Abstract
A total of 21 clones of acyclovir (ACV)-resistant (ACVr) herpes simplex virus type 1 (HSV-1) and 23 clones of penciclovir (PCV)-resistant (PCVr) HSV-1, emerging during serial passages in the presence of ACV or PCV, were isolated under conditions excluding contamination of resistant mutants in the starting virus culture, and their mutations in the thymidine kinase (TK) and DNA polymerase (DNA Pol) genes were analyzed comparatively. Mutations in the TK genes from ACVr mutants consisted of 50% single nucleotide substitutions and 50% frameshift mutations, while the corresponding figures for the PCVr mutants were 4 and 96%, respectively (P < 0.001). Eight of the 21 ACVr clones, but none of the 23 PCVr clones, had mutations in DNA Pol. Only nucleotide substitution(s) could be detected in the DNA Pol gene, as the gene is essential for virus replication. Therefore, the results for the DNA Pol mutants are concordant with those for the TK mutants in that a single nucleotide substitution was commonly observed in the ACVr, but not in the PCVr, mutants. These results clearly point to differential mutation patterns between ACVr and PCVr HSV-1 clones.
As the number of patients treated with acyclovir (ACV) has increased, increasing numbers of ACV-resistant (ACVr) herpes simplex virus (HSV) and varicella-zoster virus strains have been isolated, mainly from immunocompromised patients (8). The selectivity of ACV as an antiherpesvirus drug is based on its specific interaction with virus-encoded enzymes, thymidine kinase (TK) and DNA polymerase (DNA Pol). Therefore, the mechanisms responsible for ACV resistance are mutations in the TK and/or DNA Pol polypeptides (1). A previous large-scale clinical study on ACVr HSV strains isolated from patients infected with human immunodeficiency virus indicated that 96% of ACVr HSV mutants were low producers of, or deficient in, TK activity (TK−), with 4% being TK mutants with an altered substrate specificity. No DNA Pol mutants were isolated (12).
It is generally believed that ACVr strains arise from naturally occurring mutations during DNA replication and are selected by ACV both in vitro (cell culture experiments) and in vivo (patients) (2). This hypothesis was formed from the following observations: (i) without exposure to ACV, approximately 0.3 to 20 ACVr mutants occur in 104 PFU of clinical HSV-1 isolates (11, 19, 26, 28) and of a laboratory strain grown from one plaque (2); (ii) ACVr isolates under ACV selective pressure represent only a small proportion of the viable virions in the original virus population (19); and (iii) no ACV mutagenic activity has been detected in previous studies (3, 30).
However, there is still a possibility that ACV does influence the development of ACVr strains during ACV treatment. To address the question of whether or not ACV induces mutation, we chose penciclovir (PCV), which is similar to ACV both in structure and in its need for phosphorylation by virus TK for its anti-HSV action, as a control drug and isolated a series of ACVr and PCV-resistant (PCVr) strains emerging during serial passages of HSV-1 in the presence of ACV or PCV. Sequencing of the TK and DNA Pol genes showed differential mutation patterns in the ACVr and PCVr isolates, suggesting that one, if not both, of the drugs had a differential effect on the generation of drug-resistant mutants.
MATERIALS AND METHODS
Cells and viruses.
A human osteosarcoma TK-deficient cell line, 143B/TK−neoR, was kindly supplied by Riken Cell Bank, Tsukuba, Japan. Human embryo lung (HEL) fibroblasts, Vero, and 143B/TK−neoR cells were cultivated in Eagle's minimum essential medium (MEM) supplemented with 10% calf serum. The VR-3 strain of HSV-1 described previously (6) was obtained from the American Type Culture Collection, Rockville, Md. This virus strain was plaque purified three times, grown in HEL cells for less than three passages, and stored in small portions at −80°C.
Isolation of drug-resistant viruses.
ACVr and PCVr HSV-1 strains were isolated by serial passage of the reference VR-3 strain in the presence of increasing concentrations of ACV or PCV, as follows. Twenty PFU of the VR-3 strain were inoculated onto HEL cell monolayers in 24-well culture plates. The infected cultures were incubated until the virus cytopathic effect reached a maximal level in a medium containing 0.1 μg of ACV/ml or 0.05 μg of PCV/ml, and the concentration of the drug was increased to 0.1, 0.2, 0.5, 1.0, 2.0, and 5.0 μg/ml with each subsequent passage of the virus in HEL cells at a multiplicity of infection (MOI) of 0.01 to 0.1 PFU per cell. After a final passage in 5 μg of ACV or PCV/ml, each virus population emerging from the independent cultures was serially diluted, inoculated onto HEL cell monolayers in 24-well culture plates, and cultivated in MEM supplemented with 2% calf serum and 0.5% (wt/vol) methyl cellulose (Nakalai Tesque, Kyoto, Japan) for 2 days. One plaque was picked from each independent virus population, and then two rounds of plaque purification were performed. As a result, 21 clones of ACVr mutants and 23 clones of PCVr mutants were obtained. Each clone was grown in HEL cells and stored in small aliquots at −80°C.
