Abstract
GATA transcription factors are important regulators of both hematopoiesis (GATA-1/2/3) and cardiogenesis (GATA-4) in mammals. The transcriptional activities of the GATA proteins are modulated by their interactions with other transcription factors and with transcriptional coactivators and repressors. Recently, two related zinc finger proteins, U-shaped (USH) and Friend of GATA-1 (FOG) have been reported to interact with the GATA proteins Pannier and GATA-1, respectively, and to modulate their transcriptional activities in vitro and in vivo. In this report, we describe the molecular cloning and characterization of a third FOG-related protein, FOG-2. FOG-2 is an 1,151 amino acid nuclear protein that contains eight zinc finger motifs that are structurally related to those of both FOG and USH. FOG-2 is first expressed in the mouse embryonic heart and septum transversum at embryonic day 8.5 and is subsequently expressed in the developing neuroepithelium and urogenital ridge. In the adult, FOG-2 is expressed predominately in the heart, brain, and testis. FOG-2 associates physically with the N-terminal zinc finger of GATA-4 both in vitro and in vivo. This interaction appears to modulate specifically the transcriptional activity of GATA-4 because overexpression of FOG-2 in both NIH 3T3 cells and primary rat cardiomyocytes represses GATA-4-dependent transcription from multiple cardiac-restricted promoters. Taken together, these results implicate FOG-2 as a novel modulator of GATA-4 function during cardiac development and suggest a paradigm in which tissue-specific interactions between different FOG and GATA proteins regulate the differentiation of distinct mesodermal cell lineages.
GATA transcription factors are important regulators of development in worms, flies, and mammals (1–5). The six mammalian GATA proteins (GATA-1–6) share related CX2CX17CX2C zinc finger DNA-binding domains and bind to the consensus motif, WGATAR, as well as to related sequences such as CGATGG and AGATTA (6, 7). GATA factors can be divided into two subfamilies based on their structures and patterns of expression. GATA-1/2/3 are expressed in the hematopoietic cell lineages and play critical roles in the development of the erythroid, hematopoietic stem cell, and T cell lineages, respectively (8–10). In contrast, the structurally related proteins GATA-4/5/6 are expressed in overlapping patterns in the heart, gut, urogenital system, and smooth muscle cell lineages (11–14).
GATA-4 is an important regulator of early cardiogenesis (15, 16). GATA-4 is expressed in developing murine procardiomyocytes at embryonic day 7.5 (E7.5) before the formation of a linear heart tube (12). Many, if not all, cardiac-restricted promoters contain functionally important GATA-4 binding sites, and forced expression of GATA-4 can transactivate many of these promoters in noncardiac cells (17–20). Targeted disruption of the GATA-4 gene in mice results in early embryonic lethality because of a specific defect in ventral heart tube formation (21, 22).
The transcriptional activities of the GATA proteins are modulated by interactions both with other transcription factors and with transcriptional coactivators and repressors. For example, the C-terminal zinc finger of GATA-1 interacts specifically with the SP1 and EKLF transcription factors to synergistically activate erythroid-specific gene expression (23). Similarly, both Nkx2.5 and NFAT3 associate with the C-terminal zinc finger of GATA-4 and coactivate the transcription of cardiac-restricted genes (24–27). Recently, two related zinc finger proteins, Friend of GATA-1 (FOG) and U-shaped (USH), have been shown to interact specifically with the N-terminal zinc fingers of GATA-1 and the Drosophila GATA protein Pannier, respectively (28, 29). The interaction of USH with Pannier appears to repress the transcriptional activity of Pannier (29). In contrast, FOG, in conjunction with GATA-1, synergistically activates expression of the erythroid-specific NF–E2 promoter and, like GATA-1, is required for normal erythropoiesis in the mouse (28, 30).
FOG is not expressed in embryonic or adult cardiomyocytes (28). Thus, we postulated the existence of a distinct FOG-related protein in cardiomyocytes that would interact with and regulate the transcriptional activity of GATA-4 during cardiogenesis. In this report, we describe the cloning and characterization of such a novel FOG-related protein called FOG-2. FOG-2 contains eight zinc fingers that are structurally related to those of both FOG and USH. FOG-2 is coexpressed with GATA-4 in both the developing and adult heart and is also expressed in the brain and tissues of the urogenital system. FOG-2 interacts directly with the N-terminal zinc finger of GATA-4 in vitro and in vivo. Moreover, forced expression of FOG-2 in both fibroblasts and cardiomyocytes represses the transcriptional activity of GATA-4 on a variety of cardiac-restricted promoters. Taken together, these results implicate FOG-2 as a potentially important regulator of GATA-4 transcriptional activity in the heart.
