Abstract
GATA factors are transcriptional regulatory proteins that play critical roles in the differentiation of multiple cell types in both vertebrates and invertebrates. Recent evidence suggests that the biological activities of both mammalian and Drosophila GATA factors are controlled in part by physical interaction with multitype zinc-finger proteins, Friend of GATA-1 (FOG) and U-shaped (Ush), respectively. Here we describe a new FOG-related polypeptide, designated FOG-2, that is likely to participate in differentiation mediated by GATA factors in several tissues. Expression of FOG-2 mRNA differs from that of FOG and is largely restricted to heart, neurons, and gonads in the adult. Somewhat broader expression is evident during mouse embryonic development. Similar to FOG and Ush, FOG-2 protein interacts specifically with the amino finger of GATA factors in the yeast two-hybrid system and in mammalian cells. Remarkably, though FOG-2 is quite divergent from FOG in its primary sequence, forced expression of FOG-2 rescues terminal erythroid maturation of FOG−/− hematopoietic cells. Thus, members of the FOG family of cofactors share highly specific association with GATA factors and are substantially interchangeable with respect to some aspects of function in vivo. The interaction of GATA and FOG family members constitutes an evolutionarily conserved paradigm for transcriptional control in differentiation and organogenesis.
Members of the GATA family of zinc-finger proteins figure prominently in transcriptional regulation of cell lineage commitment and differentiation (for review, see ref. 1). Six vertebrate GATA factors have been described. Each recognizes a consensus WGATAR motif through a conserved multifunctional DNA-binding domain comprised of two zinc fingers of the Cx2Cx17Cx2C type (2, 3). Properties of the GATA-factor DNA-binding domain have been characterized most extensively in GATA-1, the founding member, and are presumed to be shared among other family members (4, 5). The carboxyl-terminal (C) finger is essential for DNA binding, whereas the amino-terminal (N) finger appears to stabilize interaction with a subset of sites, particularly those with palindromic GATA sequences (6). In rescue assays of developmentally arrested GATA-1− erythroid cells, both zinc fingers are required for terminal maturation (7).
Each GATA factor exhibits a unique spatial and temporal pattern of expression during development. Sequence similarity and expression pattern serve as a basis for separation of GATA factors into two subfamilies. GATA-1/2/3 are highly expressed in selected hematopoietic cell lineages (8). Gene-targeting experiments have revealed that each is essential for aspects of hematopoietic development (9–13). GATA-4/5/6 proteins are expressed outside the hematopoietic system, principally in heart, gut, and brain (14–19). Loss of GATA-4 in the mouse leads to early lethality because of impaired ventral morphogenesis (20, 21).
Although GATA-1 is a potent transcriptional activator in heterologous test systems (22), the capacity of GATA-1 to drive erythroid cell maturation does not require a transcriptional activation domain (7). This observation led to the hypothesis that the function of GATA-1 in vivo requires a cell-restricted cofactor. A yeast two-hybrid protein-interaction screen yielded a candidate cofactor, a multitype zinc-finger protein designated FOG (Friend of GATA-1) (23). FOG interacts specifically with the N-f of GATA-1 in vitro and in erythroid cells and is coexpressed with GATA-1 during mouse development and in erythroid and megakaryocytic lineages. Moreover, FOG cooperates with GATA-1 in GATA-1-mediated differentiation of erythroid and megakaryocytic cell lines in tissue culture. Disruption of the FOG gene in embryonic stem cells and mice results in embryonic lethality and developmental arrest in the erythroid lineage that was reminiscent of the GATA-1− erythroid phenotype (24). Unexpectedly, loss of FOG leads to a more extreme defect in megakaryocytic development. These in vivo observations are consistent with FOG acting as a cofactor for GATA-1 in erythroid maturation, but also point to GATA-1 independent roles for FOG in the megakaryocytic lineage (24).
Several features of the structure of GATA factors and the interaction of GATA-1 and FOG suggest the possible involvement of additional FOG-like proteins in the control of other vertebrate GATA factors. First, extraordinary conservation of the N-f of GATA factors suggests a critical conserved function. Indeed, the N-f of GATA-1 is highly related to the N-f of a Drosophila GATA factor, pannier (25), which has been shown to interact physically with a FOG-like protein, U-shaped (Ush). Though structurally similar, FOG and Ush are not homologous proteins and, in contrast to FOG, Ush negatively regulates pannier activity (26, 27). Second, other vertebrate GATA factors, such as GATA-4/5/6, are expressed in sites such as the heart, where FOG is not expressed. Because FOG is capable of interacting with all GATA factors tested thus far in vitro (unpublished data), it seemed likely that additional vertebrate FOG-like factors might exist.