To determine the variations in drug susceptibilities of the VR-3 clones after serial passages, 16 VR-3 clones were isolated by using the same protocol as that used for the isolation of drug-resistant virus, except that a drug-free medium was used. Briefly, after seven passages in HEL cells at a MOI of 0.01 to 0.1 PFU per cell, 16 VR-3 clones were isolated and then purified by two rounds of plaque purification.
Plaque reduction assay.
The susceptibilities of virus clones to antiherpesvirus compounds were assayed with a 50% plaque reduction assay in Vero cells, as described previously (31).
TK assay.
Confluent 143B/TK−neoR cell monolayers in 24-well tissue culture plates were either mock infected or infected with HSV-1 clones at a MOI of 1 PFU per cell. After incubation for 8 h, the cells were harvested by trypsinization, washed once with MEM, and suspended in 0.2 ml of 200 mM Tris-HCl, pH 7.4. The cells were disrupted by sonication, and the supernatant was collected after centrifugation at 10,000 × g for 10 min at 4°C. TK activity was determined by a previously described method by using [methyl-3H]thymidine as the substrate (14).
DNA sequencing.
The nucleotide sequences of the TK and DNA Pol genes of the HSV-1 isolates were determined by a PCR-directed sequencing method. Briefly, the double-stranded DNA templates for sequencing reactions were prepared by PCR by using oligonucleotide sets S1 and S6 for the TK gene, as described previously (29), and Pol-1 and Pol-R1 for the DNA Pol gene. The sequence of Pol-1 was 5′-ATCCGCCAGACAAACAAGGCCCTT (positions 62655 to 62678 of the HSV-1 DNA sequence) (17), and that of Pol-R1 was 5′-GGCTCATAGACCGGATGCTCAC (positions 66694 to 66673). PCR for the TK gene was performed with 100 μl of reaction mixture by using an Expand Hi-Fi PCR system (Roche Molecular Biochemicals, Mannheim, Germany). Initial denaturation was at 94°C for 2 min followed by 10 cycles of denaturation (20 s at 94°C), annealing (1 min at 54°C), and extension (1 min and 30 s at 72°C) and then 20 cycles of denaturation, annealing, and extension (1 min and 30 s plus 5 s/cycle extension at 72°C), with an additional 20 min at 72°C after the last cycle. PCR for the DNA Pol gene was carried out with 100 μl of reaction mixture containing 0.5 M GC-rich solution (GC-rich PCR system; Roche Molecular Biochemicals). Initial denaturation was at 94°C for 2 min followed by 10 cycles of denaturation (30 s at 95°C), annealing (1 min at 55°C), and extension (5 min at 68°C) and then 20 cycles of denaturation, annealing, and extension (5 min plus 5 s/cycle extension at 68°C), with an additional 20 min at 68°C after the last cycle. The 1,332-bp and 4,039-bp PCR products from the TK and DNA Pol genes, respectively, were purified with a QIAquick PCR purification kit (Qiagen, Hilden, Germany) and used as templates for DNA sequencing. Sequencing was performed by using a cycle sequencing method with a commercial kit (BigDye; Applied Biosystems, Foster City, Calif.) for both strands of the genes and 7 or 19 oligonucleotide primers for the TK or DNA Pol gene, respectively (17). Identified mutations were confirmed by using different PCR products as templates.
Nucleotide sequence accession numbers.
The nucleotide sequences of the TK and DNA Pol genes from the VR-3 strain have been submitted to the DDBJ database and have been assigned the accession numbers AB009254 and AB072389, respectively.
RESULTS
Establishment of experimental design.
In general, most mutations occur during DNA synthesis as mistakes by DNA Pol. Therefore, replication of the virus must be standardized through HSV-1 passages in the presence of ACV and PCV for the analysis of mutations in virus genes. The 50% inhibitory concentrations (IC50s) of ACV and PCV for the VR-3 strain in HEL cells were 0.6 and 0.4 μg/ml, respectively, and the HSV-1 yields were almost equal, 10 to 30% of the control culture, in 0.1 μg of ACV/ml and 0.05 μg of PCV/ml. Therefore, we started HSV-1 passaging from 0.1 μg of ACV/ml and 0.05 μg of PCV/ml, and the concentrations of the drugs were increased about twofold with each subsequent passage to 5 μg/ml for both ACV and PCV.
