Abstract
Aspergillus fumigatus is the predominant mold pathogen in patients who lack functional innate immunity. The A. fumigatus rhbA gene was first identified as a transcript that was upregulated when the organism was grown in the presence of mammalian cells. To gain insight into the function of rhbA in the growth and pathogenesis of A. fumigatus, we constructed a strain that lacks a functional rhbA gene. The ΔrhbA mutant showed a significant reduction in virulence compared to the virulence of the wild type in a mouse model of invasive aspergillosis. Complementation of the deletion with the wild-type gene restored full virulence. Although the ΔrhbA mutant grew as well as the wild type on solid medium containing the rich nitrogen source ammonium, the growth of the mutant was impaired on medium containing poor nitrogen sources. Like the Saccharomyces cerevisiae rhb1 mutant, the ΔrhbA mutant exhibited increased uptake of arginine. In addition, the ΔrhbA strain underwent asexual development in submerged cultures, even under ammonium-excess conditions. Growth of the mutant with poor nitrogen sources eliminated both the arginine uptake and submerged asexual development phenotypes. The mutant showed enhanced sensitivity to the TOR kinase inhibitor rapamycin. These findings establish the importance of rhbA for A. fumigatus virulence and suggest a role for rhbA in nutrient sensing.
The ability to adapt to changes in nutrient availability is an essential attribute of many successful pathogens. In fungi, nutrient-regulated signaling pathways have been clearly linked to pathogenesis in a number of species, but their contribution to the virulence of Aspergillus fumigatus is largely unexplored (14, 33). A. fumigatus is an opportunistic fungal pathogen that has emerged as a major cause of morbidity and mortality in patients who are neutropenic or lack intact innate immunity (21). As a saprophytic fungus, A. fumigatus can utilize a wide variety of carbon and nitrogen sources, and it has been hypothesized that such physiological versatility contributes to its status as the predominant mold pathogen (15).
The Rheb proteins comprise a family of Ras-related proteins that exhibit deviations from the consensus amino acid sequence in the first GTP-binding domain, as well as in the effector domain (24, 36, 41). Rheb proteins are conserved from lower eukaryotes to mammals, yet the mechanism by which they function remains elusive. In mammalian cells, Rheb interacts with Raf-1 in a cAMP-dependent manner and antagonizes Ras signaling (9, 43). In yeast, Rheb is required for the maintenance of wild-type sensitivity to the toxic arginine analog canavanine (36, 42). The canavanine hypersensitivity of Saccharomyces cerevisiae rhb1 mutants is due to increased arginine uptake mediated by CAN1p, the basic amino acid transporter (36). Deletion of rhb1 in Schizosaccharomyces pombe results in a phenotype that mimics nitrogen starvation, with cell cycle arrest and upregulation of fnx1 and mei2 (22).
The rhbA gene was first identified in a differential display reverse transcription-PCR screening analysis of transcripts upregulated when A. fumigatus was grown in cultures with human endothelial cells (26). This experimental design mimics the intimate contact between the pathogen and the endothelium during angioinvasion. The rhbA gene is the only Rheb homolog in the A. fumigatus genome, as confirmed by a search of The Institute for Genome Research genome database (http://www.tigr.org/tdb/e2k1/afu1/).
It has previously been demonstrated that nitrogen starvation in A. fumigatus is accompanied by increased expression of the Rheb homolog rhbA, suggesting that this gene plays a role in a nitrogen-regulated signaling pathway (24). Two main pathways regulating nitrogen metabolism have been identified in the fungi. Nitrogen catabolite repression (NCR) regulates the ability of an organism to discriminate between rich and poor nitrogen sources (1, 10). In the presence of a rich nitrogen source, such as ammonium, NCR prevents transcription of genes necessary for the utilization of poor nitrogen sources (34). In the presence of a poor nitrogen source, NCR is released. General control (GCN) or cross pathway control (CPC) is a means by which cells adapt to amino acid starvation, among other stresses (8, 16, 25). Many of the genes under the control of the GCN-CPC pathway encode amino acid and nucleotide biosynthetic enzymes (23).
In S. cerevisiae, gene regulation in response to nitrogen quality is mediated by a member of the target of rapamycin (TOR) family of PI-3 related kinases (29). In the presence of a rich nitrogen source, active TOR kinase mediates the cytoplasmic sequestration of Gln3p, a GATA-type transcription factor that is required for release of NCR (5). TOR kinases are specifically inhibited by the antibiotic rapamycin, and treatment of growing S. cerevisiae yeasts with rapamycin results in a rapid release of NCR and cell cycle arrest (7, 35). Whereas vegetative cultures of S. pombe are not affected by treatment with rapamycin, nitrogen-limited cells become sensitive to the drug (39), suggesting that nitrogen metabolism in S. pombe is likewise regulated by TOR kinase. TOR has also been implicated in the control of GCN-CPC-regulated genes in S. cerevisiae (37). TOR-dependent regulation of either NCR or GCN-CPC has yet to be determined in any filamentous fungus.
Although nitrogen metabolism in A. fumigatus is not well characterized, A. fumigatus mutants lacking areA, the ortholog of S. cerevisiae GLN3, are unable to grow in the presence of poor nitrogen sources, suggesting that the pathways governing NCR in both A. fumigatus and S. cerevisiae are closely related (15). Moreover, areA mutants display reduced virulence in vivo, suggesting that versatility in nitrogen utilization plays an important role in the pathogenesis of invasive pulmonary aspergillosis (IPA).