MATERIALS AND METHODS
Cloning of FOG-2.
A screen of the expressed sequence tag (EST) database at the National Center for Biotechnology Information using the FOG cDNA sequence (28) identified one mouse EST (W12035) with significant homology to FOG. PCR primers based on this EST (5′-TCTGGGTCTGAAAGCCAAAATGGC and 5′-TGATGTACTACTGTAGTGGGAGGC) were used to amplify a 321-bp fragment from mouse heart cDNA. This fragment was then used to screen 1 × 106 recombinants from an E13 mouse heart cDNA library (Stratagene). Five independent clones were obtained and sequenced by using a Perkin–Elmer automated sequencer.
Northern Analysis.
A 2-μg Poly(A)+ Northern blot containing mRNAs from multiple mouse tissues was hybridized according to the manufacturer’s instructions (CLONTECH) to a radiolabeled 452-bp HindIII–SacI fragment of the FOG-2 cDNA (bp 404 to 856) or to a control human β-actin cDNA (CLONTECH).
Subcellular Localization.
COS-7 cells were transfected with a Flag epitope-tagged FOG-2 expression construct (pcDNAFlag/FOG-2) or with pcDNA3 and assayed for FOG-2 expression by indirect immunofluorescence by using the new line anti-M2 Flag antibody (Kodak) and fluorescein isothiocyanate-conjugated goat anti-mouse IgG (17).
In Situ Hybridization.
In situ hybridizations were performed by using radiolabeled cRNA sense and antisense probes generated from the FOG-2 cDNA (bp 404–856) (13).
Glutathione S-Transferase (GST) Fusion Proteins and Binding Assays.
A BamHI fragment containing the ORF of the FOG-2 cDNA (bp 132–4,770) was cloned into the BamHI site of pcDNA3 (Invitrogen) to create pcDNA-FOG-2. A 309-bp fragment of the GATA-4 cDNA encoding the N- and C-terminal zinc fingers (aa 199–302) was cloned into the EcoRI site of pGEX4T2 to create pGST-GATA-4. GST binding assays were performed as described previously (31).
Immunoprecipitations.
pcDNA3.1FOG-2 contained the Express epitope fused in frame with the full-length FOG-2 cDNA (bp 132–4,770) cloned into the BamHI site of pcDNA3.1His(c) (Invitrogen). COS-7 cells were transfected by using lipofectamine with either pcDNA3.1FOG-2, pcDNAGATA-4 (13), or both. Nuclear extracts were prepared from the transfected cells 48 hr after transfection (17). Nuclear extract (500 μg) was immunoprecipitated with 10 μl of goat anti-GATA-4 antibody (Santa Cruz Biotechnology) and protein G Sepharose (31). Western analysis was performed by using an anti-Express antibody (Invitrogen) and peroxidase-labeled goat anti-mouse IgG (Kirkegaard & Perry Laboratories).
Yeast 2-Hybrid Assays.
pG4ZnN+C was constructed by using the PCR and primers (5′-GAGAATTCGAAGGCAGAGAGTGTGTCAATTGTGGG and 5′-CTCTTAAGGTAGAGGCCGCAGGCATTACATACAGG) to produce a DNA fragment encoding both zinc fingers of GATA-4, which was cloned into the EcoRI site of pAS2–1 (CLONTECH) to generate a GAL4 DNA-binding domain/GATA-4 fusion protein. pG4ZnN and pG4ZnC were constructed similarly by using primers (5′-GAGAATTCGAAGGCAGAGAGTGTGTCAATTGTGGG and 5′-GCGCGAATTCGGCCTACCCGGCGGGAAGCGGACAGGCGGC) and (5′-GAGAATTCTCCTGTGCCAACTGCCAGACTACC and 5′-GCGCGAATTCGGCCTACCCGGCGGGAAGCGGACAGGCGGC), respectively. pACT-FOG-2 was constructed by cloning an NcoI/BamHI fragment of the FOG-2 cDNA (987–4770 bp) into the NcoI/BamHI sites of pACT2 (CLONTECH). This construct was then transformed into the three yeast strains containing the GAL4-GATA fusion proteins and double transformants were selected by growth on tryptophan- and leucine-deficient media. Strains of yeast containing both plasmids were then tested for their ability to grow on media containing 15 mM 3-aminotriazole and lacking tryptophan, leucine, and histidine. The β-galactosidase activity of each yeast strain was determined by using a filter assay.