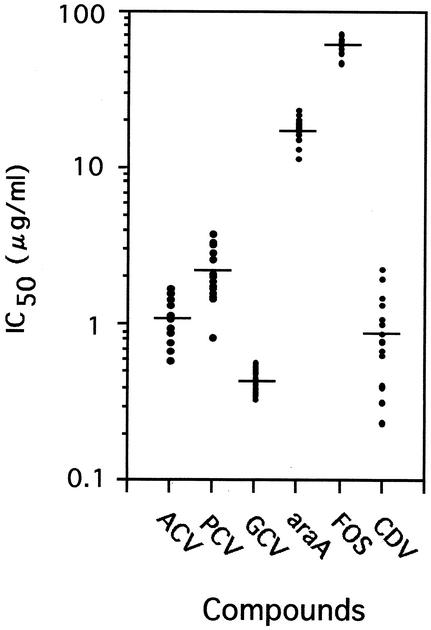
To make a standard for judging clones derived from the VR-3 strain, the susceptibilities to antiherpesvirus compounds of 16 wild-type clones, which had been isolated from the VR-3 population by using the same protocol as that used for the ACVr and PCVr clones except that passaging was carried out in a drug-free medium, were determined in Vero cells (Fig. 1). The difference in the IC50s for the wild-type clones was greatest in the case of cidofovir (CDV) and smallest in the case of foscarnet (FOS). The criterion for judging resistance to each drug was set at the average IC50 of the drug for the VR-3 strain + 2 standard deviations (SD). IC50s indicating resistance were as follows: ACV, >1.87 μg/ml; PCV, >5.75 μg/ml; ganciclovir (GCV), >0.58 μg/ml; vidarabine (ara-A), >47.6 μg/ml; FOS, >77.6 μg/ml; and CDV, >2.05 μg/ml.
FIG. 1.
Susceptibilities of 16 wild-type VR-3 clones to antiherpesvirus compounds (ACV, PCV, GCV, ara-A, FOS, and CDV). Crossbars indicate the average IC50s for the 16 clones.
Genotypic and phenotypic characterization of the ACVr HSV-1 clones.
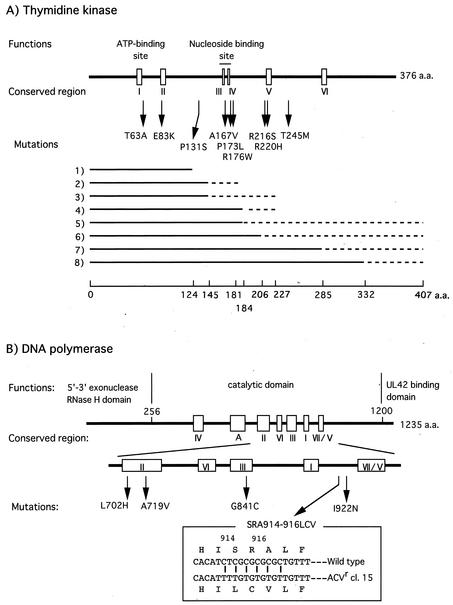
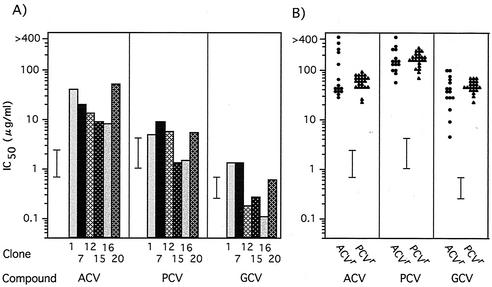
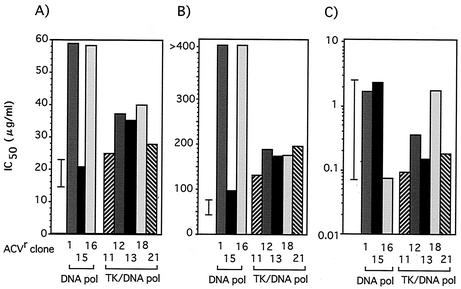
We isolated 21 ACVr VR-3 clones from 24 independent cultures passaged in the presence of increasing concentrations of ACV. The presence of mutation(s) in the TK and DNA Pol genes of these ACVr clones was then determined (Table 1; Fig. 2). Seven clones, categorized as group A, had a single nucleotide deletion or insertion in the 7-G or 6-C homopolymer nucleotide stretch known to be a mutation hot spot (15, 27), and six clones (group B) had a single nucleotide substitution resulting in an amino acid change in the TK gene. These clones carried the wild-type DNA Pol gene and were resistant to all tested deoxyguanosine analogs, ACV, PCV, and GCV, except for clone 20, which showed sensitivity to GCV (Fig. 3). In three clones (group C), mutations were observed solely in the DNA Pol gene. These clones were resistant to ACV but sensitive or marginally resistant to PCV and GCV. Five mutants, classified as group D, possessed mutations in both TK and DNA Pol genes; thus, it was difficult to identify which mutation was responsible for their drug resistance. The susceptibilities of the ACVr mutants to drugs that do not require viral TK activity for their antiherpes activity, such as ara-A, FOS, and CDV, were also determined (Fig. 4). All clones encoding wild-type DNA Pol showed sensitivity to these compounds (data not shown), while all DNA Pol mutants were CDV sensitive but resistant to FOS, though variations in the degrees of resistance were observed. Three of the mutations in the DNA Pol gene, L702H, A719V, and G841C, were observed in the region conserved between the herpesvirus DNA Pol genes (Fig. 2B). The mutation A719V was observed in three clones, and I922N was observed in two clones. We started virus passages from 20 PFU of stock virus in each culture, and the same mutations were identified in two or three of the 21 ACVr mutants, indicating that if the mutants existed in the stock virus population, two or three clones in 420 PFU (20 PFU × 21 cultures) of stock virus should carry the mutation. To clarify this possibility, 4 × 104 PFU of stock virus were tested to see whether they could form plaques in 80 μg of FOS/ml, which was the criterion for a FOS-resistant virus in this study. The results suggested that the mutations were introduced into the DNA Pol gene during passages in the presence of ACV, as no clones in 4 × 104 PFU of stock virus population were detected as FOS resistant (data not shown). The mutation in the ACVr clone 15 consisted of six nucleotide