In order to increase our understanding of Rheb function in the fungi, we characterized an rhbA deletion mutant of A. fumigatus. In this paper, we describe construction of a strain of A. fumigatus that lacks the rhbA gene and demonstrate that there was reduced virulence of the mutant in a mouse model of IPA. Our results are consistent with a model in which the mutant is defective in nitrogen signaling and metabolic diversity is intimately linked to pathogenicity.
MATERIALS AND METHODS
Strains and media.
The strain of A. fumigatus used in this study was H237, a clinical isolate with no known nutritional markers (15). Liquid cultures of H237 were grown in YG (0.5% yeast extract, 2% glucose) or Aspergillus minimal medium (11). H237 was propagated on Aspergillus minimal medium plates with 10 mM ammonium tartrate as the nitrogen source. Alternative nitrogen sources were added at a concentration of 10 mM unless otherwise specified.
Deletion of rhbA.
An rhbA deletion construct was made by digesting a 2.5-kb rhbA genomic clone (GenBank accession number AF283573) in pPT7-Blue with BglII and removing a 1-kb fragment containing three of the four rhbA coding exons (3). A BamHI-XbaI fragment containing the hygromycin resistance cassette was removed from pMAD91 (40), blunted, and then ligated into the BglII-digested, filled-in rhbA genomic clone. Sequencing revealed that the hygromycin resistance cassette is in the inverse orientation to the rhbA gene. The rhbA deletion cassette was excised with BamHI and HindIII and was introduced into H237 by protoplast transformation as previously described (44). Hygromycin-resistant colonies were screened for homologous recombination by PCR by using primers that amplify the entire genomic clone. The 5′ primer sequence was 5′-GAGGCAAAAGGACGTCGCAAACGG −3′, and the 3′ primer sequence was 5′-GGTGCAAATCGCTTCCGCTGG-3′. Homologous recombination at the desired locus was confirmed by Southern blot analysis of EcoRI-digested genomic DNA probed with exon 1.
Complementation of the rhbA deletion.
A complementation construct was built by moving a phleomycin resistance cassette as a SacI/KpnI fragment from pBCPhleo (30) into the SacI/KpnI sites of the rhbA genomic clone in pSL1180 (Pharmacia Biotech). This construct was introduced into the ΔrhbA strain by protoplast transformation. To confirm single-locus integration, a Southern blot of EcoRI-digested genomic DNA from the reconstituted strain was probed with exon 1.
Radial growth rate determination.
Conidia from the isogenic set (wild type, ΔrhbA strain, and complemented mutant) were harvested in sterile water, filtered through Miracloth (Calbiochem), and counted with a hemacytometer. A 10-μl spot containing 1 × 104 conidia was placed in the center of a plate and incubated at 37°C. Colony diameters were measured at 24 and 48 h. The growth rate (in centimeters per hour) was determined by determining the difference in colony diameter between the 24- and 48-h time points and dividing by 24. All experiments were performed in triplicate, and significance was assessed by one-way analysis of variance (ANOVA). Pairwise analyses were conducted post hoc by using Tukey's honestly significant difference test.
Amino acid uptake analysis.
To determine the uptake pattern of arginine, 30 ml of minimal medium was inoculated with 5 × 106 conidia ml−1 and incubated at 37°C and 250 rpm for 7 h. The germlings were harvested on 0.45-μm-pore-size filters, washed with sterile water, and transferred to fresh minimal medium. The culture was then equilibrated in a 37°C water bath shaking at 75 rpm for 1 h prior to the addition of 0.5 μCi of l-[U-14C]arginine at a 5:12 ratio of labeled to unlabeled arginine. Aliquots (0.5 ml) were removed at 0, 1, 3, and 5 min after the addition of the labeled amino acid, and germlings were harvested on 0.45-μm-pore-size filters on a vacuum manifold. The filters were washed with 20 ml of distilled water, dried briefly, and transferred to scintillation vials containing 10 ml of Scintiverse BD. The number of counts per minute was corrected for nonspecific uptake by subtracting the counts obtained in parallel assays performed in the presence of 100 mM sodium azide. To measure dry weight, 30-ml cultures were harvested on preweighed 0.45-μm-pore-size filters and dried at 60°C until a constant weight was obtained. The uptake data (in picomoles of arginine per milligram [dry weight]) were plotted versus time, and the initial velocity of uptake was calculated by using linear regression analysis.
Virulence analysis.
Inocula were prepared from Aspergillus minimal medium plates containing 10 mM ammonium tartrate as the nitrogen source that were incubated at 37°C for 48 to 72 h. Conidia were harvested in saline, filtered through sterile Miracloth, and counted with a hemacytometer. To ensure that the inoculum was viable, a dilution of the inoculum was plated in triplicate, and the CFU were counted.
Female CF-1 outbred mice (Charles River) weighing 20 to 24 g were immunosuppressed by intraperitoneal injection of cyclophosphamide (150 mg/kg; Sigma Chemical Company, St. Louis, Mo.) on days −3, −1, 3, 6, and 9 and with a single dose of cortisone acetate (200 mg/kg; Sigma Chemical Company) given subcutaneously on day −1 (32). Tetracycline was provided in the water at a concentration of 500 μg ml−1 to prevent bacterial infection.