Transfections.
p1.0 BNP GH contained a 1-kb fragment of the rat B-type natriuretic peptide (BNP) promoter prepared by the PCR from rat genomic DNA (primers: 5′-CGTCTAGATGTATACCCCACAGCCTCATTTCC and 5′-CAGGATCCTGTCCTGCTCTCTCTTGTCTCGCC) and cloned into the HindIII-BamHI sites of p0GH (Nichols Institute). Primary neonatal cardiomyocytes (17) or NIH 3T3 cells were transfected with p1.0BNP GH, pcDNAGATA-4, and pcDNAFOG-2 as well as pVRβ-gal (32) to normalize for transfection efficiencies and pcDNA3 to ensure equal amounts of DNA in each transfection. Forty-eight hrs after transfection, cell media and lysates were assayed for human growth hormone, β-galactosidase, and total protein by using commercially available kits (Nichols Institute, Promega, and Bio-Rad). Growth hormone levels were normalized to β-galactosidase activity per mg protein.
RESULTS
Identification, Cloning, and Subcellular Localization of FOG-2.
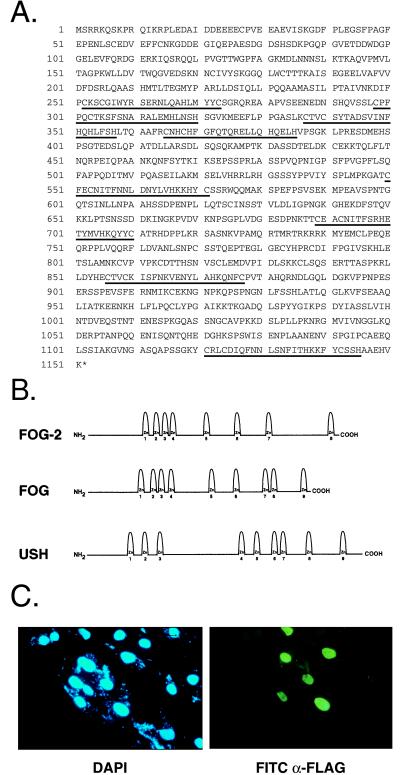
To identify novel cardiac FOG-related proteins, we screened the National Center for Biotechnology Information EST database with the coding sequence of murine FOG. One mouse EST (GenBank accession no. W12035) was identified that displayed 63% sequence identity to the region encoding zinc fingers 1 and 2 of FOG (bp 1,145 to 1,336). Screening of an E13 mouse heart cDNA library with a PCR fragment derived from this EST identified five clones, the largest of which contained a cDNA insert of 4.7 kb. By DNA sequence analysis, this clone contained a 3,453-bp ORF that was initiated by a consensus Kozak sequence (GCCGAAATGT). Conceptual translation of this ORF revealed an 1,151 amino acid polypeptide that contained an N-terminal acidic region (aa 17–111), a potential nuclear localization signal (aa 735–739), and eight zinc finger motifs that were structurally related to and organized in a similar pattern to those of both FOG and USH (Fig. 1B). Zinc fingers 1, 5, 6, and 8 were most highly conserved among the three proteins. Because of its homology to FOG and USH, we named this protein FOG-2.
Figure 1.
Predicted amino acid sequence and subcellular localization of FOG-2. (A) Predicted amino acid sequence of FOG-2. Zinc finger motifs are underlined. (B) Schematic illustration of the structures of FOG-2, FOG, and USH. Zinc fingers are numbered. (C) Subcellular localization of FOG-2. COS-7 cells transfected with pcDNAFlag/FOG-2 were assayed by indirect immunofluorescence by using an anti-FLAG antibody (Right) or stained with 4′,6-diamidino-2-phenylindole to visualize nuclei (Left).
We have shown previously that GATA-4 is localized to the nucleus (17). To determine the subcellular localization of FOG-2, COS-7 cells were transfected with an expression vector encoding a Flag epitope tag fused in frame to FOG-2. Indirect immunofluorescence by using an anti-Flag mAb demonstrated that FOG-2 was localized to the nucleus of the transfected cells (Fig. 1C). This immunofluorescence reflected specific expression of the FLAG-tagged FOG-2 protein because control cells transfected with an expression construct lacking the FOG-2 cDNA displayed only background fluorescence (data not shown). Thus, FOG-2, like GATA-4, is localized predominantly to the nucleus.
Tissue-Restricted and Developmentally Regulated Expression of FOG-2.
