Abstract
The Tar protein of Escherichia coli is unique among known bacterial chemoreceptors in that it generates additive responses to two very disparate ligands, aspartate and maltose. Aspartate binds directly to the periplasmic (extracytoplasmic) domain of Tar. Maltose first binds to maltose-binding protein (MBP). MBP then assumes a closed conformation in which it can interact with the periplasmic domain of Tar. MBP residues critical for binding Tar were identified in a screen of mutations that cause specific defects in maltose chemotaxis. Mutations were introduced into a plasmid-borne malE gene that encodes a mutant form of MBP in which two engineered Cys residues spontaneously generate a disulfide bond in the oxidizing environment of the periplasmic space. This disulfide covalently crosslinks the NH3-terminal and COOH-terminal domains of MBP and locks the protein into a closed conformation. Double-Cys MBP confers a dominant-negative phenotype for maltose taxis, and we reasoned that third mutations that relieve this negative dominance probably alter residues that are important for the initial interaction of MBP with Tar. The published three-dimensional structures of MBP and the periplasmic domain of E. coli Tar were docked in a computer simulation that juxtaposed the residues in MBP identified in this way with residues in Tar that have been implicated in maltose taxis. The resulting model of the MBP-Tar complex exhibits good complementarity between the surfaces of the two proteins and supports the idea that aspartate and MBP may each initiate an attractant signal through Tar by inducing similar conformational changes in the chemoreceptor.
The homodimeric chemoreceptors of enteric bacteria constitute a well-established system for studying transmembrane-signaling by cell-surface receptors (1). The Tar receptor of Escherichia coli has the unusual property of generating attractant responses to two very different ligands, the acidic amino acid aspartate and the disaccharide maltose (2). Aspartate binds near the membrane-distal apex of the elongated periplasmic domain of the receptor (3). Maltose must first bind to maltose-binding protein (MBP) (4) in the cleft between its NH3-terminal and COOH-terminal globular domains, which are joined by a flexible hinge (5). Maltose stabilizes a closed form of MBP, in which regions required for interaction with Tar (6, 7) are predicted (5) to assume the correct spatial relationship to interact simultaneously with Tar. In the open form of MBP (8), which predominates in the absence of ligand, these regions are in the wrong spatial relationship to interact simultaneously with Tar.
E. coli cells can sense maltose in the presence of saturating amounts of aspartate and vice versa, and the responses to aspartate and maltose are at least partially additive (9). Aspartate binds at one of two rotationally symmetric sites at the subunit interface of the Tar homodimer and interacts with different residues in each subunit (3). Binding of aspartate to Tar is strongly negatively cooperative (10), and E. coli Tar binds only one aspartate at physiological concentrations of the amino acid. Aspartate-induced signals are processed asymmetrically within the Tar dimer (11), and in vitro (12) and in vivo (13, 14) studies have shown that aspartate can signal through a dimer lacking one cytoplasmic domain.
In contrast to the situation with aspartate, the structure of the MBP–Tar complex has not been determined experimentally. One of the difficulties has been the low affinity of Tar for MBP, with a Kd for the interaction estimated from in vivo measurements at 250 μM MBP (15). In a cell fully induced with maltose, the periplasmic concentration of MBP is around 1 mM. Therefore, this low affinity does not impede maltose taxis. It has, however, hampered biochemical analysis of the MBP–Tar interaction.
Stoddard and Koshland (16) modeled the MBP–Tar complex by a binary-docking method. Residues altered by mutations known to disrupt maltose taxis (6) were used to select two octapeptides for the procedure. One peptide is in the NH3-terminal domain of MBP and the other is in the COOH-terminal domain. An energy-minimization program was used to dock the peptides independently to the periplasmic domain of E. coli Tar, which was homology modeled from the published structure of Salmonella Tar (3) by reference to the predicted amino acid sequences of Salmonella (17) and E. coli Tar (18). When the structure of the closed form of E. coli MBP (5) was superimposed on the docked peptides, they occupied approximately their proper position with the protein.