changes from C to T at every other nucleotide from positions 2739 to 2749, resulting in three amino acid substitutions, SRA to LCV, at positions 914 to 916. To clarify whether these six nucleotide substitutions had been accumulated over six passages in ACV or had been induced as a single event, we tried to isolate the ACVr mutants from the virus stocks of earlier passages. ACVr mutants were observed and isolated from among 104 virions after the third passage, when virus was cultured in 0.5 μg of ACV/ml, but not before the second passage, when virus was cultured in 0.2 μg of ACV/ml. The mutant isolated after the third passage carried a mutant DNA Pol gene identical to that of ACVr clone 15, isolated after the sixth passage, when 5 μg of ACV/ml was used. This finding indicated that the six nucleotide substitutions were introduced over the course of at least three passages.
TABLE 1.
Genotypic and phenotypic characterization of 21 ACVr clones
| Group (no. of clones) | Clone(s) | Mutationa
|
TK activity (%)c | |
|---|---|---|---|---|
| TK | DNA Polb | |||
| A (7) | 2, 4, 8, 14 | +G within 7 Gs (430-436) | − | |
| 9, 17 | −C within 6 Cs (548-553) | − | ||
| 10 | +C within 6 Cs (548-553) | − | ||
| B (6) | 19 | A187G (T63A) | − | |
| 6 | G247A (E83K) | − | ||
| 20 | C500T (A167V) | − | ||
| 3 | C518T (P173L) | − | ||
| 7 | G619A (R220H) | 2.1 ± 0.6 | ||
| 5 | C734T (T245M) | − | ||
| C (3) | 1 | G2521T (G841C) | 88.2 ± 6.3 | |
| 15 | *(SRA914-916LCV) | 107.5 ± 3.1 | ||
| 16 | T2105A (L702H) | 109.0 ± 4.5 | ||
| D (5) | 11 | +G within 7 Gs (430-436) | T2765A (I922N) | − |
| 13 | +G within 7 Gs (430-436) | C2156T (A719V) | − | |
| 12 | C391T (P131S) | C2156T (A719V) | − | |
| 18 | C526T (R176W) | C2156T (A719V) | − | |
| 21 | C646A (R216S) | T2765A (I922N) | − | |
Each nucleotide insertion or deletion is shown as + or −, respectively, and, its nucleotide position in the TK gene is shown (in parentheses). Each nucleotide substitution and its position are shown in the form A187G (where A is replaced by G at nucleotide 187), and the corresponding amino acid substitution and its position are shown in parentheses.
Six nucleotide changes from C to T were observed at positions 2739 to 2749 (indicated by *), as shown in Fig. 2.
TK activity of less than 0.5% of that of the wild-type strain is shown as (−). The results are expressed as the means ± standard errors of results from triplicate experiments.
FIG. 2.
Mutations in the HSV-1 TK gene (A) and DNA Pol gene (B). (A) Predicted polypeptides synthesized from the frameshift mutant genes are shown by solid and dotted lines. Solid lines indicate the N-terminal part consisting of wild-type amino acid (a.a.) sequences, and dotted lines show the C-terminal site after frameshift mutations. (1) Deletion of a C within three Cs (371 to 373); (2) deletion of a G within seven Gs (430 to 436); (3) insertion of a G within seven Gs (430 to 436); (4) insertion of a C within six Cs (548 to 553); (5) deletion of a C within six Cs (548 to 553); (6) deletion of a G within five Gs (615 to 619); (7) deletion of a G within four Gs (853 to 856); (8) deletion of a C within three Cs (997 to 999). cl., clone.
FIG. 3.
Susceptibilities of 21 ACVr clones and 23 PCVr clones to deoxyguanosine analogs. The IC50s (μg/ml) for ACVr clones showing sensitivity (A) or resistance (B) to compounds are summarized. The averages ± 2 SD of the IC50s (μg/ml) of compounds for the wild-type VR-3 clones, described in Fig. 1, are represented by bars.
FIG. 4.
Susceptibilities of ACVr mutants possessing mutations in the DNA Pol genes to ara-A (A), FOS (B), and CDV (C). The averages ± 2 SD of the IC50s (μg/ml) for the wild-type VR-3 clones, described in Fig. 1, are represented by bars.
