., M. A. Edelstein, F. Higa, and S. Falkow. 1999. Discovery of virulence genes of Legionella pneumophila by using signature tagged mutagenesis in a guinea pig pneumonia model. Proc. Natl. Acad. Sci. USA 96:8190-8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edelstein, P. H., and M. A. C. Edelstein. 1989. WIN 57273 is bactericidal for Legionella pneumophila grown in alveolar macrophages. Antimicrob. Agents Chemother. 33:2132-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edelstein, P. H., and M. A. C. Edelstein. 1993. Comparison of three buffers used in the formulation of buffered charcoal yeast extract medium. J. Clin. Microbiol. 31:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganz, T. 2002. Antimicrobial polypeptides in host defense of the respiratory tract. J. Clin. Investig. 109:693-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groisman, E. A., C. Parra-Lopez, M. Salcedo, C. J. Lipps, and F. Heffron. 1992. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc. Natl. Acad. Sci. USA 89:11939-11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo, L., K. B. Lim, C. M. Poduje, M. Daniel, J. S. Gunn, M. Hackett, and S. I. Miller. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95:189-198. [DOI] [PubMed] [Google Scholar]
- 18.Harder, J., U. Meyer-Hoffert, L. M. Teran, L. Schwichtenberg, J. Bartels, S. Maune, and J. M. Schroder. 2000. Mucoid Pseudomonas aeruginosa, TNF-α, and IL-1β, but not IL-6, induce human β-defensin-2 in respiratory epithelia. Am. J. Respir. Cell Mol. Biol. 22:714-721. [DOI] [PubMed] [Google Scholar]
- 19.Higa, F., and P. H. Edelstein. 2001. Potential virulence role of the Legionella pneumophila ptsP ortholog. Infect. Immun. 69:4782-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horwitz, M. A., and S. C. Silverstein. 1980. Influence of the Escherichia coli capsule on complement fixation and on phagocytosis and killing by human phagocytes. J. Clin. Investig. 65:82-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horwitz, M. A., and S. C. Silverstein. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J. Clin. Investig. 66:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horwitz, M. A., and S. C. Silverstein. 1981. Interaction of the Legionnaires' disease bacterium (Legionella pneumophila) with human phagocytes. I. L. pneumophila resists killing by polymorphonuclear leukocytes, antibody, and complement. J. Exp. Med. 153:386-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kisich, K. O., L. Heifets, M. Higgins, and G. Diamond. 2001. Antimycobacterial agent based on mRNA encoding human β-defensin 2 enables primary macrophages to restrict growth of Mycobacterium tuberculosis. Infect. Immun. 69:2692-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kodaka, H., A. Y. Armfield, G. L. Lombard, and V. R. Dowell, Jr. 1982. Practical procedure for demonstrating bacterial flagella. J. Clin. Microbiol. 16:948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez, E., B. Bartolome, and F. de LaCruz. 1988. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ alpha reporter gene of pUC8/9 and pUC18/19 plasmids. Gene 68:159-162. [DOI] [PubMed] [Google Scholar]
- 26.Matthews, M., and C. R. Roy. 2000. Identification and subcellular localization of the Legionella pneumophila IcmX protein: a factor essential for establishment of a replicative organelle in eukaryotic host cells. Infect. Immun. 68:3971-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer, R. D., P. H. Edelstein, B. D. Kirby, M. H. Louie, M. E. Mulligan, A. A. Morgenstein, and S. M. Finegold. 1980. Legionnaires' disease: unusual clinical and laboratory features. Ann. Intern. Med. 93:240-243. [DOI] [PubMed] [Google Scholar]
- 28.Moffat, J. F., P. H. Edelstein, D. P. Regula, Jr., J. D. Cirillo, and L. S. Tompkins. 1994. Effects of an isogenic Zn-metalloprotease-deficient mutant of Legionella pneumophila in a guinea-pig pneumonia model. Mol. Microbiol. 12:693-705. [DOI] [PubMed] [Google Scholar]
- 29.Morales, V. M., A. Bäckman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 30.Nolte, F. S., C. A. Conlin, and M. A. Motley. 1986. Electrophoretic and serological characterization of the lipopolysaccharides of Legionella pneumophila. Infect. Immun. 52:676-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parra-Lopez, C., R. Lin, A. Aspedon, and E. A. Groisman. 1994. A Salmonella protein that is required for resistance to antimicrobial peptides and transport of potassium. EMBO J. 13:3964-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perrin, D. D., and B. Dempsey. 1974. Buffers for pH and metal ion control, p. 45. Chapman & Hall, New York, N.Y.
- 33.Robey, M., W. O'Connell, and N. P. Cianciotto. 2001. Identification of Legionella pneumophila rcp, a pagP-like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection. Infect. Immun. 69:4276-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy, C. R., K. H. Berger, and R. R. Isberg. 1998. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28:663-674. [DOI] [PubMed] [Google Scholar]
- 35.Roy, C. R., and R. R. Isberg. 1997. Topology of Legionella pneumophila DotA: an inner membrane protein required for replication in macrophages. Infect. Immun. 65:571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroder, J. M., and J. Harder. 1999. Human beta-defensin-2. Int. J. Biochem. Cell Biol. 31:645-651. [DOI] [PubMed] [Google Scholar]
- 37.Schutte, B. C., and P. B. McCray, Jr. 2002. β-Defensins in lung host defense. Annu. Rev. Physiol. 64:709-748. [DOI] [PubMed] [Google Scholar]
- 38.Starner, T. D., W. E. Swords, M. A. Apicella, and P. B. McCray, Jr. 2002. Susceptibility of nontypeable Haemophilus influenzae to human β-defensins is influenced by lipooligosaccharide acylation. Infect. Immun. 70:5287-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu, C., S. Menard, J. D. Dubreuil, and J. M. Fairbrother. 1996. Detection and localization of the EaeA protein of attaching and effacing Escherichia coli O45 from pigs using a monoclonal antibody. Microb. Pathog. 21:205-213. [DOI] [PubMed] [Google Scholar]
- 40.Zuckman, D. M., J. B. Hung, and C. R. Roy. 1999. Pore-forming activity is not sufficient for Legionella pneumophila phagosome trafficking and intracellular growth. Mol. Microbiol. 32:990-1001. [DOI] [PubMed] [Google Scholar]