Here we describe the isolation and characterization of a FOG homologue, a multifinger nuclear protein we term FOG-2. Like FOG and Ush, FOG-2 interacts specifically with the N-f of GATA factors. FOG-2 mRNA is expressed in heart, nervous tissue, and gonads in the adult mouse and during embryonic development, and thus is potentially available to GATA proteins other than GATA-1. Although FOG-2 resembles FOG in overall structure and has related zinc fingers, it is quite divergent in primary sequence. In an effort to test the function of FOG-2 in vivo, we have asked whether FOG-2 can substitute for FOG in cooperating with GATA-1 in erythroid cells. Remarkably, we find that FOG-2 is competent to restore terminal erythroid maturation in a FOG−/− cell line. Taken together, our results extend the paradigm of GATA–FOG protein interaction and suggest that FOG-2 will serve as a critical cofactor for specific vertebrate GATA factors in nonhematopoietic tissues. The apparent interchangeability of FOG-2 and FOG in some aspects implies that FOG-like cofactors may function in a generic fashion to couple GATA factors bound to DNA target sites to the transcriptional machinery.
MATERIALS AND METHODS
All recombinant DNA work was performed by using standard techniques (28). Details of plasmid construction and oligonucleotide sequences are available on request.
cDNA Cloning.
AA231039 expressed sequence tag (EST) identified in a blast search was obtained from the I.M.A.G.E. Consortium (Research Genetics, Huntsville, AL) and was used as probe in a T2 RNase protection assay (29). Total RNA from various mouse tissues was isolated by standard procedures (28). Poly(A)+ RNA of E14.5 day mouse embryonic brain was purified on oligo(dT) columns (Stratagene) and was used to isolate a full-length cDNA corresponding to expressed sequence tag. Overlapping clones, obtained by a combination of long-range PCR (Marathon, CLONTECH) and RACE (GIBCO/BRL), were sequenced on both strands by standard methods and assembled into a full-length sequence.
Yeast Two-Hybrid System.
To examine the interaction of FOG-2 with GATA baits, a PCR-generated fragment encoding FOG-2 (aa 641-1096) was cloned into the GAL4 activation domain plasmid pGAD10 (CLONTECH). The amino finger (N-f) of murine GATA-4 was cloned into the GAL4 DNA-binding domain plasmid pGBT9. The fusions of the GAL4 DNA-binding domain with the N-f of GATA-1/2/3 have been described (23). The FOG-2 and GATA N-f plasmids were cotransformed into HF7c yeast cells by using the lithium acetate method (30). Several colonies from each cotransformation were restreaked onto Trp−Leu−His− dextrose plates and assayed for β-galactosidase activity.
Immunoprecipitations and Western Blot Analysis.
FOG-2 cDNA was subcloned into the expression vector pCS2+ (30). The hemagglutinin(HA)-GATA-4 expression plasmid was also generated in pCS2+ by subcloning full-length GATA-4 cDNA in-frame with oligonucleotides encoding amino-terminal HA tag. 293 cells were transfected with 8 μg of plasmids by using Lipofectin (GIBCO/BRL). After 48 hr, cells were lysed on plates as described (28). For immunoprecipitations, lysates were incubated with 50 μl of polyclonal FOG antiserum or 10 μl of α-HA antiserum (Santa Cruz Biotechnology) for 1 hr at 4°C. Complexes were precipitated with 30 μl of protein A Sepharose (Pharmacia), washed four times, boiled for 10 min in loading buffer, and resolved by 7.5% SDS/PAGE. Proteins were transferred to nitrocellulose membrane that was incubated with α-FOG-2 antibody and developed by enhanced chemiluminescence (Santa Cruz Biotechnology).
Northern Blot Analysis.
Multiple Tissue Northern blots (CLONTECH) were hybridized sequentially to the radiolabeled FOG-2 cDNA probe (1–927 bps) and a control probe.
In Situ Hybridization.
The partial FOG-2 cDNA inserts were subcloned into pBluescript KS (Stratagene). Antisense RNA probes were prepared by in vitro transcription of the pBSKS templates by using T7 polymerase with digoxigenin-UTP (Boehringer Mannheim). Whole-mount in situ hybridizations were performed as described (31). Whole-mount hybridized embryos were subsequently embedded in paraffin and sectioned at 7 μm. In situ hybridizations of frozen sections (14 μm) were performed essentially as described (31).
Immunofluorescence Analysis of FOG-2 Expression.
For immunofluorescence staining, cells were grown on minichamber slides (Lab-Tek) and were fixed and permeabilized according to standard procedures (28). Cerebellular granule cells prepared from P6 rats as described (32) were cultured for 2 days before fixation. After blocking, cells were incubated with preimmune or FOG-2 antibodies for 4 hr at room temperature, followed by fluorescein-conjugated goat anti-rabbit antibodies (Vector Laboratories). For nuclei counterstaining, cover slips were mounted in Vectorshield with propidium iodide (Vector Laboratories).
Rescue of FOG−/− Cells.