At zero time, mice were anesthetized with 3.5% isofluorane. Once a mouse was sedated, a 20-μl drop containing the desired inoculum was placed on its nares. Mice were monitored twice daily for 14 days postinoculation, and moribund mice were sacrificed. Cumulative mortality curves were generated, and statistical significance was assessed by Kruskall-Wallis analysis (ANOVA on Ranks). Pairwise analyses were performed post hoc by using Dunn's procedure. Animal studies were carried out in compliance with the University of Cincinnati Institutional Animal Care and Use Committee regulations.
Image analysis.
The average lesion area was determined by using lung sections from mice inoculated with the isogenic set that were sacrificed on day 7 postinoculation. Tissue was fixed in 10% buffered formalin and submitted to the University of Cincinnati Comparative Pathology core facility. Tissue sections were assigned anonymous code numbers (by D.L.H.S.) and were analyzed by an investigator (J.C.P.) blinded to the code. Images of silver-stained lesions were captured with an SGI computer (Silicone Graphics), and the lesion area was measured by using ScionImage image analysis software. The lesion border was defined as the perimeter of the region containing visible fungal mass (hyphae) but not including surrounding inflammation and necrosis. All lesions present in three consecutive 5-μm sections from each of three mice per strain were measured. The average lesion size was computed, and the data were analyzed by one-way ANOVA. Pairwise analysis was performed post hoc by using the least significant difference test. Fungal burden was determined by dividing the sum of all lesion areas in each section by the total area of the section.
RESULTS
rhbA gene is not essential in A. fumigatus.
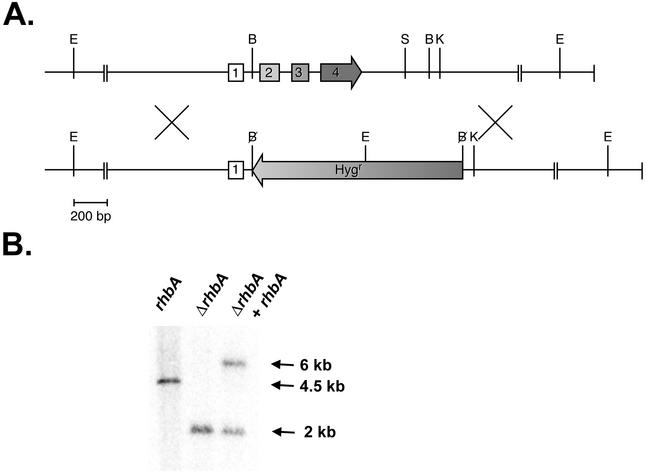
To determine if the rhbA gene is essential in A. fumigatus, a deletion construct that replaces three of the four rhbA coding exons with the hygromycin resistance gene was designed and introduced into A. fumigatus protoplasts (Fig. 1A). Hygromycin-resistant colonies were screened for homologous recombination by PCR (data not shown). One putative homologous recombinant was identified and confirmed by probing a Southern blot with exon 1, taking advantage of an EcoRI site present in the hygromycin resistance gene (Fig. 1B). The blot was also probed with a fragment of the hygromycin resistance gene and the pT7-Blue backbone to ensure that there was a single integration of the construct at the rhbA locus (data not shown).
FIG. 1.
(A) Genomic organization of the rhbA gene (top) and the rhbA deletion construct (bottom). The rhbA deletion construct was built by replacing the BglII fragment of the rhbA genomic clone with the hygromycin resistance gene. The restriction sites include EcoRI (E), BglII (B), SacI (S), and KpnI (K) sites. Numbers indicate the four coding exons. (B) Southern blot of genomic DNA from the wild type (rhbA), the ΔrhbA strain (ΔrhbA), and the reconstituted strain (ΔrhbA + rhbA) cut with EcoRI and probed with an rhbA exon 1 probe. Homologous recombination of the deletion construct at the rhbA locus results in the introduction of an EcoRI site that reduces the size of the hybridizing band. Introduction of the complementation construct at a nonhomologous locus results in maintenance of the ΔrhbA band and the appearance of a larger band corresponding to the integrated complementation construct.
To demonstrate that any phenotype seen in the ΔrhbA strain was due to deletion of rhbA alone, a complemented strain was constructed by introducing the complementation construct into protoplasts of the ΔrhbA strain and selecting for phleomycin-resistant colonies. Single-locus integration of the complementation construct was confirmed by Southern blot probing with exon 1 (Fig. 1B).