New data from additional mutagenesis studies of MBP (7) and Tar (19, 20) suggested that it was time to reexamine the docking problem. In particular, the previous model predicted that MBP should interact with regions of Tar in which extensive mutagenesis (19) has not identified residue substitutions that selectively disrupt maltose taxis. Furthermore, the actual crystal structure of E. coli Tar has become available (21). We describe here a genetic approach to determining which residues in MBP are specifically involved in binding to Tar. Using that information together with genetic data that suggest which residues in Tar are required for signaling in response to MBP, we have performed a computer-directed docking of MBP to Tar. The structural model of the MBP–Tar complex obtained in this way differs significantly from the complex proposed by Stoddard and Koshland. It provides a good conformational fit of the two proteins, and it is consistent with results from mutagenic analyses. Finally, the model provides a plausible mechanism for MBP-induced signaling by means of Tar that is consistent with and similar to the intrasubunit mechanism proposed to explain transmembrane signaling initiated by aspartate (22).
MATERIALS AND METHODS
Strains and Plasmids.
Strain YZ8 (7) is a derivative of strain MC4100 that carries an internal nonpolar deletion of the malE gene. It is constitutive for expression of genes of the mal regulon because it contains the malTc-1 mutation. Strain YZ11 (23) is like YZ8 except that it contains a full-length malE gene with a signal-sequence mutation that decreases the periplasmic content of MBP to 23% of normal (15). The plasmid-borne malE gene expressing double-Cys MBP (DC-MBP) (23) was modified by primer-directed site-specific mutagenesis to introduce residue substitutions that cause defects in the maltose-taxis activities of MBP. The plasmid encodes ampicillin resistance.
Assays for DC-MBP Crosslinking and Chemotaxis.
Cells were grown at 35°C in MMA minimal medium (24) with 0.4% glycerol/0.1% maltose/0.2% casamino acids (Difco)/1 μg/ml thiamine/25 μg/ml ampicillin. SDS/PAGE under nonreducing conditions (23) was performed on osmotic shock fluid prepared from overnight cultures of strain YZ8 containing plasmids expressing the various mutant forms of DC-MBP. Capillary assays with exponential-phase cells (OD578 nm of 0.5–0.7) of strain YZ11 containing the mutant malE plasmids were done according to the method of Adler (25) as described previously (23).
Nomenclature.
Residue substitutions are denoted with the single-letter amino acid code and the residue number. Thus, G69C indicates that Gly is replaced by Cys at residue 69. The α-helices of MBP (5) are designated with Roman numerals, and the long α-helices of Tar are designated with Arabic numerals, numbering from the NH3-terminus to COOH-terminus of each protein.
Computerized Docking Simulation.
The most recent high-resolution structures for E. coli Tar (21) and MBP (26) were initially docked manually by using the spock software (27) to align residues that were candidates for interaction based on the genetic analysis described here. Steric overlap was held to a minimum during this initial phase of the docking. Specifically, we aligned MBP and Tar such that residues Thr-53 and Asp-55 of MBP were brought as close as possible to residues Tyr-143′ and Asn-145′ in subunit T′ of Tar, residue Arg-367 of MBP was brought as close as possible to residues Tyr-143 and Asn-145 in subunit T of Tar, and the apical loop between helices 1 and 2 of subunit T of Tar fit into the substrate-binding cleft of MBP. Visual inspection of the manually docked model revealed that the steric collisions still present could be eliminated by reorientation of the side chains. To allow the side chains to repack, we refined the model using the molecular dynamics facilities of the amber software (28). Molecular dynamics calculations were performed by moving Tar about 2 Å away from MBP to eliminate all overlap and by using amber to simulate the docking. All simulations were carried out by using a distance-dependent dielectric field at an initial temperature of 300 K with a time step of 0.001 ps and by using the default force field prescribed by amber. During the molecular dynamics simulations, the backbone coordinates of MBP were fixed (i.e., held at their initial values), whereas the side chains were free to move. For Tar, both the backbone and side-chain coordinates were allowed to move, but restraints were placed on Tar so that backbone dihedral angles would not deviate by more than 0.1 degree without an energy penalty. Also, the (i, i + 4) O-N hydrogen-bond distances were not allowed to deviate by more than 0.1 Å without an energy penalty. These restraints permit the flexible apical loops of Tar to move while preserving the helical nature of the core of Tar. Finally, a “docking” pseudo-NMR restraint was applied to pull residues Tyr-143′ and Asn-145′ in subunit T′ of Tar toward residues Thr-53 and Asp-55 of MBP and to pull residues Tyr-143 and Asn-145 in subunit T of Tar toward residue Arg-367 of MBP. The docking restraints were relative and were designed to decrease the distance between the interacting residues by 2 Å over the course of the simulation, which lasted 10,000 iterations, or 10 ps. The pseudo-NMR docking restraints were gradually increased in strength over the first 2.5 ps and then released over the next 2.5 ps. For the last 5.0 ps, the simulation was run without docking restraints but with the backbone-position constraints for MBP and with the torsion-angle and distance restraints for Tar still in effect. The final model is an energy-minimized form of the result of this simulation. Further simulations were run on the docked complex with all constraints and restraints released, allowing both MBP and Tar to move, but no significant differences from the original model were observed.