A hematopoietic cell line was derived from FOG−/− embryonic stem (24) cells by in vitro differentiation followed by immortalization with HOX-11. Details of the generation and characterization of this cell line will be published elsewhere. cDNAs encoding FOG or FOG-2 were cloned into the retroviral expression vector MFG-IRES-Zeo and the resultant constructs were transfected into the retroviral producer cell line BOSC 23. After 24 hr, FOG−/− cells at 1 × 106 cells per ml were cocultivated with the confluent monolayer of transfected BOSC 23 cells in 3 ml of Isocove’s modified Dulbecco’s medium containing 15% heat-inactivated fetal calf serum/10 ng/ml interleukin-3/2 μg per ml polybrene/100 units per ml penicillin/100 mg/ml streptomycin/2 mM glutamine. After 48 hr, the cells in suspension were removed and incubated in fresh media (without polybrene) for an additional 24 hr. The cells were then subjected to selection for 6 days in medium containing 1.5 mg/ml of Zeocin (Invitrogen). After selection, cells were washed once in phosphate-buffered saline and stained with o-diansidine (dimethoxybenzidine) as previously described (33). Cytospin preparations of the cells were made and the cells were counterstained with May–Grunwald stain.
RESULTS
Cloning of a Full-Length FOG-2 cDNA.
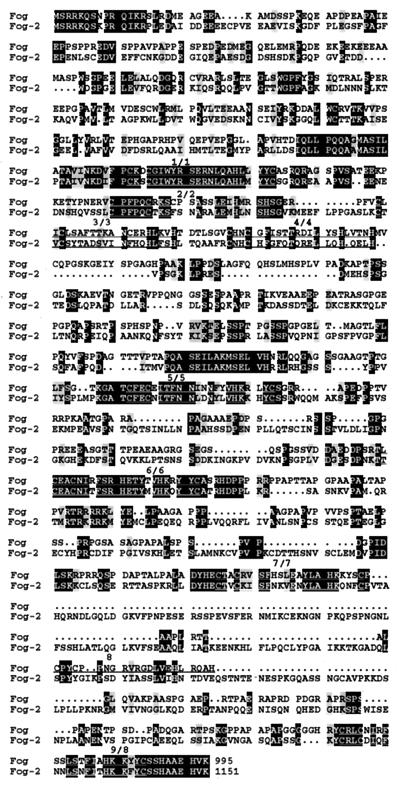
To identify new FOG-like or vertebrate Ush-like molecules, we performed a blast search of the expressed sequence tag database using the mouse FOG sequence as a query. A mouse expressed sequence tag (AA231039) contained two zinc-finger motifs exhibiting significant homology to fingers five and six of FOG. This expressed sequence tag was obtained from I.M.A.G.E. Consortium and was used as an RNase protection probe. Embryonic brain was identified as containing the highest level of RNA transcripts (data not shown). Hence, embryonic brain RNA was used to isolate additional cDNAs corresponding to AA231039 and to assemble a full-length clone (see Materials and Methods). Conceptual translation of the full-length sequence yielded an 1,151 amino acid ORF, designated FOG-2. Comparison of the primary sequences of FOG and FOG-2 (Fig. 1) reveals several interesting features. The arrangement of C2HC and C2H2 finger motifs is similar with respect to each other and to the protein termini. A high degree of similarity is evident between some of the fingers; only short stretches of homology are present outside the fingers. Although the nature and arrangement of the finger motifs in FOG and FOG-2 resemble those in Ush (23, 27), the FOG proteins are more similar to each other than either of them is to Ush. Finally, if only nonfinger regions are considered, little sequence similarity exists among all three proteins (Fig. 1 and data not shown).
Figure 1.
Sequence alignment of the FOG proteins. Sequences were aligned by using the program pileup (GCG, version 8.0) and shaded by using the program boxshade. The darker shading represents identity at a given residue, whereas light shading represents amino acid similarity between the proteins. The positions of the zinc finger regions are indicated.
Association of FOG and GATA Proteins in Yeast and Mammalian Cells.
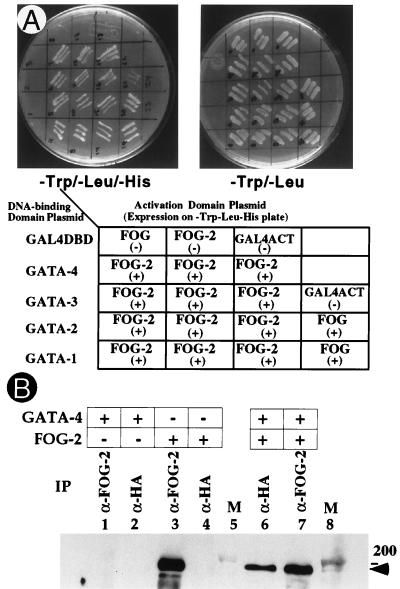
The minimal interaction domains of FOG and GATA proteins include finger six of FOG and the N-f of GATA (23). Based on conservation of the corresponding finger motif in FOG-2, we sought a similar association between FOG-2 and the GATA proteins. Physical interaction between FOG-2 and GATA proteins was assessed first by the yeast two-hybrid system. Specifically, the activation domain of GAL4 was fused to the carboxyl-terminal portion of FOG-2 (aa 641-1096) and was coexpressed with the GAL4 DNA-binding domain fused to the N-fs of GATA-1, 2, or 3. Analogous to the interaction of FOG with these factors, FOG-2 also associated with all three GATA family members (ref. 23; Fig. 2A).