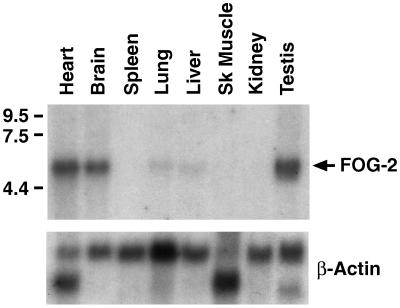
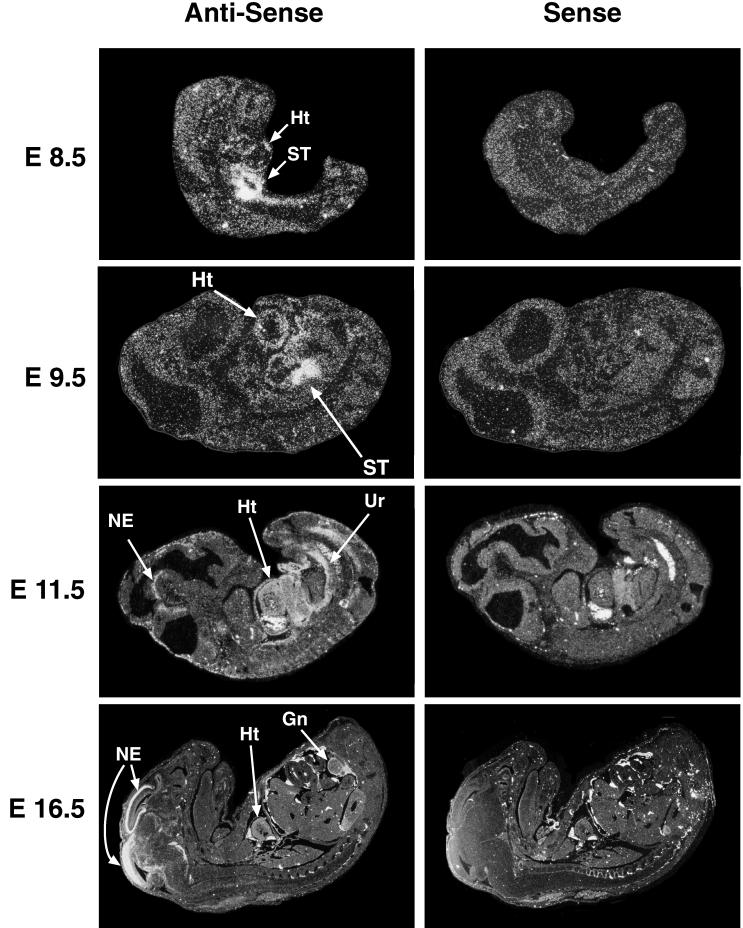
Northern blot analysis of adult mouse tissues by using a FOG-2 cDNA probe revealed a single predominant mRNA species of approximately 5 kb that is expressed predominantly in heart, brain, and testis (Fig. 2). Significantly lower levels of FOG-2 mRNA were detected in the lung and liver. To examine the pattern of FOG-2 expression during mouse embryogenesis, we performed in situ hybridizations using radiolabeled FOG-2 cRNA probes (Fig. 3). FOG-2 expression was first detectable at approximately E8.5 in the developing ventral heart tube and septum transversum. Cardiac expression persisted throughout the remainder of embryonic development with expression in the atria as well as in all layers of the ventricles (endocardium, myocardium, and pericardium). FOG-2 expression in the neuroepithelium of the developing midbrain and hindbrain was first detected at E11.5 and increased in intensity between E12 and E16.5. FOG-2 was also expressed in the urogenital ridge beginning at E11.5 and subsequently localized to the gonads by E16.5. This pattern of FOG-2 expression overlaps with the patterns of expression of GATA-4/5/6 in the developing heart, GATA-3 in the brain, and GATA-1/4 in the gonads (12–14, 33, 34).
Figure 2.
Northern analysis of FOG-2 expression. A Northern blot containing 2 μg of Poly(A)+ RNA from different adult mouse tissues was hybridized to a radiolabeled FOG-2 cDNA probe (Upper) or a β-actin cDNA probe (Lower). RNA size in kb is shown to the left of the autoradiogram. Sk muscle = skeletal muscle.
Figure 3.
In situ hybridization analysis of FOG-2 expression in mouse embryos. Sense and antisense FOG-2 cRNA probes were hybridized to sections of E8.5 to E16.5 mouse embryos. The positive signals seen in the livers, atrial cavities, and dorsal aorti of the E11.5 embryos with both sense and antisense probes represent epifluorescence of erythrocytes. Heart (Ht), Septum Transversum (ST), Neural Epithelium (NE), Urogenital Ridge (Ur), and Gonad (Gn) are shown by arrows.
FOG-2 Associates with GATA-4 in Vitro and in Vivo.