RESULTS
Determination of MBP Residues Directly Involved in Binding to Tar.
In DC-MBP a disulfide bond forms spontaneously in the oxidizing environment of the periplasmic space (23). This crosslink locks the protein in a closed conformation, and DC-MBP confers a dominant-negative phenotype for maltose taxis when it is overexpressed in wild-type cells. Negative dominance is more pronounced in strain YZ11 (23), in which a defective leader peptide decreases the level of wild-type MBP to 23% of the normal level (15). We infer that crosslinked DC-MBP competes with wild-type MBP for binding to Tar. Substitutions at third positions in DC-MBP should relieve negative dominance if they disrupt binding of DC-MBP to Tar. Because DC-MBP remains in a closed form even without maltose, effects of the third substitutions on maltose binding by MBP do not complicate the analysis.
DC-MBP containing third substitutions was present at about the same level as the original DC-MBP in strain YZ8 (ΔmalE) and exhibited the same extent (≈80–90%) of crosslinking (Fig. 1). Maltose taxis was quantified in capillary assays by using strain YZ11 harboring plasmids expressing the different triple-mutant proteins. Substitutions T53I and D55N in the NH3-terminal domain and R367A in the COOH-terminal domain of MBP completely relieved dominance (Fig. 2). The other substitutions tested provided partial or no relief. We conclude that T53I, D55N, and R367A specifically impair the ability of DC-MBP to bind to Tar.
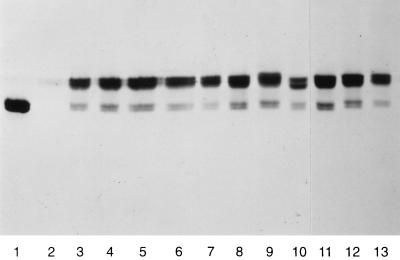
Figure 1.
Immunoblot analysis of osmotic shock fluid from strain YZ8 (ΔmalE; 7) expressing plasmid-encoded mutant DC-MBP. Samples for SDS/PAGE were run under nonreducing conditions with samples from equal numbers of cells expressing: (1) wild-type MBP; (2) no MBP (vector plasmid only); (3) DC-MBP; 4–13, DC-MBP with (4) E38K; (5) D41N; (6) K46Q; (7) T53I; (8) D55N; (9) Y210S; (10) R344A; (11) R354; (12) E359K; (13) R367A. The two upper bands represent crosslinked DC-MBP; the lower two bands represent noncrosslinked proteins. The doublets in both the upper and lower bands are also seen with wild-type MBP (lane 1), which contains no Cys residues. DC-MBP forms a single band during SDS/PAGE run under reducing conditions (23). We conclude that the upper and lower doublets represent two differently oxidized forms of crosslinked and noncrosslinked MBP, respectively.
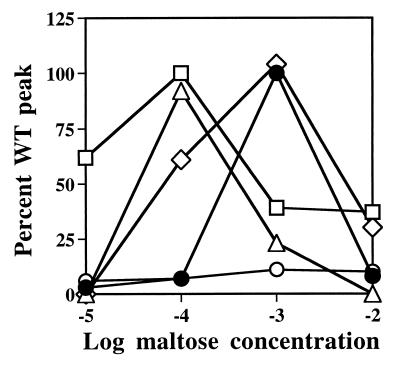
Figure 2.