Figure 2.
Interaction of FOG-2 with GATA family members in yeast and in mammalian cells. (A) HF7c yeast cells were cotransformed with plasmids encoding GAL4 DNA-binding domain (DBD) and activation domain (ACT) fusion proteins as indicated. Three independent GATA-FOG-2 double transformants were replica plated on Trp-Leu- and Trp-Leu-His- plates along with indicated negative and positive controls. (B) Coimmunoprecipitation of FOG-2 with HA-GATA-4 in transfected 293 cells. Cells were transfected with plasmids expressing indicated proteins and nuclear lysates were immunoprecipitated with α-HA or α-FOG-2 antibody. Western blot analysis was performed with α-FOG-2 antibody. The position of FOG-2 is indicated.
Because FOG-2 appears to be coexpressed with GATA-4/5/6 proteins in vivo (see below), we also tested interaction of FOG-2 with the N-f-GAL4 DNA-binding domain fusion of GATA-4. Coexpression of FOG-2 and GATA-4 fusion proteins resulted in an interaction visualized by growth on Trp−Leu−His− media and by a β-galactosidase filter assay (Fig. 2A and data not shown).
To examine interaction between FOG-2 and GATA-4 in mammalian cells, expression vectors encoding full-length FOG-2 and HA-epitope-tagged GATA-4 were transiently transfected into 293 (human embryonic kidney) cells. Interactions were analyzed subsequently by an immunoprecipitation-Western assay. As shown in Fig. 2B, FOG-2 was detected in immunoprecipitates from cells transfected with FOG-2 (lanes 3 and 7), but not from cells transfected with HA-GATA-4 alone (lanes 1 and 2). FOG-2 efficiently coprecipitated with HA-GATA-4 (Fig. 2B, lane 6), demonstrating an in vivo association between the two proteins.
These findings establish that FOG-2, like FOG, physically interacts with the N-f of vertebrate GATA factors. Indeed, the precise specificity of FOG-2 for interaction with determinants in the N-f of GATA factors is also retained. Substitutions within the N-f of GATA-1 that selectively impair FOG interaction and retain the DNA-binding properties of GATA-1 also disrupt interaction with FOG-2 (ref. 34; unpublished observations). Thus, FOG-2 and FOG exhibit remarkably similar specificity for GATA-factor N-f interaction.
Expression of FOG-2 mRNA During Mouse Embryonic Development and in the Adult.
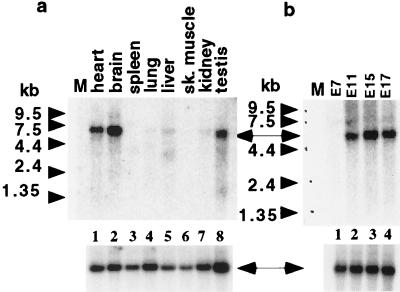
The expression pattern of FOG-2 transcripts was analyzed by Northern blot analysis, RNase protection assays, and in situ hybridization. Northern blot analysis revealed a single RNA species of ≈5 kb in the heart, brain, and testis in adult mouse (Fig. 3a, lanes 1, 2, and 8). Of particular note, FOG-2 mRNA was not detected in spleen and liver, sites in which FOG is expressed (ref. 23; lanes 3 and 5). mRNA for FOG-2 and FOG is coexpressed in testis (lane 8), and neither is appreciably expressed in adult lung, kidney, or skeletal muscle (ref. 23; Fig. 3a, lanes 4, 6, and 7).
Figure 3.
Northern blot analysis of adult (a) and embryonic (b) tissues of the mouse using FOG-2 cDNA. CLONTECH multiple-tissue Northern (MTN) blot membranes (a, CL7762–1; b, CL7763–1) were hybridized with a P32-labeled 0.9-kb BamHI-XbaI fragment of pCS2+FOG-2 (Upper). Position of the FOG-2 mRNA band is indicated by an arrow. To ensure the presence of RNA in every lane, blots were rehybridized for a ubiquitously expressed gene (Lower).