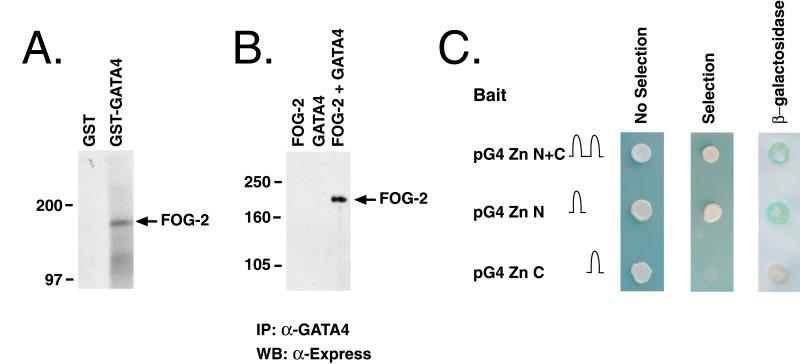
Both FOG and USH have been shown to associate specifically with the N-terminal zinc fingers of GATA-1 and Pannier, respectively (28, 29). Given the structural similarities between FOG-2, FOG, and USH (Fig. 1B), and the finding that FOG-2 is coexpressed with GATA-4 in murine cardiomyocytes, it was of interest to determine whether FOG-2 could associate physically with GATA-4 in vitro and in vivo. In an initial series of experiments, we performed in vitro binding assays using purified bacterially expressed GST/GATA-4 fusion proteins and in vitro translated 35S-labeled FOG-2. As shown in Fig. 4A, FOG-2 associated with GST/GATA-4 but failed to associate with GST protein alone. To determine whether FOG-2 could also interact with GATA-4 in mammalian cells, we performed immunoprecipitation experiments using nuclear extracts prepared from COS-7 cells that had been cotransfected with plasmids encoding FOG-2 and GATA-4. COS-7 cells were used in these experiments because they lack endogenous expression of GATA-4 (data not shown). As shown in Fig. 4B, FOG-2 and GATA-4 could be specifically coimmunoprecipitated from cells cotransfected with GATA-4 and FOG-2 expression vectors. The association between GATA-4 and FOG-2 required the expression of both proteins because it was not seen in immunoprecipitates from COS-7 cells transfected with either GATA-4 or FOG-2 expression vectors alone, despite the finding by Western blot analysis that the FOG-2 and FOG-2 + GATA-4 transfected cells expressed equivalent levels of FOG-2 (Fig. 4B and data not shown).
Figure 4.
A physical association between FOG-2 and GATA-4. (A) FOG-2 interacts with GST-GATA-4 in vitro. 35S-labeled in vitro translated FOG-2 was incubated with purified GST or GST-GATA-4 fusion proteins and the complexes were affinity purified by using glutathione-Sepharose beads and resolved by SDS/PAGE. Size markers in kDa are shown to the left of the autoradiogram. (B) FOG-2 interacts with GATA-4 in mammalian cells. COS-7 cells were transfected with an Express epitope-tagged FOG-2 expression construct (FOG-2), a GATA-4 expression construct (GATA4), or both (FOG-2 + GATA4). Nuclear extracts prepared from the transfected cells were immunoprecipitated with an α-GATA-4 antibody. Immune complexes were resolved by SDS/PAGE and analyzed by Western blot by using an α-Express antibody. Molecular masses in kDa are shown to the left of the autoradiogram. (C) FOG-2 interacts with GATA-4 in yeast. Yeast 2 hybrid assays using expression vectors encoding the N-terminal (pG4 Zn N), C-terminal (pG4 Zn C), or both (pG4 Zn N + C) zinc fingers of GATA-4 fused to the DNA binding domain of GAL4 and an expression vector encoding FOG-2 fused to the activation domain of GAL4. Note that all transformants grew under nonselective conditions (No selection). Positive interactions are indicated by yeast growth under selection conditions (Selection) and blue staining indicating β-galactosidase activity (β-galactosidase).
To confirm the interaction between FOG-2 and GATA-4 in eukaryotic cells and to better map the sites of GATA-4 that interact with FOG-2, we performed a series of yeast 2-hybrid assays using plasmids encoding (1) the DNA-binding domain of GAL4 fused to different regions of the GATA-4 cDNA, and (2) a fragment of the FOG-2 cDNA (aa 236–1,151) fused to the transactivation domain of GAL4. As shown in Fig. 4C, FOG-2 interacted with the fusion protein produced by pG4ZnN+C that encodes both the C- and N-terminal zinc fingers of GATA-4 and with the fusion protein encoded by pG4ZnN that contains only the N-terminal zinc finger of GATA-4. In contrast, FOG-2 failed to interact with the fusion protein encoded by pG4ZnC containing only the C-terminal zinc finger of GATA-4. In similar experiments, FOG-2 was also shown to bind specifically to the N-terminal zinc finger of GATA-3 (data not shown). Thus, using three different assays, we were able to demonstrate a specific interaction between FOG-2 and GATA-4 both in vitro and in vivo and to map this interaction to the N-terminal zinc finger of GATA-4.