Genotypic and phenotypic characterization of HSV-1 PCVr clones.
Twenty-three PCVr HSV-1 clones were isolated from the VR-3 strain by using the same method as that employed for the isolation of the ACVr mutants, except that the initial concentration of PCV was 0.05 μg/ml. Twenty-two clones had a single nucleotide deletion or insertion in the G or C homopolymer nucleotide stretch, and just one clone had a single nucleotide substitution (Table 2; Fig. 2). No mutations in the DNA Pol genes of the 23 PCVr clones were observed. All PCVr mutants were highly resistant to the deoxyguanosine analogs, ACV, PCV, and GCV (Fig. 3B), but sensitive to ara-A, FOS, and CDV (data not shown).
TABLE 2.
Genotypic and phenotypic characterization of 23 PCVr clonesa
| Group (no. of clones) | Clone(s) | TK mutationb |
|---|---|---|
| A (22) | 3-5, 13, 15, 17-22 | −C within 6 Cs (548-553) |
| 2, 8, 12, 16 | −G within 7 Gs (430-436) | |
| 10, 23 | +G within 7 Gs (430-436) | |
| 1, 14 | −C within 3 Cs (997-999) | |
| 7 | −C within 3 Cs (371-373) | |
| 11 | −G within 5 Gs (615-619) | |
| 6 | −G within 4 Gs (853-856) | |
| B (1) | 9 | C761A (S254Stop) |
Symbols are as indicated in Table 1, footnote a.
No mutations in the DNA Pol genes were observed, and TK activity was less than 0.5% of that of the wild-type strain.
DISCUSSION
A low level of replication error is typically associated with DNA synthesis and, in the case of herpesviruses, occasionally introduces a single base mutation into the TK and/or DNA Pol genes, thus resulting in drug resistance in the viruses. This naturally occurring mutation is thought to be a major cause of drug-resistant viruses, and its frequency is estimated at a few virions per 104 PFU, though some variations in frequencies have been observed between HSV-1 and HSV-2 and their associated strains (11, 19, 26, 28). On the other hand, the majority of ACVr HSVs have been reported to occur in immunocompromised patients undergoing a prolonged course of ACV chemotherapy (mean duration, 43.5 days) in clinical studies (18). It is likely that a long period is necessary for the selection of ACVr during ACV therapy, but that ACVr mutant induction can occur during prolonged ACV therapy is undeniable. Therefore, we analyzed the influence of ACV and PCV on the induction of virus mutation while taking the following points into consideration. (i) To prevent contamination of spontaneous ACVr and PCVr mutants in the starting material, the passages of the HSV-1 VR-3 strain were started with 20 PFU of stock virus in this experiment. Less than 5 PFU of ACVr mutants existed in 104 PFU of our virus stock (data not shown); therefore, contamination of ACVr mutants was less than 1%. (ii) To estimate the influence of ACV on virus mutation, we compared it with that of PCV, which is analogous to ACV both in structure and in its need for virus TK for antiviral activity.
The differences in the mutations observed between ACVr and PCVr isolates were well beyond our expectations. Mutations in the TK genes from ACVr mutants consisted of 50% single nucleotide substitutions and 50% frameshift mutations, while the corresponding figures for TK gene mutations in the PCVr mutants were 4 and 96%, respectively (Fisher's exact test; P < 0.001). All the ACVr and PCVr clones in which resistance was caused by a mutation in the TK gene were cross resistant to PCV and ACV, respectively (Fig. 3). Thus, the differences in mutations were attributed to differences in the mutagenic effects of ACV and PCV on replicating DNA rather than to differences in the selection of spontaneous mutants by ACV or PCV during the virus passages. In the case of the DNA Pol gene, only nucleotide substitutions, not frameshift mutations, are compatible with survival as the gene is essential for virus replication. DNA Pol mutations were observed in 8 of 21 ACVr clones but not in the 23 PCVr clones. This result was in accord with TK gene mutation results, where ACVr mutants, unlike PCVr mutants, showed a high frequency of single nucleotide substitutions. The major reason for the absence of DNA Pol mutations in PCVr clones was that DNA Pol mutations never confer more than a three- to fourfold increase in resistance to PCV, as previously described and as shown in this study (Fig. 3A) (20).
Except in the R220H TK mutant, we could not detect TK activity. However, some ACVr TK− mutants showed susceptibility or a lower resistance to PCV (clones 12 and 20), indicating that these mutant TKs might express low-level TK activity and phosphorylate PCV to PCV monophosphate. PCV has an affinity for HSV-1 TK that is 100 times higher than that of ACV (the K[infi]i values for PCV and ACV are 1.5 and 173 μM, respectively [5, 16, 34]), resulting in a pool of PCV-triphosphate that is approximately 200-fold larger than that of ACV-triphosphate in HSV-infected cells (34, 35). Thus, mutations in the TK gene which lead to a complete loss of PCV phosphorylation activity were required for PCVr.