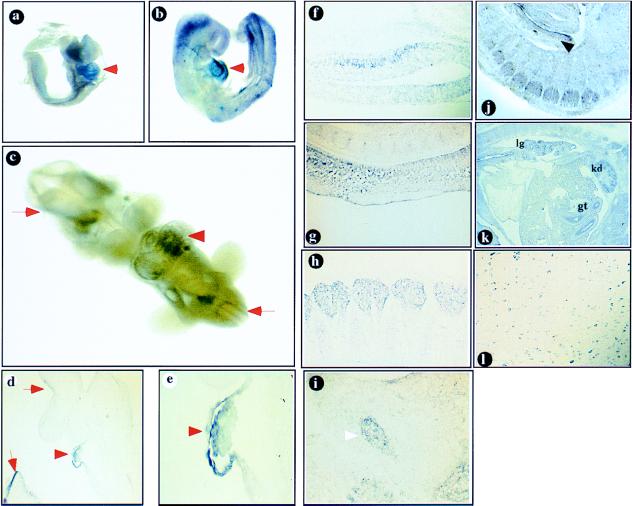
During embryonic development, FOG-2 mRNA is first detected between day E7 and E11, as revealed by Northern blot analysis (Fig. 3b). Whole-mount in situ RNA hybridization analysis detected high levels of FOG-2 expression in the heart region of E8.5, E9.5, and E10.5 embryos (Fig. 4 a–e). Examination of FOG-2 expression by in situ hybridization of sagittal sections from older embryos (E11.5 and beyond) demonstrated localized expression in the developing central and peripheral nervous system, including the developing hemispheres (specifically within the walls of the mesencephalon and the pontine flexure), the otic vesicle, the ganglia (trigeminal, facio-acoustic, and dorsal root), olfactory epithelium, and neuroepithelium of the spinal cord (Fig. 4 f–i and data not shown). FOG-2 expression was also detected in the developing lung, kidney, gut, and urogenital ridge (Fig. 4 j–k). Expression of FOG-2 persists in the nervous system at later stages (Fig. 4l).
Figure 4.
(a–e) Whole-mount RNA in situ hybridization analysis of FOG-2 expression. FOG-2 is expressed in the developing mouse heart (arrowheads) in E8.5 (a), E9.5 (b, d, e), and E10.5 (c) embryos. The expression in the developing nervous system is shown by arrows (c–e). The embryo in b was sectioned to demonstrate the expression in the developing pericardium and myocardium (d, ×100, e, ×400).(f–l) In situ hybridization of FOG-2 antisense RNA probe to mouse tissue sections.(f–i) sagittal sections of an E11.5 embryo. Expression is detected in the midbrain neurons (f), spinal cord (g), dorsal root (h), and trigeminal (i, arrowhead) ganglia. Expression in the urogenital ridge is indicated by an arrow (j). (k) Sagittal section of an E15.5 embryo. Expression is detected in the developing lung (lg), kidney (kd), and gut (gt). (l) FOG-2 expression in the postnatal (P21) brainstem.
The pattern of FOG-2 expression in the E11.5-day embryo resembles that described for GATA-3 (35). In addition, GATA-3 is abundantly expressed in T-lymphocytes (36) and is critical to both early T-cell development and the generation of Th2 T-cells (12, 13, 37). Though expression of FOG in T cells and T cell lines has been documented, preliminary analysis of FOG−/− cells suggests that FOG is not required for T cell development (23, 24). To assess a potential role of FOG-2 in lymphopoiesis, we examined FOG-2 expression in T-lymphocyte cell lines (R1.1 and RLM 11). No FOG-2 mRNA was detected in these cell lines, despite abundant GATA-3 expression (data not shown). Little, if any, FOG-2 message was detected in several other hematopoietic cell lines assayed, representing myeloid, lymphoid, macrophage, and mast lineages (data not shown).
Thus, FOG-2 transcripts are present in multiple nonhematopoietic sites in a pattern overlapping the domains of expression of several GATA factors.
Nuclear Localization and Expression of FOG-2 Protein in PC12 Cells and Granular Neurons of the Cerebellum.
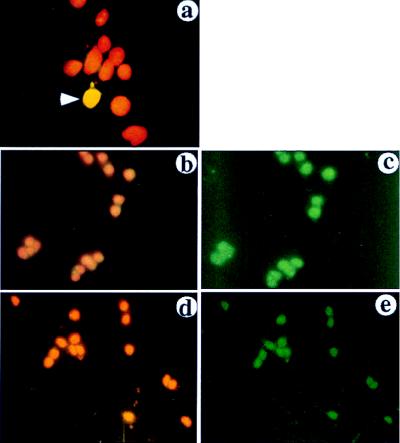
To confirm expression of FOG-2 protein in cells of neuronal origin, we performed an immunofluorescence analysis using anti-FOG-2 antibody. Specificity of the antibody was first assessed in 293 cells transfected with a FOG-2 expression vector. Nuclear staining is evident in cells expressing FOG-2 cDNA but not in untransfected cells (Fig. 5a) or in cells incubated with preimmune antibody (data not shown). Anti-FOG-2 antibody revealed the presence of FOG-2 in pheochromocytoma PC12 cells (Fig. 5 b–c) and in differentiated cerebellular granule neurons (Fig. 5 d–e).
Figure 5.
Immunofluorescence analysis of FOG-2 expression. Cells were stained with α-FOG-2 antibody followed by a fluorescein isothiocyanate-conjugated antibody (green), and nuclei were counterstained with propidium iodide (red). Staining was visualized through a triple-filter (4′,6-diamidino-2-phenylindole/fluorescein isothiocyanate-conjugated/Texas Red, Omega Engineering, Stamford, CT) to detect both the FOG-2 protein and the nuclear staining (a, b, and d) or a single filter to detect FOG-2 only (c and e). (a) 293 cells transiently transfected with FOG-2 expression plasmid. Arrowhead indicates a transfected cell in the field. (b and c) PC12 cells. (d–e) Granule neurons of cerebellum.