FOG-2 Represses GATA-4-Dependent Transcription in Cultured Fibroblasts and Cardiomyocytes.
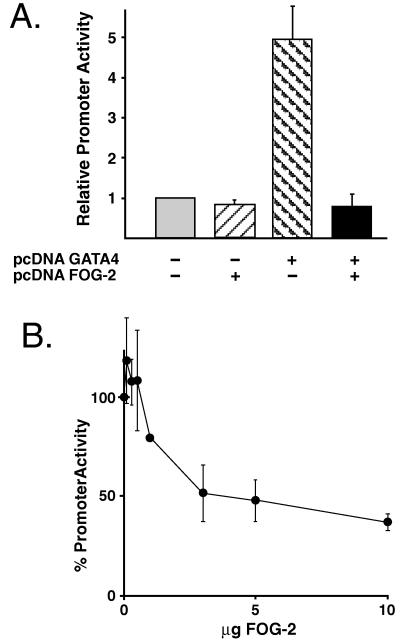
FOG-related proteins have been reported to function as both coactivators and repressors of GATA-dependent transcription (28, 29). Thus, it was of interest to study the effects of the FOG-2/GATA-4 interaction on GATA-4-dependent transcription. In an initial series of experiments, NIH 3T3 cells were transfected with a human growth hormone (hGH) reporter construct under the control of the GATA-dependent BNP promoter along with GATA-4 and FOG-2 expression plasmids (Fig. 5A). Expression of FOG-2 alone had little effect on the basal activity of the BNP promoter in NIH 3T3 cells. In contrast, and consistent with previous reports, we observed an approximately 5-fold transactivation of the BNP promoter by GATA-4 alone (19). Cotransfection of equivalent amounts of GATA-4 and FOG-2 expression plasmids completely inhibited this GATA-4-dependent activation of the BNP promoter (Fig. 5A). Similar and complete inhibition of GATA-4-dependent transactivation by FOG-2 was observed by using reporter constructs containing 3 kb of the cardiac troponin I promoter or 124 bp of the cardiac troponin C promoter (data not shown). This inhibition of GATA-4-dependent transcription did not reflect a generalized inhibition of transcription by FOG-2 because FOG-2 expression did not inhibit the activity of the pVRβ-gal reporter plasmid that was included as a control in each transfection (data not shown). To demonstrate directly that the GATA-4–FOG-2 interaction is required for the ability of FOG-2 to inhibit GATA-4-dependent transcription, we produced a mutant GATA-4 protein containing a single amino acid substitution in the N-terminal zinc finger of GATA-4 (E215 → K), which specifically blocks the ability of GATA-4 to interact with FOG-2. Overexpression of this mutant GATA-4 protein in NIH 3T3 cells transactivated the BNP promoter to the same degree as wild-type GATA-4. However, unlike wild-type GATA-4, the transcriptional activity of the (E215 → K) mutant was not inhibited by overexpression of FOG-2 (data not shown).
Figure 5.
FOG-2 represses GATA-4-mediated transcription from the BNP promoter. (A) NIH 3T3 cells were transfected with a BNP promoter–hGH reporter construct along with GATA-4 and FOG-2 expression constructs as indicated. (B) Primary neonatal rat cardiomyocytes were cotransfected with the BNP promoter–hGH reporter construct along with increasing amounts of a FOG-2 expression plasmid. The data are presented as relative promoter activity normalized to the activity of the reporter plasmid alone in each cell type. Each point represents the mean ± SEM from at least three independent transfections.
To determine the effects of FOG-2 on GATA-4-dependent transcription in cardiomyocytes, we cotransfected primary neonatal rat cardiomyocytes with the rat BNP promoter–hGH reporter construct and increasing amounts of the FOG-2 expression plasmid. Expression of FOG-2 resulted in a dose-dependent inhibition of BNP promoter activity in cardiomyocytes (Fig. 5B). Similar results were obtained by using a cardiac troponin C promoter–hGH reporter construct (data not shown). Thus, FOG-2 by interacting with the N-terminal zinc finger of GATA-4 appears to repress GATA-4-dependent transcription from a variety of cardiac-restricted promoters in both cultured fibroblasts and cardiomyocytes.
DISCUSSION
In this report, we have identified and characterized a zinc finger protein, FOG-2, which is structurally related to both the GATA-1-associated protein, FOG, and the Pannier-associated protein, USH. FOG-2 is coexpressed with GATA-4 in both embryonic and adult cardiomyocytes, interacts specifically with the N-terminal zinc finger of GATA-4, and represses GATA-4-dependent transcription in both cultured fibroblasts and neonatal cardiomyocytes. Together, these findings suggest a general paradigm in which evolutionarily conserved and tissue-specific interactions between different FOG and GATA proteins regulate the development of distinct mesodermal cell lineages.