Capillary assays with cells of strain YZ11 (23) expressing plasmid-encoded mutant forms of DC-MBP. This strain contains wild-type MBP at 23% of the induced level from a single chromosomal gene not containing a mutation affecting the pre-MBP leader peptide (15). The ordinate values represent the accumulations of cells in capillaries containing the concentrations of maltose indicated on the abscissa. These responses were normalized to the response of each strain to 1 mM l-aspartate. This concentration gives the peak accumulation to this alternative attractant sensed by Tar. The response of strain YZ11 containing the vector plasmid pBR322, and therefore no competing DC-MBP, was set to 100%, which corresponds to about 40,000 cells per capillary. Symbols: (•), vector plasmid; (○) DC-MBP; (▵), T53I DC-MBP; (□) D55N DC-MBP; (◊), R367A DC-MBP. The shift in the peak from 1 mM to 0.1 mM maltose with T53I and D55N DC-MBP presumably occurs because these proteins compete with wild-type MBP in maltose transport. Inhibition of maltose uptake increases periplasmic maltose concentrations and heightens sensitivity in maltose taxis (4).
Selection of Tar Residues for Use in the Docking Simulation.
Complementation analysis of tar mutations that selectively disrupt maltose taxis indicates that MBP binds across the Tar dimer and interacts with different residues in each subunit (20). Further analysis of the effects of different substitutions on maltose taxis has shown that residue Tyr-143 must be intact in the subunit of Tar that is primarily responsible for signaling in response to binding of MBP (29). Therefore, we chose Tyr-143 and its near neighbor Asn-145, which is also essential for the response to maltose, as the Tar residues to use in the docking simulation with MBP.
Computer-Assisted Docking of MBP and Tar.
The spock program (26) was used to dock E. coli MBP in its closed conformation (27) with the ligand-free form of the periplasmic domain of E. coli Tar (21). Candidate residues in each protein were brought close to each other in various pairwise combinations while endeavoring to avoid steric overlap. Inspection of the docked complex generated in this way indicated the surface complementarity between the two proteins was excellent. The manually docked complex was then refined by using the molecular dynamics and energy-minimization capabilities of the amber software (28).
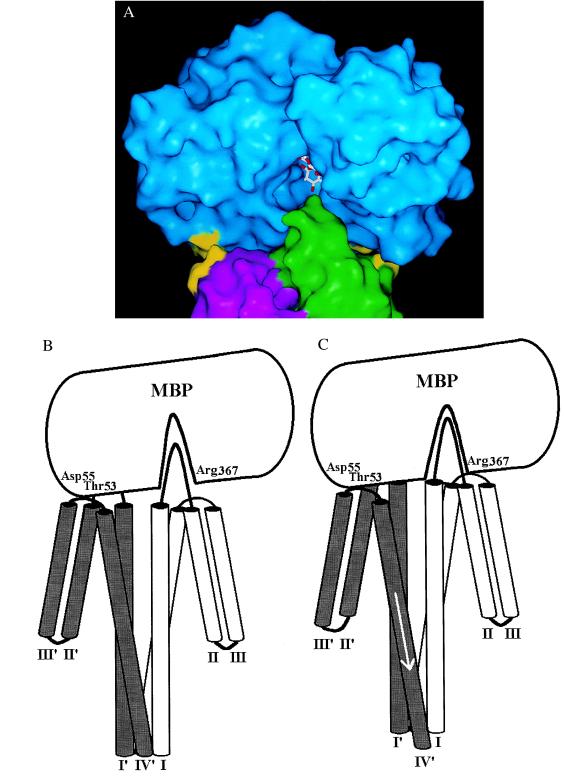
The computer-generated model of the complex is shown in Fig. 3A. It differs substantially from the Stoddard and Koshland model, in which MBP binds asymmetrically to Tar with its long axis (between the NH3-terminal and COOH-terminal domains) tilted at about 45° to the vertical axis of the Tar dimer. Our modeled complex exhibits pseudosymmetry. MBP sits at the apical tip of the Tar dimer with its long axis roughly perpendicular to the vertical axis of the Tar dimer. A more highly magnified view (Fig. 3B) reveals that the antiparallel COOH-terminal helices XIII and XIV of MBP pack against the turn between helices 3 and 4 of Tar in subunit T. Helix XIII lies in a groove between this turn and the apical loop of subunit T, and helix II in the NH3-terminal domain of MBP lies in the equivalent furrow between the corresponding turn and apical loop of subunit T′. The apical loop of subunit T extends into the substrate-binding cleft of MBP but does not collide with bound maltose. The apical loop of subunit T′ does not extend as far into the cleft. The disulfide bond of DC-MBP fits above (Fig. 3B) and between (Fig. 3C) the apical loops of T and T′. Residues in both proteins altered by mutations causing impaired maltose taxis are in close proximity, as is seen particularly well in the apical view of the complex in which portions of MBP not involved in contact to Tar have been “clipped” (Fig. 3C).