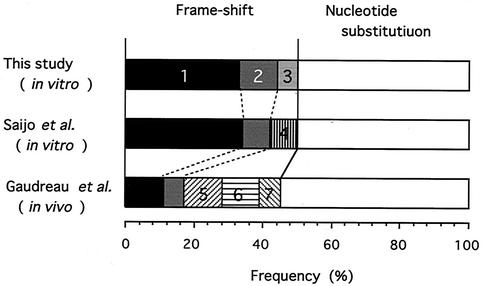
Several groups have published the nucleotide sequences of mutant TK and DNA Pol genes obtained from ACVr strains from both cell culture experiments and patients. We chose two studies reporting sequencing of a relatively large number of mutant TK genes of ACVr HSV-1 and compared their results with ours (Fig. 5). Saijo et al. analyzed 24 mutant clones isolated in vitro from a high titer of stock virus; thus, most clones had emerged by spontaneous mutation and had been selected by ACV (24). The paper published by Gaudreau et al. described the genotypic characterization of 22 ACVr HSV-1 isolates from immunocompromised patients, which would be a mixture of ACV-induced and ACV-selected TK mutants (10). The frequencies of single nucleotide substitutions and frameshift mutations observed in these three studies were almost identical, meaning that PCV influenced the mutation pattern to induce frameshift in the TK gene more than ACV did.
FIG. 5.
Ratio of mutations in the TK gene of the ACVr HSV-1 strain. The total numbers of ACVr HSV-1 clones in this study and those in the studies of Saijo et al. and Gaudreau et al. are 21, 24, and 29, respectively (10, 24). The mutations consisting of a single nucleotide insertion or deletion in the homopolymer repeat are shown as follows: 1, insertion of a G within seven Gs at positions 430 to 436; 2, deletion of a C within six Cs at positions 543 to 553; 3, insertion of a C within six Cs at positions 548 to 553; 4, deletion of a C within four Cs at positions 1061 to 1064; 5, deletion of a C within four Cs at positions 666 to 669; 6, deletion of a C within five Cs at positions 460 to 464; and 7, deletion of an A within four As at positions 184 to 187.
Recently, Sarisky et al. reported the characteristics of ACVr and PCVr HSV strains (25). They isolated 14 ACVr clones and 12 PCVr clones of the HSV-1 SC-16 strain and 8 ACVr clones and 9 PCVr clones of the HSV-2 SB5 strain in vitro by serial passage of the virus in the presence of increasing concentrations of ACV and PCV, respectively, and then characterized them. Surprisingly, their conclusions were the opposite of ours; that is, ACVr isolates commonly showed frameshift mutations while PCVr isolates contained mostly nucleotide substitutions in their TK genes. This discrepancy may result from differences in the conditions under which the mutations arose in the TK genes. In the present study, drug-resistant mutants emerged in the presence of the drug during passages with a limited number of virions as the starting material, while in the previous study, the drug-resistant isolates originated from spontaneous mutants preexisting in 7 × 104 PFU of the virus population. Over half of the isolates in the previous study possessed multiple mutations, with a maximum of six mutations in the TK gene from one clone, indicating that the mutants had been developing and accumulating in the virus population during several passages prior to analysis and were selected and isolated from that population as drug resistant.
We could not clarify the molecular mechanism for the differences in the effects of ACV and PCV on mutation, but differences in the modes of inhibition of DNA synthesis between these compounds might be the key point. ACV monophosphate is incorporated into the 3′ end of replicating DNA by viral DNA Pol as a chain terminator and inhibits further DNA chain elongation in both infected cells and in vitro enzymatic studies (4, 9). An in vitro study with purified DNA Pol showed that PCV also acts as chain terminator, but no study has described how PCV inhibits DNA synthesis in herpesvirus-infected cells (7). There is a possibility that PCV is incorporated into replicating DNA internally in infected cells, because (i) PCV possesses a hydroxy group in its side chain, like GCV, which can form phosphodiester linkages in DNA, and (ii) GCV can be efficiently incorporated into DNA in mammalian cells expressing HSV-1 TK, though the drug inhibits DNA elongation due to chain termination in vitro (13, 21-23, 33). Many-sided studies are required to understand the mechanisms for the effects of PCV on both the inhibition of DNA synthesis and the induction of frameshifts (32).
Acknowledgments
We thank Yukihiro Nishiyama (Nagoya University School of Medicine, Japan) and Seiji Yasumura (Fukushima Medical University, Japan) for their helpful comments.