Functional Interchangeability of FOG-2 and FOG in Erythroid Differentiation of FOG−/− Cells.
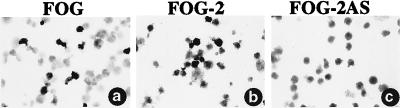
GATA-1 and FOG cooperate in promoting GATA-1-induced erythroid maturation of a GATA-1− erythroid cell line (23). The use of altered specificity mutants of GATA-1 and FOG also demonstrates that FOG is required for the expression of the majority of GATA-1 target genes in erythroid cells (34). These cellular tests of the effects of FOG on GATA-1 action, which define a positive role for the cofactor, contrast with transient reporter assays of short promoters in which FOG often antagonizes GATA-1-mediated transactivation (data not shown). To establish a functional assay for FOG-2 and compare its properties with those of FOG, we have used a rescue system in which test constructs are introduced into immortalized FOG−/− hematopoietic cells. FOG−/− hematopoietic cells of this line fail to complete erythroid maturation. Retroviral transfer of FOG cDNA into these cells restores differentiation, as shown by staining for hemoglobin accumulation with benzidine (A.B.C. and S.H.O., unpublished work; Fig. 6a, 20% benzidine-positive cells). Given the overall similarity of FOG and FOG-2, we asked whether FOG-2 is also able to cooperate with GATA-1 and rescue erythroid maturation. Expressible FOG-2 cDNA (Fig. 6b, 33.6% benzidine-positive cells) but not antisense FOG-2 cDNA (Fig. 6c, 0.2% benzidine-positive cells), promoted erythroid differentiation of FOG−/− cells at approximately the same frequency as wild-type FOG cDNA (Fig. 6a). Thus, FOG-2 is capable not only of interaction with the N-f of various GATA factors (see above), but is also competent to cooperate with a particular GATA protein such as GATA-1 to activate gene expression and induce terminal differentiation. FOG and FOG-2, therefore, share functional as well as structural properties.
Figure 6.
Rescue of erythropoiesis of FOG−/− cells by FOG-2. FOG−/− hematopoietic cells were infected with retroviruses packaged with the expression vector MFG-IRES-Zeo containing cDNAs encoding FOG (a), FOG-2 (b), or FOG-2 AS (FOG-2 cDNA inserted into the vector in the antisense orientation) (c). Cells were stained for hemoglobin with benzidine. Cytospin preparations were made and the cells were counterstained with May–Grunwald stain.
DISCUSSION
Here we describe the identification and initial characterization of FOG-2, a new member of the multitype zinc-finger FOG/Ush family of GATA-protein cofactors. The similarities in structure, N-f-interaction specificity, and function of FOG and FOG-2 reinforce the emerging view that modulation of GATA-factor activity through association with FOG-like proteins is likely to be a recurring theme in transcriptional control and differentiation. Various pairwise combinations of GATA- and FOG-factors may be used to execute similar steps at different times and in different tissues during development.
Prior work has shown that loss of FOG is lethal to the developing mouse because of a failure of erythroid cell development. In addition, megakaryocytes are not formed in the absence of FOG (24). The largely nonoverlapping patterns of expression of FOG and FOG-2 predict nonredundant functions for FOG-2 in vivo. The expression pattern of FOG-2, as revealed by in situ hybridization, is complex and suggests that FOG-2 may function in concert with more than a single GATA factor during development. Indeed, expression of FOG-2 during the development of the nervous system resembles that of GATA-3 (35), whereas expression in the developing heart more closely approximates that of the GATA-4/5/6 subfamily (15, 17).
A potential role for FOG-2 as a cofactor for various GATA factors during heart development is of particular interest. Functionally relevant GATA-binding sites have been identified in the regulatory regions of several cardiac muscle-specific genes, including atrial natriuretic factor (17), cardiac troponin C (38), α-myosin heavy chain (39), b-type natriuretic peptide (40), and cardiac troponin I (41). In addition, expression of the NK-type homeobox gene Nkx2–5, the earliest known marker of the cardiac lineage in vertebrates (42, 43), is controlled in part by a GATA-dependent enhancer (44). It has also been shown that Nkx2–5 and GATA-4 bind adjacent sites within the atrial natriuretic factor gene promoter, interact with each other, and cooperate to activate atrial natriuretic factor gene transcription (45, 46). Targeted mutation of either Nkx2–5 or GATA-4 leads to early embryonic lethality in mice. Loss of Nkx2–5 impairs looping morphogenesis (47), while GATA-4 deficiency prevents heart tube formation because of a failure of ventral morphogenesis (20, 21). In neither instance is initial specification of cardiomyocytes perturbed. The circumscribed defects resulting from loss of these factors might be accounted for in part by potential compensation by other family members in affected cells. For example, GATA-6 levels are increased in the absence of GATA-4 (20, 21). If multiple GATA factors use a common cofactor, such as FOG-2, loss of the cofactor might be expected to produce a more profound defect in cellular commitment and differentiation. Future experiments should reveal what role FOG-2 plays in the transcription hierarchy of heart development.