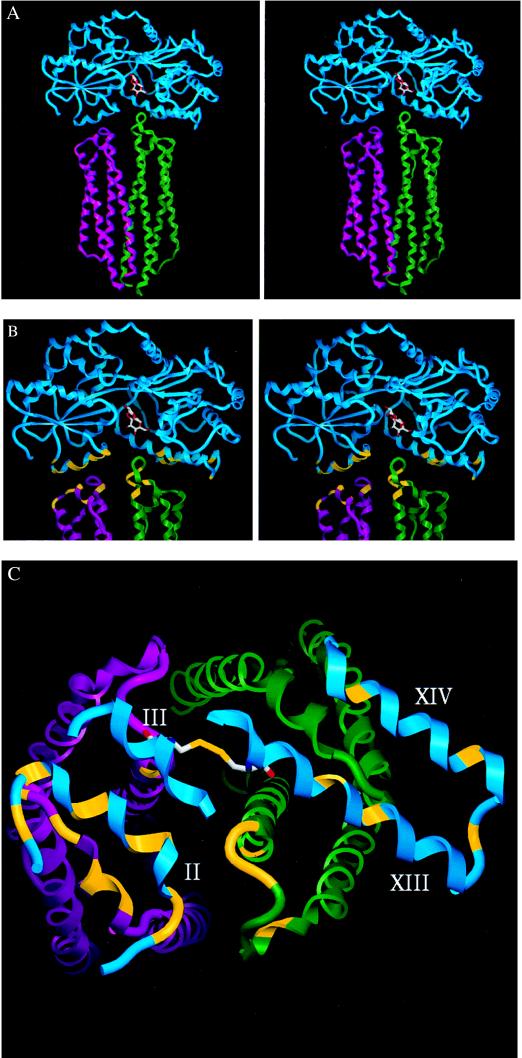
Figure 3.
MBP–Tar complex with the α-carbon-backbone structure of proteins. MBP is cyan, Tar subunit T is green, and subunit T′ is magenta. (A) Stereo view of our model for the docking of MBP (28) to the periplasmic domain of E. coli Tar (21). Maltose is shown in the binding cleft. (B) Stereo closeup of the complex. A modeled disulfide bond is shown between Cys-69 and Cys-337 (23). Sites in MBP (7) and Tar (19) at which mutations interfere with maltose taxis are indicated in yellow. For MBP: D41N, E45K, K46N/Q, Q49K/R, V50A/F, T53I/A, D55N in the N-terminal domain (Left) and A342D/P, T345I/P, R354A, E359K, and R367A/C in the C-terminal domain. Tar substitutions are distributed according to their complementation pattern (20): R73K, M75K/R, M76K/R, D77H, and S83R in subunit T and M75′K/R, Y143′S, N145′K, G147′R, Y149′S, and F150′S in subunit T′. (C) Apical view of the complex, looking down the vertical axis of the Tar dimer. MBP has been clipped to leave only regions important for interaction with Tar. Residues and disulfide bond as in B.
Evaluation of the Docking Model.
Salmonella Tar does not interact productively with MBP (30, 31). When we tried to dock E. coli MBP onto Salmonella Tar in the orientation shown in Fig. 3, the longer apical loops of the Salmonella receptor dimer did not fit into the substrate-binding cleft of MBP. Although the apical loops are predicted to be quite flexible, this mismatch could create steric hindrance that would prevent MBP from binding, which might explain the inability of Salmonella Tar to mediate maltose taxis.