REFERENCES
- 1.Balfour, H. H. 1983. Resistance of herpes simplex to acyclovir. Ann. Intern. Med. 98:404-406. [DOI] [PubMed] [Google Scholar]
- 2.Coen, D. M. P., A. Schaffer, P. A. Furman, P. M. Keller, and M. H. St. Clair. 1982. Biochemical and genetic analysis of acyclovir-resistant mutants of herpes simplex virus type 1. Am. J. Med. 73(Suppl. 1A):351-360. [DOI] [PubMed] [Google Scholar]
- 3.Collins, P. 1988. Viral sensitivity following the introduction of acyclovir. Am. J. Med. 85(Suppl. 2A):129-134. [PubMed] [Google Scholar]
- 4.Derse, D., Y.-C. Cheng, P. A. Furman, M. H. St. Clair, and G. B. Elion. 1981. Inhibition of purified human and herpes simplex virus-induced DNA polymerases by 9-(2-hydroxyethoxymethyl)guanine triphosphate. J. Biol. Chem. 256:11447-11451. [PubMed] [Google Scholar]
- 5.Detema, R., A. C. Ericson, H. J. Field, A. Larsson, and K. Stenber. 1987. Critical determinants of antiherpes efficacy of buciclovir and related acyclic guanosine analogs. Antivir. Res. 7:303-316. [DOI] [PubMed] [Google Scholar]
- 6.Dowdle, W. R., A. J. Nahmias, R. W. Harwell, and F. P. Pauls. 1967. Association of antigenic type of herpesvirus hominis with site of viral recovery. J. Immunol. 99:974-980. [PubMed] [Google Scholar]
- 7.Earnshaw, D. L., T. H. Bacon, S. J. Darlison, K. Edmonds, R. M. Parkins, and R. A. Vere Hodge. 1992. Penciclovir: mode of antiviral action of penciclovir in MRC-5 cells infected with herpes simplex virus (HSV-1), HSV-2, and varicella-zoster virus. Antimicrob. Agents Chemother. 36:2747-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Englund, J. A., M. E. Zimmerman, E. M. Swierkosz, J. L. Goodman, D. R. Scholl, and H. H. Balfour, Jr. 1990. Herpes simplex virus resistant to acyclovir. A study in a tertiary care center. Ann. Intern. Med. 112:416-422. [DOI] [PubMed] [Google Scholar]
- 9.Furman, P. A., P. V. McGuirt, P. M. Keller, J. A. Fyfe, and G. B. Elion. 1980. Inhibition by acyclovir of cell growth and DNA synthesis of cells biochemically transformed with herpesvirus genetic information. Virology 102:420-430. [DOI] [PubMed] [Google Scholar]
- 10.Gaudreau, A., E. Hill, H. H. Balfour, A. Erice, Jr., and G. Boivin. 1998. Phenotypic and genotypic characterization of acyclovir-resistant herpes simplex viruses from immunocompromised patients. J. Infect. Dis. 178:297-303. [DOI] [PubMed] [Google Scholar]
- 11.Hasegawa, T., T. Kawana, T. Okuda, M. Horii, T. Tsukada, and K. Shiraki. 2001. Susceptibility to acyclovir of herpes simplex virus isolates obtained between 1977 and 1996 in Japan. J. Med. Virol. 63:57-63. [PubMed] [Google Scholar]
- 12.Hill, E. L., G. A. Hunter, and M. N. Ellis. 1991. In vitro and in vivo characterization of herpes simplex virus clinical isolates recovered from patients infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 35:2322-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ilsley, D. D., S. H. Lee, W. H. Miller, and R. D. Kuchta. 1995. Acyclic guanosine analogs inhibit polymerase α, δ, and ɛ with very different potencies and have unique mechanisms of action. Biochemistry 34:2504-2510. [DOI] [PubMed] [Google Scholar]
- 14.Jamieson, A. T., G. A. Gentry, and J. H. Subak-Sharp. 1974. Induction of both thymidine and deoxycytidine kinase activity by herpes viruses. J. Gen. Virol. 24:465-480. [DOI] [PubMed] [Google Scholar]
- 15.Kit, S., M. Sheppard, H. Ichimura, S. Nusinoff-Lehrman, M. N. Ellis, J. A. Fyfe, and H. Otsuka. 1987. Nucleotide sequence changes in thymidine kinase gene of herpes simplex virus type 2 clones from an isolate of a patient treated with acyclovir. Antimicrob. Agents Chemother. 31:1483-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsson, A., K. Stenberg, A. C. Ericson, U. Haglund, W. A. Yisak, N. G. Johansson, B. Oberg, and R. Detema. 1986. Mode of action, toxicity, pharmacokinetics, and efficacy of some new antiherpesvirus guanosine analogs related to buciclovir. Antimicrob. Agents Chemother. 30:598-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGeoch, D. J. M., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 18.Nugier, F., J. N. Colin, M. Aymard, and M. Langlois. 1992. Occurrence and characterization of acyclovir-resistant herpes simplex virus isolates: report on a two-year sensitivity screening survey. J. Med. Virol. 36:1-12. [DOI] [PubMed] [Google Scholar]