The pattern of expression of FOG-2 suggests that it may interact with and modulate the activities of different GATA factors in different tissues. Coexpression of FOG-2 with GATA-4/5/6 in the heart suggests the possibility of important functional interactions between these proteins in both embryonic and adult cardiomyocytes. Similarly, FOG-2 may interact with GATA-1/4/6 in the urogenital tissues and with GATA-3 in the brain. Consistent with such a model, we have found that FOG-2 is capable of interacting with and repressing the activities of both GATA-3 and GATA-6 (data not shown). Of the eight zinc finger motifs of FOG-2, fingers 1, 5, 6, and 8 are most highly conserved among all three members of the family and may therefore be important in mediating some of the conserved functions of these proteins. For example, it has been suggested that finger 6 of FOG is sufficient for binding to GATA-1 (28). Similarly, our preliminary results suggest that either finger 1 or finger 6 of FOG-2 is sufficient for association with GATA-4 (data not shown).
Like USH, which has been reported to inhibit the transcriptional activity of Pannier, FOG-2 appears to repress the transcriptional activity of GATA-4 in both cultured fibroblasts and cardiomyocytes. This repressor activity would appear to distinguish FOG-2 and USH from FOG, which has been reported to function as a coactivator of GATA-1 activity both in vitro and in vivo (28, 30). However, the situation may be more complex than initially imagined in that we have shown that FOG, like FOG-2, represses GATA-4-dependent transcription from the BNP promoter after cotransfection of both NIH 3T3 cells and primary cardiomyocytes (data not shown). Thus, the effects of the FOG-related proteins on GATA-dependent transcription may depend on both the cell types in which they are expressed and on the promoter contexts in which they function. A definitive understanding of the function of FOG-2 in regulating cardiomyocyte-specific gene expression and cardiac development in vivo will await the analysis of FOG-2-deficient mice produced by ongoing gene-targeting experiments. Because FOG-2 would be expected to interact with all three cardiac GATA factors (GATA-4/5/6), such gene-targeting experiments, by simultaneously ablating the activity of all GATA proteins in procardiomyoctes, may also shed light on the potentially redundant roles of GATA factors in cardiomyocyte lineage determination and cardiogenesis.
The mechanism(s) by which FOG-2 represses the transcriptional activity of GATA-4 remains unclear. FOG-2 may associate with GATA-4 and prevent it from binding to its cognate sites on cardiomyocyte gene promoters. Alternatively, FOG-2 may interact with DNA-bound GATA-4, thereby directly blocking the ability of GATA-4 to activate transcription. Finally, it is possible that FOG-2 can bind one or more repressor proteins and can thereby serve as an adapter that brings such repressors together with GATA-4 on DNA. In this regard, it is of interest that FOG has recently been shown to bind the potent transcriptional repressor, mCtBP2 (35). Interestingly, FOG-2, like FOG, contains a consensus mCtBP2 binding site (PIDLS, aa 829–833). Ongoing experiments are underway to distinguish these possible mechanisms of action.
Acknowledgments
We thank Jiang Fang for her help with the in situ hybridizations and Ed Morrisey, Hon Ip, and Michael Parmacek for their critical and insightful discussions. We also thank Denise Wiler for help in preparing the figures. We thank Stuart Orkin for sharing his results on FOG-2 before publication. This work was supported in part by a grant from Roche Pharmaceuticals to E.C.S. and by a grant from the National Institutes of Health to J.M.L. (HL54592).
ABBREVIATIONS
- BNP
B-type natriuretic peptide
- USH
U-shaped
- FOG
Friend of GATA-1
- hGH
human growth hormone
- EST
expressed sequence tag
- GST
glutathione S-transferase
- En
embryonic day n
Footnotes
Data Deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF118845).