The surface of MBP around Tyr-341 has a positive charge that is the highest on the entire surface of the binding protein (data not shown). Substitutions in the apical loop that impair maltose taxis (M75K/R, M76K/R, D77H, and S83R) replace neutral or negatively charged residues with positively charged ones (19). Positively charged residues introduced into the region of Tar in close proximity to Tyr-341 of MBP might be expected to destabilize the complex. The model predicts that such mutationally introduced positive charge should be more disruptive in the apical loop of subunit T than of subunit T′ because the former is closer to Tyr-341. In the Stoddard and Koshland model, the apical loops of Tar do not enter the substrate-binding cleft of MBP, so that the effect of introduced positive charge in this region of Tar is not anticipated. Also, none of our ten random mutations in tar that cause specific defects in maltose taxis (19) alter residues in helices 2 or 3 of Tar (5, 21), which in the Stoddard and Koshland model contact the COOH-terminal domain of MBP extensively.
The complementation pattern of maltose-defective tar mutations (20) strongly supports our model. When the M76K/R, D77H, or S83R receptor is coexpressed with either the Y143S or N145K receptor, the heterodimers that form mediate good maltose taxis, although none of the homodimers do. We infer that the crucial interactions of Met-76, Asp-77, and Ser-83 of Tar with MBP and of Tyr-143 and Asn-145 of Tar with MBP involve opposing subunits of the Tar dimer. Since predictions based on surface charge suggest that M76K/R, D77H, and S83R should be tolerated less well in subunit T (Fig. 4), it follows that Y143S and N145K should be tolerated less well in subunit T′.
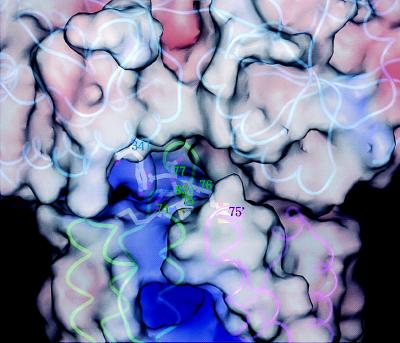
Figure 4.
Closeup view of the Tar apical loops and the MBP substrate-binding cleft. The image has been rotated 180° about the long axis of the Tar dimer relative to the image in Fig. 3 A and B because this vantage point gives a better view of the relevant residues. Polypeptide backbones are shown in cyan (MBP), green (subunit T of Tar), and magenta (subunit T′ of Tar). Molecular surfaces are color-coded by electrostatic potential. Side chains are shown for key residues. Positive potential in MBP centers on Tyr-341, which is surrounded by Met-74, Met-75, Met-76, Asp-77, and Ser-83 in T and Met-75′ in T′. Additional positive charge in this region would be expected to destabilize the complex. No substitutions at Met-74 that specifically disrupt maltose taxis have been found (19), perhaps because a positively charged residue next to Arg-73 would interfere with folding and render Tar unstable. Note the intensely positive aspartate-binding site (middle bottom).
DISCUSSION
In our model of the MBP–Tar docked complex, the surface of the NH3-terminal domain of MBP containing Thr-53 and Asp-55 is in contact with the surface occupied by Tyr-143′ and Asn-145′ in the turn between helices 3′ and 4′ of Tar (Fig. 5A). The surface of the COOH-terminal domain of MBP containing Arg-367 is less intimately associated with Tyr-143 and Asn-145 in subunit T of Tar (Fig. 5 A and B). The fit between the COOH-terminal domain of MBP and subunit T of Tar may be tighter in the true docked complex, since the movement of the polypeptide backbones was significantly restricted during the simulation. This restraint limits the extent to which the proteins can accommodate their surfaces to one another during the docking protocol.
Figure 5.