A. fumigatus rhbA is a virulence-related gene.
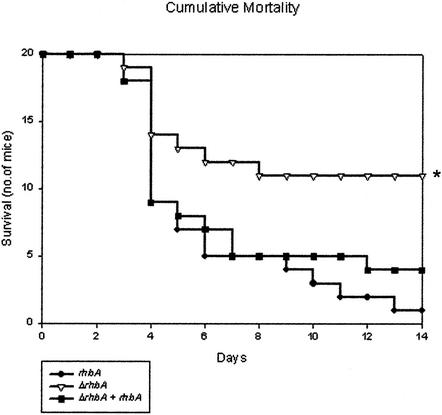
To determine the effect of rhbA deletion on the virulence of A. fumigatus, 5 × 104 conidia of the wild type, the rhbA deletion strain, or the reconstituted strain were used to inoculate immunosuppressed mice, and mortality was monitored for 14 days. The mice inoculated with the ΔrhbA strain survived significantly longer than the mice infected with the wild type or the reconstituted strain (P < 0.05) (Fig. 2). The same experiment repeated by using an inoculum containing 50-fold-fewer conidia again resulted in increased survival of mice inoculated with the ΔrhbA strain compared to the survival of the mice inoculated with either the wild type or the reconstituted strain (data not shown). Introduction of the complementation construct into the ΔrhbA strain restored wild-type virulence.
FIG. 2.
Cumulative mortality of immunosuppressed mice inoculated intranasally at zero time with 5 × 104 conidia of the wild type (rhbA), the ΔrhbA strain (ΔrhbA), or the reconstituted strain (ΔrhbA + rhbA). The asterisk indicates that the P value is <0.05.
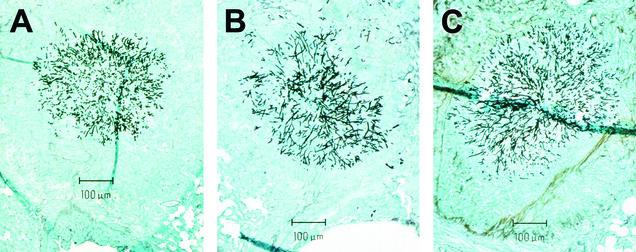
In a separate experiment, mice inoculated with each strain of the isogenic set were sacrificed on day 7 postinoculation, and the area of individual pulmonary lesions was measured in silver-stained sections by quantitative image analysis. The resulting measurements were used to calculate the average lesion area and the fungal burden for each strain of the isogenic set. The lesion area in mice inoculated with the ΔrhbA strain was 496 ± 82 μm2 (n = 48), compared with values for mice inoculated with the wild type of 1,921 ± 457 μm2 (n = 8) and values for mice inoculated with the reconstituted strain of 1,564 ± 311 μm2 (n = 13) (mean ± standard error of the mean; n is the number of lesions in a section) (P < 0.001). The lesions seen in mice inoculated with either the wild type or the complemented strain were few and large, whereas those seen in mice inoculated with the ΔrhbA strain were numerous and small. Since the number of lesions was greater in the ΔrhbA strain-inoculated mouse sections than in either the wild-type or complemented strain-inoculated mouse sections, the percentage of total sectional area that contained lesions was calculated as a measure of fungal burden. The percentage of lesion area in the ΔrhbA strain-inoculated mouse sections was less than that in sections from mice inoculated with the wild-type or complemented strain, but the difference was not significant due to high variation between mice (data not shown). Although there was a difference in average lesion area between strains, all strains had the ability to invade surrounding tissue (Fig. 3). Whereas rhbA is not required for invasive growth, the in vivo growth potential of the ΔrhbA strain is reduced. Taken together, these data support the hypothesis that rhbA plays a role in the pathogenesis of IPA.
FIG. 3.
Similar-size lesions from lung sections of mice inoculated with the isogenic set. (A) Wild type; (B) ΔrhbA mutant; (C) reconstituted strain. The sections were stained with silver stain.
rhbA deletion mutant exhibits a reduced growth rate on poor nitrogen sources.
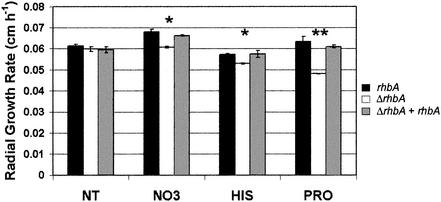
To determine if the reduced growth potential seen in mouse lungs was limited to in vivo growth, the in vitro growth rates of the isogenic set were measured on various solid media. The rhbA deletion mutant exhibited a significantly reduced growth rate on minimal medium containing the poor nitrogen sources proline, histidine, and nitrate compared to the growth rate of either the wild type or the reconstituted deletion strain (Fig. 4). On minimal medium containing the rich nitrogen source ammonium, however, the ΔrhbA strain grew at a rate similar to the growth rates of the wild type and the reconstituted mutant. Similarly, the ΔrhbA strain exhibited the wild-type growth rate, 0.082 ± 0.001 cm h−1, compared with a growth rate of 0.086 ± 0.001 cm h−1 for the mutant, on Sabouraud dextrose agar, a medium containing a pancreatic digest of casein and neopeptone as sources of nitrogen.
FIG. 4.
Radial growth rates of the wild type (rhbA) (closed bar), the ΔrhbA strain (ΔrhbA) (open bar), and the reconstituted strain (ΔrhbA + rhbA) (gray bar) on Aspergillus minimal medium with various nitrogen sources at a concentration of 10 mM. NT, ammonium tartrate; NO3, sodium nitrate; HIS, histidine; PRO, proline. One asterisk indicates that the P value is <0.05; two asterisks indicate that the P value is <0.001.
Deletion of rhbA results in increased uptake of arginine that is nitrogen source dependent.