The role of FOG-2 in modulating GATA factor transcriptional activity in diverse tissue environments may be quite complex. Whereas our data indicate that FOG acts to positively regulate the activity of GATA-1 in transcription, evidence has been provided to suggest that Ush negatively controls the activity of Drosophila pannier (26, 27). In transient transfection assays using short promoters containing GATA sites, FOG, as well as FOG-2, appears to blunt GATA-1 transactivation (unpublished observations). Given in vivo evidence pointing to cooperation, rather than antagonism, of GATA-1 and FOG, we are led to conclude that the transcriptional effects of these interactions are highly context dependent. As such, we have sought to examine the function of FOG-like proteins within an intact cell system where other components are appropriately represented. In this regard, the ability of FOG-2 to restore erythroid maturation in a FOG−/ − hematopoietic cell line is particularly relevant. Despite considerable divergence in primary sequence, FOG-2 is efficient in the rescue of differentiation. Because FOG-2 is not normally expressed within the erythroid lineage, these findings point to shared properties of FOG and FOG-2 rather than an intrinsic role for FOG-2 in hematopoietic development. Although they demonstrate that FOG-2 and GATA-1 are able to cooperate in transcriptional control, they do not exclude a role for FOG-2 as a negative regulator of GATA-factor function in another context.
Our findings underscore the modulation of GATA-factor function through FOG-like protein interaction as a recurring paradigm in development. It is likely that FOG-like molecules act as complex adapters that link DNA-bound GATA factors to the transcriptional machinery. In this regard, we envision that FOG and FOG-2, though quite divergent in primary sequence, are able to recruit possibly shared components to a GATA–FOG complex to assemble an active transcriptional complex. The considerable interchangeability of GATA factors and FOG-like factors in various cell systems suggests that the GATA–FOG complex exhibits inherent versatility, which has been exploited in multiple ways during development.
Acknowledgments
We thank David Wilson for pBSK-GATA-4 plasmid and members of the Orkin laboratory for supplying some of the plasmids and reagents used in this study, for discussions, and for critical reading of this manuscript.
ABBREVIATIONS
- FOG
Friend of GATA-1
- Ush
U-shaped
- HA
hemagglutinin
- aa
amino acids
- N
amino terminal
- C
carboxyl terminal
- N-f
amino finger
- EST
expressed sequence tag
Footnotes
Data Deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF107306).
References
- 1.Orkin S H. Curr Opin Genet Dev. 1996;6:597–602. doi: 10.1016/s0959-437x(96)80089-x. [DOI] [PubMed] [Google Scholar]
- 2.Ko L, Engel J. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merika M, Orkin S H. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans T, Felsenfeld G. Cell. 1989;58:877–885. doi: 10.1016/0092-8674(89)90940-9. [DOI] [PubMed] [Google Scholar]
- 5.Tsai S F, Martin D I, Zon L I, D’Andrea A D, Wong G G, Orkin S H. Nature (London) 1989;339:446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- 6.Trainor C D, Omichinski J G, Vandergon T L, Gronenborn A M, Clore G M, Felsenfeld G. Mol Cell Biol. 1996;16:2238–2247. doi: 10.1128/mcb.16.5.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss M J, Yu C, Orkin S H. Mol Cell Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orkin S H. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- 9.Pevny L, Simon M C, Robertson E, Klein W H, Tsai S-F, D’Agati V, Orkin S H, Costantini F. Nature (London) 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 10.Weiss M J, Keller G, Orkin S H. Genes Dev. 1994;8:1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- 11.Tsai F-Y, Keller G, Kuo F C, Weiss M J, Chen J-Z, Rosenblatt M, Alt F, Orkin S H. Nature (London) 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 12.Pandolfi P P, Roth M E, Karis A, Leonard M W, Dzierzak E, Grosveld F G, Engel J D, Lindenbaum M H. Nat Genet. 1995;11:40–44. doi: 10.1038/ng0995-40. [DOI] [PubMed] [Google Scholar]
- 13.Ting C-N, Olson M C, Barton K P, Leiden J M. Nature (London) 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- 14.Arceci R J, King A A J, Simon M C, Orkin S H, Wilson D B. Mol Cell Biol. 1993;13:2235–2246. doi: 10.1128/mcb.13.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelley C, Blumberg H, Zon L I, Evans T. Development (Cambridge, UK) 1993;118:817–827. doi: 10.1242/dev.118.3.817. [DOI] [PubMed] [Google Scholar]