MBP signaling. (A) The interacting protein surfaces in the complex. Residues Thr-53, Asp-55, and Arg-367 of MBP and Tyr-143, Asn-145, Tyr-143′, and Asn-145′ of Tar are indicated in yellow. Note the predicted close contact of Thr-53 and Asp-55 of MBP with Tyr-143′ and Asn-145′ of Tar. (B) Cartoon of the docked complex shown in Fig. 5A. (C) Cartoon in Fig. 5B modified to allow close packing between Arg-367 of MBP and the turn between helices 3 and 4 of subunit T. This snug fit requires that Thr-53 and Asp-55 of MBP displace Tyr-143′ and Asn-145′, and thus helices 3′ and 4′ of T′. We propose that this displacement may be a downward movement of helix 4′ along its axis to initiate transmembrane signaling, as shown.
If the fit between helices XIII and XIV of MBP and the turn between helices 3 and 4 of subunit T of Tar is in fact more snug in the actual complex, the surface of MBP containing residues Thr-53 and Asp-55 may collide with residues Tyr-143′ and Asn-145′ in the turn between helices 3′ and 4′. This interaction could push helix 4′ downward along its helical axis (Fig. 5C) to initiate a transmembrane signal. Various crystallographic and biophysical studies have suggested that when aspartate binds, its contacts with residues Tyr-149, Gln-152, and Thr-154 in helix 4 push that helix downward by about 1.6 Å (22). Thus, a small molecule like aspartate binding at the subunit interface of the Tar dimer and a relatively large protein like MBP (40 kDa) binding across the apex of the Tar dimer may initiate transmembrane signaling by causing a similar conformational change. In this regard it is relevant to note that a similar displacement of helix 4 of the Trg chemoreceptor has been reported to occur when Trg binds its ligand ribose-binding protein (32), although no model for the binding protein/Trg complex has been proposed.
Additive responses to aspartate and maltose (9) can potentially be explained by the ability of aspartate and maltose to signal through opposing subunits of the Tar dimer (28). We know from that study that Ser, and presumably other amino acids (19, 20), cannot substitute for Tyr-143 in the subunit through which MBP signals. When MBP binds as shown in Fig. 5, we predict that interaction between Thr-53 and Asp-55 of MBP with Tyr-143′ and Asn-145′ of Tar will initiate a signal through helix 4′ of subunit T′ of Tar. So, aspartate should be able to bind in the “opposite” orientation (using the terminology of ref. 28) so as to interact with Tyr-149, Gln-152, and Thr-154. The signal would be transmitted through helix 4 of subunit T. Thus, the E. coli Tar dimer may be able to undergo a similar and simultaneous conformational change in response to aspartate in one subunit and to maltose-bound MBP in the other subunit.
This paper is dedicated to Winfried Boos on the occasion of his 60th birthday. We thank J. Sacchettini for the original model of the MBP–Tar complex and for critically reading the manuscript. Financial support was provided by National Institutes of Health grant GM39736 to M.D.M.
ABBREVIATIONS
- MBP
maltose-binding protein
- DC-MBP (double-Cys MBP)
MBP containing engineered Cys residues at position 69 (G69C) in the NH3-terminal domain and position 337 (S337C) in the COOH-terminal domain
References
- 1.Stock J B, Surette M G. In: Escherichia coli and Salmonella. Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1103–1129. [Google Scholar]
- 2.Springer M S, Goy M F, Adler J. Proc Natl Acad Sci USA. 1977;74:3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milburn M V, Privé G G, Milligan D L, Scott W G, Jancarik J, Koshland D E, Jr, Kim S-H. Science. 1991;254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 4.Hazelbauer G L. J Bacteriol. 1975;122:206–214. doi: 10.1128/jb.122.1.206-214.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spurlino J C, Lu G Y, Quiocho F A. J Biol Chem. 1991;266:5202–5219. doi: 10.2210/pdb1mbp/pdb. [DOI] [PubMed] [Google Scholar]