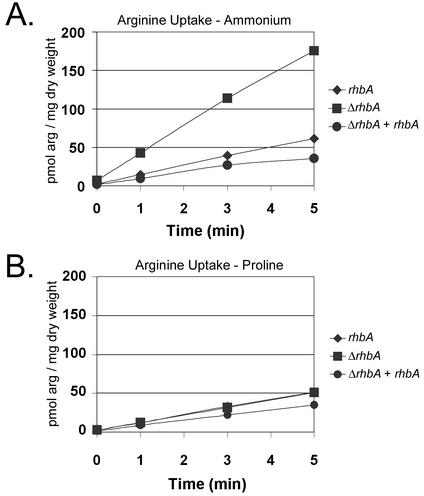
In S. cerevisiae, deletion of RHB1 results in hypersensitivity to the toxic arginine analog canavanine that is due to increased arginine uptake (36). It has previously been demonstrated that expression of A. fumigatus rhbA is able to complement the canavanine hypersensitivity of an S. cerevisiae rhb1 mutant, suggesting that these genes have overlapping functions (24). To determine if the ΔrhbA strain might exhibit a phenotype similar to that of the S. cerevisiae rhb1 mutant, we assessed the pattern of uptake of arginine in the isogenic set. In minimal medium containing ammonium, the slope of the line generated by plotting the amount of arginine taken up per milligram (dry weight) versus time was greatly increased in the ΔrhbA strain (Fig. 5A). Calculation of the initial velocities from the plots revealed that the ΔrhbA strain exhibited a nearly threefold-higher initial velocity of arginine uptake than either the wild type or the complemented strain (data not shown). The magnitude of this result is similar to that seen in the S. cerevisiae rhb1 mutant. In the presence of a poor nitrogen source (10 mM sodium nitrate or 10 mM proline), however, the rates of arginine uptake were similar for all members of the isogenic set (Fig. 5B).
FIG. 5.
(A) l-[U- 14C]arginine uptake in germlings of the wild type (rhbA), the ΔrhbA strain (ΔrhbA), and the complemented strain (ΔrhbA + rhbA) after incubation for 1 h in fresh minimal medium with 10 mM ammonium tartrate as the nitrogen source. (B) l-[U-14C]arginine uptake in germlings of the wild type, the ΔrhbA strain, and the complemented strain after incubation for 1 h in fresh minimal medium with 10 mM proline as the nitrogen source. Each graph is representative of several experiments.
ΔrhbA strain undergoes asexual development in submerged culture that is nitrogen source dependent.
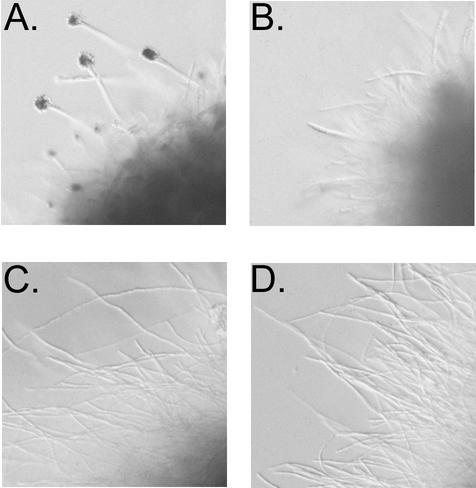
Asexual development in Aspergillus species is dependent on several factors, including the acquisition of competence and the presence of an air-water interface (2). Under nutrient deprivation conditions, asexual development can be induced in a submerged culture (31). After 18 to 24 h of growth in minimal medium containing 10 to 40 mM ammonium tartrate, gross observation of cultures of the ΔrhbA mutant revealed the production of a vibrant green pigment. Microscopic observation of the cultures revealed the presence of conidiophores and masses of dark green conidia (Fig. 6A). After a similar amount of time in rich medium (YG) or minimal medium with a poor-quality nitrogen source (10 mM sodium nitrate or 10 mM proline), the hyphae appeared normal with no pigment or signs of development (swollen vesicles or conidiophores) (Fig. 6C).
FIG. 6.
Photomicrographs of the wild type (B and D) and the ΔrhbA strain (A and C) taken from submerged cultures after 24 h of incubation in minimal medium containing 40 mM ammonium tartrate (A and B) or 40 mM sodium nitrate (C and D) as the nitrogen source.
ΔrhbA strain is hypersensitive to the TOR kinase inhibitor rapamycin.
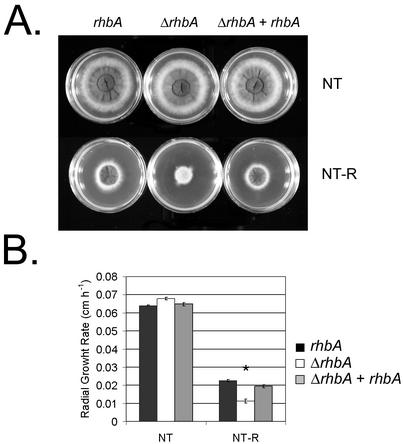
TOR kinase regulates the response to nitrogen availability in both S. cerevisiae and S. pombe (7, 19). To determine if the rhbA deletion phenotype might be due to aberrations in TOR kinase signaling, the sensitivity of the isogenic set to rapamycin was assessed (Fig. 7). The growth rate of the rhbA deletion strain was less than one-half that of the wild type or the reconstituted strain when the organisms were grown in the presence of rapamycin on ammonium-containing medium. The magnitude of growth inhibition seen on medium containing poor nitrogen sources was similar to that on ammonium, suggesting that rapamycin sensitivity cannot be ablated by the release of NCR (data not shown). This hypersensitivity to rapamycin is compatible with the explanation that loss of rhbA function may alter signaling through a TOR kinase pathway.