- 19.Parris, D. S., and J. E. Harrington. 1982. Herpes simplex virus variants restraint to high concentrations of acyclovir exist in clinical isolates. Antimicrob. Agents Chemother. 22:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelosi, E., G. B. Mulanba, and D. M. Coen. 1998. Penciclovir and pathogenesis phenotypes of drug-resistant herpes simplex virus mutants. Antivir. Res. 37:17-28. [DOI] [PubMed] [Google Scholar]
- 21.Reardon, J. E. 1989. Herpes simplex virus type 1 and human DNA polymerase interactions with 2′-deoxyguanosine 5′-triphosphate analogues. J. Biol. Chem. 264:19039-19044. [PubMed] [Google Scholar]
- 22.Reid, R., E. C. Mar, E. S. Huang, and M. D. Topal. 1988. Insertion and extension of acyclic dideoxy and ara nucleotides by herpesviridae human α and β polymerases: a unique inhibition mechanism for 9-(1, 3-dihydroxy-2-propoxymethyl)guanine triphosphate. J. Biol. Chem. 263:3898-3904. [PubMed] [Google Scholar]
- 23.Rubsam, L. Z., B. L. Davidson, and D. S. Shewach. 1998. Superior cytotoxicity with ganciclovir compared with acyclovir and 1-β-D-arabinofuranosylthymine in herpes simplex virus-thymidine kinase-expressing cells: a novel paradigm for cell killing. Cancer Res. 58:3873-3882. [PubMed] [Google Scholar]
- 24.Saijo, M., T. Suzutani, E. De Clercq, M. Niikura, A. Maeda, S. Morikawa, and I. Kurane. 2002. Genotypic and phenotypic characterization of the thymidine kinase of ACV-resistant HSV-1 derived from an acyclovir-sensitive herpes simplex virus type 1 strain. Antivir. Res. 56:253-262. [DOI] [PubMed] [Google Scholar]
- 25.Sarisky, R. T., M. R. Quail, P. E. Clark, T. T. Nguyen, W. S. Halsey, R. J. Wittrock, J. O'Leary Bartus, M. M. Van Horn, G. M. Sathe, S. Van Horn, M. D. Kelly, T. H. Bacon, and J. J. Leary. 2001. Characterization of herpes simplex viruses in culture for resistance to penciclovir or acyclovir. J. Virol. 75:1761-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarisky, R. T., T. T. Nguyen, K. E. Duffy, R. J. Wittrock, and J. J. Leary. 2000. Difference in incidence of spontaneous mutations between herpes simplex virus types 1 and 2. Antimicrob. Agents Chemother. 44:1524-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasadeusz, J. J., F. Tufaro, S. Safrin, K. Schubert, M. M. Hubinette, P. K. Cheung, and S. L. Sacks. 1997. Homopolymer mutational hot spots mediate herpes simplex virus resistance to acyclovir. J. Virol. 71:3872-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin, Y. K., G. Y. Cai, A. Weinberg, J. J. Leary, and M. J. Levin. 2001. Frequency of acyclovir-resistant herpes simplex virus in clinical specimens and laboratory isolates. J. Clin. Microbiol. 39:913-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzutani, T., S. Koyano, M. Takada, I. Yoshida, and M. Azuma. 1995. Analysis of the relationship between cellular thymidine kinase activity and virulence of thymidine kinase-negative herpes simplex virus types 1 and 2. Microbiol. Immunol. 39:787-794. [DOI] [PubMed] [Google Scholar]
- 30.Suzutani, T., and H. Machida. 1992. Analysis of toxic and mutagenic activities of antiherpesvirus nucleosides against HeLa cells and herpes simplex virus type 1. Mutat. Res. 267:125-131. [DOI] [PubMed] [Google Scholar]
- 31.Suzutani, T., H. Machida, and T. Sakuma. 1988. Efficacies of antiherpesvirus nucleosides against two strains of herpes simplex virus type 1 in Vero and human embryo lung fibroblast cells. Antimicrob. Agents Chemother. 32:1046-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomicic, M. T., E. Bey, P. Wutzler, R. Thust, and B. Kaina. 2002. Comparative analysis of DNA breakage, chromosomal aberrations and apoptosis induced by the anti-herpes purine nucleoside analogues aciclovir, ganciclovir and penciclovir. Mutat. Res. 505:1-11. [DOI] [PubMed] [Google Scholar]
- 33.Tomicic, M. T., R. Thust, and B. Kaina. 2002. Ganciclovir-induced apoptosis in HSV-1 thymidine kinase expressing cells: critical role of DNA breaks, bcl-2 decline and caspase-9 activation. Oncogene 21:2141-2153. [DOI] [PubMed] [Google Scholar]
- 34.Vere Hodge, R. A., and Y.-C. Cheng. 1993. The mode of action of penciclovir. Antivir. Chem. Chemother. 4(Suppl. 1):13-24. [Google Scholar]
- 35.Vere Hodge, R. A., and R. M. Perkins. 1989. Mode of action of 9-(4-hydroxy-3-hydroxymethylbut-1-yl)guanine (BRL 39123) against herpes simplex virus in MRC-5 cells. Antimicrob. Agents Chemother. 33:223-229. [DOI] [PMC free article] [PubMed] [Google Scholar]