- 6.Kossmann M, Wolff C, Manson M D. J Bacteriol. 1988;170:4516–4521. doi: 10.1128/jb.170.10.4516-4521.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Conway C, Rosato M, Suh Y, Manson M D. J Biol Chem. 1992;267:22813–22830. [PubMed] [Google Scholar]
- 8.Sharff A J, Rodseth L E, Spurlino J C, Quiocho F A. Biochemistry. 1992;31:10657–10663. doi: 10.1021/bi00159a003. [DOI] [PubMed] [Google Scholar]
- 9.Mowbray S L, Koshland D E., Jr Cell. 1987;50:171–180. doi: 10.1016/0092-8674(87)90213-3. [DOI] [PubMed] [Google Scholar]
- 10.Biemann H P, Koshland D E., Jr Biochemistry. 1994;33:629–634. doi: 10.1021/bi00169a002. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Park H, Inouye M. J Mol Biol. 1993;232:493–498. doi: 10.1006/jmbi.1993.1405. [DOI] [PubMed] [Google Scholar]
- 12.Milligan D L, Koshland D E., Jr Science. 1991;254:1651–1654. doi: 10.1126/science.1661030. [DOI] [PubMed] [Google Scholar]
- 13.Tatsuno I, Homma M, Oosawa K, Kawagishi I. Science. 1996;274:423–425. doi: 10.1126/science.274.5286.423. [DOI] [PubMed] [Google Scholar]
- 14.Gardina P J, Manson M D. Science. 1996;274:425–426. doi: 10.1126/science.274.5286.425. [DOI] [PubMed] [Google Scholar]
- 15.Manson M D, Boos W, Bassford P J, Jr, Rasmussen B A. J Biol Chem. 1985;260:9727–9733. [PubMed] [Google Scholar]
- 16.Stoddard B L, Koshland D E., Jr Nature (London) 1992;358:774–776. doi: 10.1038/358774a0. [DOI] [PubMed] [Google Scholar]
- 17.Russo A F, Koshland D E., Jr Science. 1983;220:1016–1020. doi: 10.1126/science.6302843. [DOI] [PubMed] [Google Scholar]
- 18.Krikos A, Mutoh N, Boyd A, Simon M I. Cell. 1983;33:615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- 19.Gardina P, Conway C, Kossmann M, Manson M. J Bacteriol. 1992;174:528–536. doi: 10.1128/jb.174.5.1528-1536.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardina P J, Bormans A F, Hawkins M A, Meeker J W, Manson M D. Mol Microbiol. 1997;23:1181–1191. doi: 10.1046/j.1365-2958.1997.3001661.x. [DOI] [PubMed] [Google Scholar]
- 21.Bowie J U, Pakula A A, Simon M I. Acta Crystallogr D. 1995;51:145–154. doi: 10.1107/S0907444994010498. [DOI] [PubMed] [Google Scholar]
- 22.Chervitz S A, Falke J J. Proc Natl Acad Sci USA. 1996;93:2545–2550. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Mannering D E, Davidson A L, Yao N, Manson M D. J Biol Chem. 1996;271:17881–17889. doi: 10.1074/jbc.271.30.17881. [DOI] [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. p. 432. [Google Scholar]
- 25.Adler J. J Gen Microbiol. 1973;74:77–91. doi: 10.1099/00221287-74-1-77. [DOI] [PubMed] [Google Scholar]
- 26.Christopher J A. spock, The Structural Properties Observation and Calculation Kit. Texas A&M University, College Station, TX: Center for Macromolecular Design; 1988. [Google Scholar]
- 27.Quiocho F A, Spurlino J C, Rodseth L E. Structure (London) 1997;5:997–1015. doi: 10.1016/s0969-2126(97)00253-0. [DOI] [PubMed] [Google Scholar]
- 28.Pearlman D A, Case D A, Caldwell J W, Ross W S, Cheatham T E, III, Ferguson D M, Seibel G L, Singh U C, Weiner P K, Kollman P A. amber 4.1. San Francisco: University of California; 1995. [Google Scholar]
- 29.Gardina P J, Bormans A F, Manson M D. Mol Microbiol. 1998;29:1147–1154. doi: 10.1046/j.1365-2958.1998.00964.x. [DOI] [PubMed] [Google Scholar]
- 30.Dahl M K, Manson M D. J Bacteriol. 1985;164:1057–1063. doi: 10.1128/jb.164.3.1057-1063.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizuno T, Mutoh N, Panasenko S M, Imae Y. J Bacteriol. 1986;165:890–895. doi: 10.1128/jb.165.3.890-895.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughson A G, Hazelbauer G L. Proc Natl Acad Sci USA. 1996;93:11546–11551. doi: 10.1073/pnas.93.21.11546. [DOI] [PMC free article] [PubMed] [Google Scholar]