FIG. 7.
(A) Representative plates showing the wild type (rhbA), the ΔrhbA strain (ΔrhbA), and the reconstituted strain (ΔrhbA + rhbA) after 48 h of growth on Aspergillus minimal medium with (NT-R) and without (NT) 5 ng of rapamycin ml−1. (B) Radial growth rate of the wild type (black bar), the ΔrhbA strain (open bar), and the reconstituted strain (gray bar) on Aspergillus minimal medium with (NT-R) and without (NT) 5 ng of rapamycin ml−1. Ammonium tartrate was the nitrogen source. The asterisk indicates that the P value is <0.005.
DISCUSSION
Rheb family members have been identified in multiple eukaryotic species, but their precise function is unclear. In the mammalian system, Rheb has been reported to antagonize Ras signaling and to be affected by the activation state of the cAMP-protein kinase A signaling pathway (41, 43). In the two yeast systems in which Rheb has been studied, S. cerevisiae and S. pombe, functional Rheb is required for maintenance of wild-type sensitivity to the arginine analog canavanine (36, 42).
Immunosuppressed mice inoculated with the ΔrhbA strain survived significantly longer than immunosuppressed mice inoculated with the wild type, implicating rhbA in aspects of the pathogenic process. The reduced average lesion area seen in mouse lungs inoculated with the ΔrhbA strain suggests that the mutant has a reduced ability to grow in vivo. On solid medium containing poor-quality nitrogen, the mutant also exhibited a lower growth rate. The in vivo reduction in lesion area may result from a decreased ability of the organism to utilize available nutrients. The reduced virulence of the A. fumigatus areA mutant, which is severely impaired in utilization of poor nitrogen sources, also supports the hypothesis that nutritional versatility plays a role in pathogenesis (15).
The rhbA transcript was first identified in a differential display reverse transcription-PCR screening analysis for transcripts upregulated during coculture with human endothelial cells (26). It was proposed that transcripts obtained from this analysis were candidates for virulence-related genes, as intimate contact with endothelial cells is a prerequisite for disseminated disease. By demonstrating that rhbA contributes to the virulence of the organism, we validated the model in which the transcript was identified, prompting further study of the genes encoding other upregulated transcripts.
As an aid to understanding the function of rhbA in A. fumigatus, we are able to draw upon the data generated by Rheb deletion in the model yeasts S. cerevisiae and S. pombe. The S. cerevisiae rhb1 mutant exhibits increased uptake of arginine that is complemented by expression of both the S. pombe rhb1 gene and the A. fumigatus rhbA cDNA (24, 36). The S. pombe rhb1 mutant displays a phenotype similar to that seen when the organism is nitrogen starved (22).
Like the S. cerevisiae rhb1 mutant, the ΔrhbA strain exhibited increased uptake of arginine when it was grown in the presence of ammonium, corroborating the hypothesis that these two genes have overlapping functions. The uptake phenotype, however, was reversed by growth on poor-quality nitrogen. In the S. cerevisiae rhb1 mutant, transcription of arginine permease was reported to be unaltered compared to transcription in the wild type, suggesting that translation, transporter activity, or localization, rather than transcription, is the cause of the increased arginine uptake (36). It is possible that release of NCR by the presence of a poor nitrogen source exerts overriding regulation on the function of the arginine permease in A. fumigatus.
Deletion of rhb1 from S. pombe results in cell cycle arrest and accumulation of fnx1 and mei2 mRNA (22). The response of A. fumigatus to nitrogen starvation has not been explored; however, it has been reported that wild-type A. nidulans can be induced to undergo asexual sporulation in submerged culture in response to either carbon or nitrogen starvation (31). The asexual development in submerged culture seen in the ΔrhbA strain may indicate that the mutant is inappropriately initiating a starvation response similar to that seen in S. pombe rhb1 mutants. Growth in medium containing poor-quality nitrogen ablated the development aspect of the ΔrhbA phenotype. These data are consistent with a model in which release of NCR could override the starvation response pathway. Furthermore, the fungal lesions in the lungs of mice inoculated with the ΔrhbA strain showed no indication of asexual development, which is consistent with the hypothesis that in vivo growth involves a response to poor-quality nitrogen.
In both S. cerevisiae and S. pombe, the response to nitrogen limitation is regulated by the activity of TOR kinase. In S. cerevisiae, TOR kinase activity is decreased in response to the absence of rich nitrogen, allowing the release of NCR (7, 35). TOR also affects translation initiation, the stability of nutrient transporters, and the autophagic response (4, 18, 27). In contrast, TOR activity is required for cell cycle arrest in response to nitrogen starvation in S. pombe (39).