- 16.Laverriere A C, MacNeill C, Mueller C, Poelmann R E, Burch J B, Evans T. J Biol Chem. 1994;269:23177–23184. [PubMed] [Google Scholar]
- 17.Grepin C, Dagnino L, Robitaille L, Haberstroh L, Antakly T, Nemer M. Mol Cell Biol. 1994;14:3115–3129. doi: 10.1128/mcb.14.5.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrisey E E, Ip H S, Tang Z, Lu M M, Parmacek M S. Dev Biol. 1997;183:21–36. doi: 10.1006/dbio.1996.8485. [DOI] [PubMed] [Google Scholar]
- 19.Morrisey E E, Ip H S, Lu M M, Parmacek M S. Dev Biol. 1996;177:309–322. doi: 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- 20.Kuo C T, Morrisey E E, Anandappa R, Sigrist K, Lu M M, Parmacek M S, Soudais C, Leiden J M. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 21.Molkentin J D, Lin Q, Duncan S A, Olson E N. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 22.Martin D I K, Orkin S H. Genes Dev. 1990;4:1886–1898. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- 23.Tsang A C, Visvader J E, Turner C A, Fujiwara Y, Yu C, Weiss M J, Crossley M, Orkin S H. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 24.Tsang A P, Fujiwara Y, Hom D B, Orkin S H. Genes Dev. 1998;12:1176–1188. doi: 10.1101/gad.12.8.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramina P, Heitzler P, Haenlin M, Simpson P. Development (Cambridge, UK) 1993;119:1277–1291. doi: 10.1242/dev.119.4.1277. [DOI] [PubMed] [Google Scholar]
- 26.Haenlin M, Cubadda Y, Blondeau F, Heitzler P, Lutz Y, Simpson P, Ramain P. Genes Dev. 1997;11:3096–3108. doi: 10.1101/gad.11.22.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cubadda Y, Heitzler P, Ray R P, Bourouis M, Ramain P, Gelbart W, Simpson P, Haenlin M. Genes Dev. 1997;11:3083–3095. doi: 10.1101/gad.11.22.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- 29.Tevosian S G, Shih H H, Mendelson K G, Sheppard K A, Paulson K E, Yee A S. Genes Dev. 1997;11:383–396. doi: 10.1101/gad.11.3.383. [DOI] [PubMed] [Google Scholar]
- 30.Rupp R A, Snider L, Weintraub H. Genes Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- 31.Wilkinson D G. In Situ Hybridization. Oxford: IRL; 1992. [Google Scholar]
- 32.Rio C, Rieff H I, Qi P, Khurana T S, Corfas G. Neuron. 1997;19:39–50. doi: 10.1016/s0896-6273(00)80346-3. [DOI] [PubMed] [Google Scholar]
- 33.Orkin S H, Harosi F I, Leder P. Proc Natl Acad Sci USA. 1975;72:98–102. doi: 10.1073/pnas.72.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crispino, J. D., Lodish, M. B., MacKay, J. P. & Orkin, S. H. (1998) Mol. Cell, in press. [DOI] [PubMed]
- 35.George K M, Leonard M W, Roth M E, Lieuw K H, Kioussis D, Grosveld F, Engel J D. Development (Cambridge, UK) 1994;120:2673–2686. doi: 10.1242/dev.120.9.2673. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto M, Ko L J, Leonard M W, Beug H, Orkin S H, Engel J D. Genes Dev. 1990;4:1650–1662. doi: 10.1101/gad.4.10.1650. [DOI] [PubMed] [Google Scholar]
- 37.Zheng W, Flavell R A. Cell. 1997;89:587–596. [Google Scholar]
- 38.Ip H S, Wilson D B, Heikinheimo M, Tang Z, Ting C N, Simon M C, Leiden J M, Parmacek M S. Mol Cell Biol. 1994;14:7517–7526. doi: 10.1128/mcb.14.11.7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molkentin J D, Kalvakolanu D V, Markham B E. Mol Cell Biol. 1994;14:4947–4957. doi: 10.1128/mcb.14.7.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thuerauf D J, Hanford D S, Glembotski C C. J Biol Chem. 1994;269:17772–17775. [PubMed] [Google Scholar]
- 41.Murphy A M, Thompson W R, Peng L F, Jones N L. Biochem J. 1997;322:393–401. doi: 10.1042/bj3220393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lints T J, Parsons L M, Hartley L, Lyons I, Harvey R P. Development (Cambridge, UK) 1993;119:419–431. doi: 10.1242/dev.119.2.419. [DOI] [PubMed] [Google Scholar]
- 43.Komuro I, Izumo S. Proc Natl Acad Sci USA. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Searcy R D, Vincent E B, Liberatore C M, Yutzey K E. Development (Cambridge, UK) 1998;125:4461–4470. doi: 10.1242/dev.125.22.4461. [DOI] [PubMed] [Google Scholar]
- 45.Durocher D, Charron F, Warren R, Schwartz R J, Nemer M. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee Y, Shioi T, Kasahara H, Jobe S M, Wiese R J, Markham B E, Izumo S. Mol Cell Biol. 1998;18:3120–3129. doi: 10.1128/mcb.18.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lyons I, Parsons L M, Hartley L, Li R, Andrews J E, Robb L, Harvey R P. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]