References
- 1.Rehorn K P, Thelen H, Michelson A M, Reuter R. Development (Cambridge, UK) 1996;122:4023–4031. doi: 10.1242/dev.122.12.4023. [DOI] [PubMed] [Google Scholar]
- 2.Ramain P, Heitzler P, Haenlin M, Simpson P. Development (Cambridge, UK) 1993;119:1277–1291. doi: 10.1242/dev.119.4.1277. [DOI] [PubMed] [Google Scholar]
- 3.Orkin S H. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- 4.Simon M C. Nat Genet. 1995;11:9–11. doi: 10.1038/ng0995-9. [DOI] [PubMed] [Google Scholar]
- 5.Zhu J, Hill R J, Heid P J, Fukuyama M, Sugimoto A, Priess J R, Rothman J H. Genes Dev. 1997;11:2883–2896. doi: 10.1101/gad.11.21.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merika M, Orkin S H. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko L J, Engel J D. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pevny L, Simon M C, Robertson E, Klein W H, Tsai S F, D’Agati V, Orkin S H, Costantini F. Nature (London) 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 9.Tsai F Y, Keller G, Kuo F C, Weiss M, Chen J, Rosenblatt M, Alt F W, Orkin S H. Nature (London) 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 10.Ting C N, Olson M C, Barton K P, Leiden J M. Nature (London) 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- 11.Laverriere A C, MacNeill C, Mueller C, Poelmann R E, Burch J B, Evans T. J Biol Chem. 1994;269:23177–84. [PubMed] [Google Scholar]
- 12.Heikinheimo M, Scandrett J M, Wilson D B. Dev Biol. 1994;164:361–373. doi: 10.1006/dbio.1994.1206. [DOI] [PubMed] [Google Scholar]
- 13.Morrisey E E, Ip H S, Lu M M, Parmacek M S. Dev Biol. 1996;177:309–322. doi: 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- 14.Morrisey E E, Ip H S, Tang Z, Lu M M, Parmacek M S. Dev Biol. 1997;183:21–36. doi: 10.1006/dbio.1996.8485. [DOI] [PubMed] [Google Scholar]
- 15.Olson E N, Srivastava D. Science. 1996;272:671–676. doi: 10.1126/science.272.5262.671. [DOI] [PubMed] [Google Scholar]
- 16.Fishman M C, Chien K R. Development (Cambridge, UK) 1997;124:2099–2117. doi: 10.1242/dev.124.11.2099. [DOI] [PubMed] [Google Scholar]
- 17.Ip H S, Wilson D B, Heikinheimo M, Tang Z, Ting C N, Simon M C, Leiden J M, Parmacek M S. Mol Cell Biol. 1994;14:7517–7526. doi: 10.1128/mcb.14.11.7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grepin C, Dagnino L, Robitaille L, Haberstroh L, Antakly T, Nemer M. Mol Cell Biol. 1994;14:3115–3129. doi: 10.1128/mcb.14.5.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molkentin J D, Kalvakolanu D V, Markham B E. Mol Cell Biol. 1994;14:4947–4957. doi: 10.1128/mcb.14.7.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thuerauf D J, Hanford D S, Glembotski C C. J Biol Chem. 1994;269:17772–17775. [PubMed] [Google Scholar]
- 21.Molkentin J D, Lin Q, Duncan S A, Olson E N. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 22.Kuo C T, Morrisey E E, Anandappa R, Sigrist K, Lu M M, Parmacek M S, Soudais C, Leiden J M. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 23.Merika M, Orkin S H. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee Y, Shioi T, Kasahara H, Jobe S M, Wiese R J, Markham B E, Izumo S. Mol Cell Biol. 1998;18:3120–3129. doi: 10.1128/mcb.18.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sepulveda J L, Belaguli N, Nigam V, Chen C Y, Nemer M, Schwartz R J. Mol Cell Biol. 1998;18:3405–3415. doi: 10.1128/mcb.18.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molkentin J D, Lu J R, Antos C L, Markham B, Richardson J, Robbins J, Grant S R, Olson E N. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durocher D, Charron F, Warren R, Schwartz R J, Nemer M. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsang A P, Visvader J E, Turner C A, Fujiwara Y, Yu C, Weiss M J, Crossley M, Orkin S H. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 29.Haenlin M, Cubadda Y, Blondeau F, Heitzler P, Lutz Y, Simpson P, Ramain P. Genes Dev. 1997;11:3096–3108. doi: 10.1101/gad.11.22.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsang A P, Fujiwara Y, Hom D B, Orkin S H. Genes Dev. 1998;12:1176–1188. doi: 10.1101/gad.12.8.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bassuk A G, Anandappa R T, Leiden J M. J Virol. 1997;71:3563–3573. doi: 10.1128/jvi.71.5.3563-3573.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tripathy S K, Svensson E C, Black H B, Goldwasser E, Margalith M, Hobart P M, Leiden J M. Proc Natl Acad Sci USA. 1996;93:10876–80. doi: 10.1073/pnas.93.20.10876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.George K M, Leonard M W, Roth M E, Lieuw K H, Kioussis D, Grosveld F, Engel J D. Development (Cambridge, UK) 1994;120:2673–2686. doi: 10.1242/dev.120.9.2673. [DOI] [PubMed] [Google Scholar]
- 34.Viger R S, Mertineit C, Trasler J M, Nemer M. Development (Cambridge, UK) 1998;125:2665–2675. doi: 10.1242/dev.125.14.2665. [DOI] [PubMed] [Google Scholar]
- 35.Turner J, Crossley M. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]