Altered sensitivity to rapamycin is a hallmark phenotype of defective TOR kinase signaling in S. cerevisiae, as evidenced by deletion or mutation of numerous TOR effectors (5, 12, 17, 38). Treatment of S. cerevisiae yeasts with rapamycin leads to rapid release of NCR, whereas the phenotypic consequences of treating the S. pombe yeasts with rapamycin are evident only under nitrogen-limited conditions (39). As with deletion of rhbA in A. fumigatus, deletion of S. cerevisiae RHB1 also results in rapamycin hypersensitivity (Panepinto, unpublished data), supporting the assertion that the ΔrhbA phenotype may result from an aberrance in TOR-dependent nutrient signaling.
The ability to adapt to changes in nutrient availability is an important attribute of pathogenic microorganisms. Preventing Histoplasma capsulatum from acquiring calcium by deletion of CBP1 results in an avirulent phenotype (28). Mutants of A. fumigatus that are auxotrophic for para-aminobenzoic acid are also avirulent (6), as are uracil auxotrophs of Candida albicans and A. fumigatus (13, 20). The decreased virulence of both the ΔrhbA and ΔareA mutants suggests that versatility in nitrogen utilization contributes to the growth of A. fumigatus in vivo. Understanding the signaling pathways regulating the metabolic versatility of A. fumigatus may uncover additional genes that are important for the pathogenesis of invasive aspergillosis.
Acknowledgments
We thank Jay Card for his assistance with photography and Sue Heffelfinger and Richard Monitone for their assistance with the image analysis.
This work was supported by NIH grants AI41119 (to J.C.R.) and AI48746 (to D.S.A.).
Editor: T. R. Kozel
REFERENCES
- 1.Arst, H. N., and D. J. Cove. 1973. Nitrogen metabolite repression in Aspergillus nidulans. Mol. Gen. Genet. 126:111-141. [DOI] [PubMed] [Google Scholar]
- 2.Axelrod, D., M. Gealt, and M. Pastushok. 1973. Gene control of developmental competence in Aspergillus nidulans. Dev. Biol. 34:9-15. [DOI] [PubMed] [Google Scholar]
- 3.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, B. A. Rapp, and D. L. Wheeler. 2002. GenBank. Nucleic Acids Res. 30:17-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berset, C., H. Trachsel, and M. Altmann. 1998. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95:4264-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertram, P. G., J. H. Choi, J. Carvalho, W. Ai, C. Zeng, T.-F. Chan, and X. F. S. Zheng. 2000. Tripartite regulation of Gln3p by TOR, Ure2p and phosphatases. J. Biol. Chem. 275:35727-35733. [DOI] [PubMed] [Google Scholar]
- 6.Brown, J., A. Aufauvre-Brown, J. Brown, J. M. Jennings, H. Arst, Jr., and D. Holden. 2000. Signature-tagged and directed mutagenesis identify PABA synthetase as essential for Aspergillus fumigatus pathogenicity. Mol. Microbiol. 36:1371-1380. [DOI] [PubMed] [Google Scholar]
- 7.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, D. J. DiComo, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13:3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carsiotis, M., R. F. Jones, and A. C. Wesseling. 1974. Cross-pathway regulation: histidine-mediated control of histidine, tryptophan and arginine biosynthetic enzymes in Neurospora crassa. J. Bacteriol. 119:893-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, G. J., M. S. Kinch, K. Rogers-Graham, S. M. Sebti, A. D. Hamilton, and C. J. Der. 1997. The ras-related protein rheb is farnesylated and antagonizes ras signaling and transformation. J. Biol. Chem. 272:10608-10615. [DOI] [PubMed] [Google Scholar]
- 10.Cooper, T. G. 1982. Nitrogen metabolism in Saccharomyces cerevisiae. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 11.Cove, D. J. 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113:51-56. [DOI] [PubMed] [Google Scholar]
- 12.Cutler, N. S., X. Pan, J. Heitman, and M. E. Cardenas. 2001. The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol. Biol. Cell 12:4103-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Enfert, C., M. Diaquin, A. Delit, N. Wushcer, J. P. Debeaupuis, M. Huerre, and J. P. Latge. 1996. Attenuated virulence of uridine-uracil auxotrophs of Aspergillus fumigatus. Infect. Immun. 64:4401-4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Souza, C., and J. Heitman. 2001. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 3:349-364. [DOI] [PubMed] [Google Scholar]
- 15.Hensel, M., H. N. Arnst, A. Aufauvre-Brown, and D. W. Holden. 1998. The role of the Aspergillus fumigatus areA gene in invasive pulmonary aspergillosis. Mol. Gen. Genet. 258:553-557. [DOI] [PubMed] [Google Scholar]
- 16.Hinnebusch, A. 1984. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc. Natl. Acad. Sci. USA 81:6442-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacinto, E., B. Guo, K. T. Arndt, T. Schmelzle, and M. N. Hall. 2001. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell 8:1017-1026. [DOI] [PubMed] [Google Scholar]
- 18.Kamada, Y., T. Funakoshi, S. Takahiro, K. Nagano, M. Ohsumi, and Y. Ohsumi. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150:1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawai, M., A. Nakashima, M. Ueno, T. Ushimaru, K. Aiba, H. Doi, and M. Uritani. 2001. Fission yeast tor1 function in response to various stresses including nitrogen starvation, high osmolarity and high temperature. Curr. Genet. 39:166-174. [DOI] [PubMed] [Google Scholar]
- 20.Kirsch, D. R., and R. R. Whitney. 1991. Pathogenicity of Candida albicans auxotrophic mutants in experimental infections. Infect. Immun. 59:3297-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latge, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mach, K. E., K. A. Furge, and C. F. Albright. 2000. Loss of Rhb1, a rheb-related GTPase in fission yeast, causes growth arrest with a terminal phenotype similar to that caused by nitrogen starvation. Genetics 155:611-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panepinto, J. C., B. G. Oliver, T. W. Amlung, D. S. Askew, and J. C. Rhodes. 2002. Expression of the Aspergillus fumigatus rheb homologue, rhbA, is induced by nitrogen starvation. Fungal Genet. Biol. 36:207-214. [DOI] [PubMed] [Google Scholar]
- 25.Piotrowska, M. 1980. Cross-pathway regulation of ornithine carbamoyltransferase synthesis in Aspergillus nidulans. J. Gen. Microbiol. 116:335-340. [Google Scholar]
- 26.Rhodes, J. C., B. G. Oliver, D. S. Askew, and T. W. Amlung. 2001. Identification of genes of Aspergillus fumigatus up-regulated during growth on endothelial cells. Med. Mycol. 39:253-260. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt, A., T. Beck, A. Koller, J. Kunz, and M. Hall. 1998. The TOR nutrient signaling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 17:6924-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sebghati, T. S., J. T. Engle, and W. E. Goldman. 2000. Intracellular parasitism by Histoplasma capsulatum: fungal virulence and calcium dependence. Science 290:1311-1312. [DOI] [PubMed] [Google Scholar]
- 29.Shamji, A. F. 2000. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 10:1574-1581. [DOI] [PubMed] [Google Scholar]
- 30.Silar, P. 1995. Two new easy to use vectors for transformations. Fungal Genet. News Lett. 42:73. [Google Scholar]
- 31.Skromne, I., O. Sanchez, and J. Aguirre. 1995. Starvation stress modulates the expression of the Aspergillus nidulans brlA regulatory gene. Microbiology 141:21-28. [DOI] [PubMed] [Google Scholar]
- 32.Smith, J. M., C. M. Tang, S. VanNoorden, and D. W. Holden. 1994. Virulence of Aspergillus fumigatus double mutants lacking restrictocin and an alkaline protease in a low-dose model of invasive pulmonary aspergillosis. Infect. Immun. 62:5247-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Talbot, N. J., H. R. K. McCafferty, M. Ma, K. More, and J. E. Hamer. 1997. Nitrogen starvation of the rice blast fungus Magnaporthe grisea may act as an environmental cue for disease symptom expression. Physiol. Mol. Plant Pathol. 50:179-195. [Google Scholar]
- 34.ter Schure, E. G., N. A. W. van Riel, and C. T. Verrips. 2000. The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 24:67-83. [DOI] [PubMed] [Google Scholar]
- 35.Thomas, G., and M. N. Hall. 1997. TOR signaling and control of cell growth. Curr. Opin. Cell Biol. 9:782-787. [DOI] [PubMed] [Google Scholar]
- 36.Urano, J., A. P. Tabancay, W. Yang, and F. Tamanoi. 2000. The Saccharomyces cerevisiae rheb G-protein is involved in regulating canavanine resistance and arginine uptake. J. Biol. Chem. 275:11198-11206. [DOI] [PubMed] [Google Scholar]
- 37.Valenzuela, L., C. Aranda, and A. Gonzalez. 2001. TOR modulates GCN4-dependent expression of genes turned on by nitrogen limitation. J. Bacteriol. 183:2331-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Merwe, G. K., T. G. Cooper, and H. J. J. van Vuuren. 2001. Ammonia regulates VID30 expression and Vid30p function shifts nitrogen metabolism toward glutamate formation especially when Saccharomyces cerevisiae is grown in low concentrations of ammonium. J. Biol. Chem. 276:28659-28666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weisman, R., M. Choder, and Y. Koltin. 1997. Rapamycin specifically interferes with the developmental response of fission yeast to starvation. J. Bacteriol. 179:6325-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woods, J. P., E. L. Heinecke, and W. E. Goldman. 1998. Electrotransformation and expression of bacterial genes encoding hygromycin phosphotransferase and beta-galactosidase in the pathogenic fungus Histoplasma capsulatum. Infect. Immun. 66:1697-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamagata, K., L. K. Sanders, W. E. Kaufmann, W. Yee, C. A. Barnes, D. Nathans, and P. F. Worley. 1994. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel ras-related protein. J. Biol. Chem. 269:16333-16339. [PubMed] [Google Scholar]
- 42.Yang, W., and F. Tamanoi. 2000. Protein farnesylation is critical for maintaining normal cell morphology and canavanine resistance in Schizosaccharomyces pombe. J. Biol. Chem. 275:429-438. [DOI] [PubMed] [Google Scholar]
- 43.Yee, W. M., and P. F. Worley. 1997. Rheb interacts with Raf-1 kinase and may function to integrate growth factor- and protein kinase A-dependent signals. Mol. Cell. Biol. 17:921-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yelton, M. M., J. E. Hamer, and W. E. Timberlake. 1984. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc. Natl. Acad. Sci. USA 81:1470-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]