Abstract
Tropheryma whipplei, the agent of Whipple's disease, grows fastidiously only in cell cultures without plaque production, and only three strains have been passaged. The formation of bacterial clumps in the supernatant precludes enumeration of viable bacteria and MIC determination. We evaluated the bacteriostatic effects of fluoroquinolones against two T. whipplei isolates by measuring the inhibition of the DNA copy number increase by real-time quantitative PCR. The analysis of the T. whipplei genome database allowed the identification not only of the gyrA gene but also the parC gene encoding the alpha subunit of the natural fluoroquinolone targets DNA gyrase (GyrA) and topoisomerase IV (ParC), respectively. The parC gene was detected in actinobacteria for the first time. High ciprofloxacin MICs (4 and 8 μg/ml) were correlated with the presence in T. whipplei GyrA and ParC sequences with an alanine residue at positions 83 and 80 (Escherichia coli numbering), respectively. Alanines at these positions have previously been associated with increased fluoroquinolone resistance in E. coli and mycobacteria. However, the MIC of levofloxacin was low (0.25 μg/ml). The same T. whipplei GyrA and ParC sequences were found in two other cultured strains and in nine uncultured tissue samples from Whipple's disease patients, allowing one to speculate that T. whipplei is naturally relatively resistant to fluoroquinolones.
Tropheryma whipplei is the agent of Whipple's disease. Because T. whipplei cannot be grown in axenic medium, the present methodologies for antibiotic susceptibility testing of extracellular bacteria were not applicable. T. whipplei is extremely fastidious and resisted reproducible culture until 2000, when it was propagated in human fibroblasts (25). To the best of our knowledge, only three strains have been passaged more than five times (unpublished data). In the fibroblast cell system, it does not produce cytopathic effects such as plaque formation, and bacterial growth results in the formation of ropes of cells in the cell culture supernatant (18). These ropes consist of large clumps of bacteria that are very difficult to disperse without altering cell viability. The formation of similar cords has been described for Mycobacterium tuberculosis (2, 33), which, together with T. whipplei, belongs to the phylum Actinobacterium, the members of which are gram-positive organisms with high G+C contents.
To overcome these technical difficulties, the antibiotic susceptibilities of three human strains of T. whipplei were determined by a strict molecular approach. The MICs of two fluoroquinolone compounds (which were used as examples) for T. whipplei were determined by a quantitative PCR-based method that measured the inhibition of the increase in the number of bacterial DNA copies, as reported for rickettsiae (27).
At the same time, two type II topoisomerase genes were identified in the T. whipplei genome These corresponded to putative gyrA and parC genes. We tentatively correlated fluoroquinolone susceptibility with specific target gene sequences and detected T. whipplei gyrA and parC in uncultured samples from patients with Whipple's disease by PCR and sequencing. Specific mutations in DNA sequences leading to amino acid substitutions have been associated with resistance both by experimental mutagenesis techniques and in clinical strains. A well-characterized example includes the fluoroquinolones and type II topoisomerase-mediated resistance. Type II topoisomerases, including DNA gyrase and topoisomerase IV, are natural targets for fluoroquinolones (14). Fluoroquinolone resistance has been associated in many bacterial species with the presence of specific amino acids at critical positions in the quinolone resistance-determining region (QRDR) of GyrA and ParC, the proteins encoded by gyrA and parC, respectively (4, 6, 14, 21, 24, 34). This allows MIC determination by genomic detection of resistance to be replaced by techniques such as DNA hybridization, PCR sequencing, and PCR-restriction fragment length polymorphism analysis (20). The purpose of this work was to evaluate the susceptibilities of T. whipplei to quinolones and to determine the correlation of resistance with a DNA target enzyme. The detection of these target DNAs was later performed with uncultured samples from patients with Whipple's disease.
MATERIALS AND METHODS
T. whipplei strains.
Twelve T. whipplei strains were studied (Table 1), including three isolates obtained in our laboratory in an HEL cell culture system and nine uncultured strains whose DNA was extracted from biopsy samples from Whipple's disease patients. The T. whipplei Twist strain, isolated from a cardiac valve sample (18, 25), was cocultivated with MRC5 fibroblast cells until its 35th passage before antibiotic susceptibility testing. T. whipplei Endo2, grown from a cardiac valve sample from another endocarditis patient, was tested after 11 passages in cell culture. T. whipplei Slow2, grown from a duodenal biopsy specimen, was tested after 16 passages. The absence of Mycoplasma sp. contamination of cell cultures was checked weekly by using the Mycoplasma detection kit (Boehringer Mannheim GmbH, Mannheim, Germany).
TABLE 1.
T. whipplei strain and patient characteristics
| Patients sex/age (yr) | Strain (genotype)a | Symptoms | Biopsy sample | Antibiotic(s) taken before sampling |
|---|---|---|---|---|
| Male/42 | Twist (2A) | Endocarditis | Cardiac valve | Gentamicin + ampicillin for 4 wk, ciprofloxacin several for 4 weeks |
| Female/74 | Slow2 (1A) | Diarrhea, arthralgia | Duodenum | None |
| Male/63 | Endo2 (1A) | Endocarditis | Cardiac valve | Gentamicin + amoxicillin for 3 wk |
| Male/70 | Unc (ND) | Fever, weight loss, lymphadenopathy, encephalitis | Duodenum | None |
| Male/63 | Unc (1A) | Diarrhea, weight loss | Duodenum | None |
| Male/69 | Unc (1A) | Diarrhea, arthralgia | Duodenum | None |
| Female/33 | Unc (1A) | Fever, weight loss, lymphadenopathy | Duodenum | Co-trimoxazole for 1 year |
| Male/47 | Unc (1A) | Fever, diarrhea, weight loss | Duodenum | Co-trimoxazole for 1 year |
| Male/60 | Unc (2A) | Diarrhea, arthralgia | Duodenum | None |
| Female/69 | Unc (ND) | Arthralgia, pulmonary embolism | Duodenum | None |
| Male/58 | Unc (2A) | Diarrhea, arthralgia | Duodenum | None |
| Male/64 | Unc (2A) | Endocarditis | Cardiac valve | None |
ND, not determined; Unc, uncultured.
Antibiotic solutions.
The antibiotics tested were ciprofloxacin (Bayer Pharma, Sens, France) and levofloxacin (Hoechst-Marion-Roussel, Romainville, France) at twofold serial concentrations ranging from 0.25 to 8 μg/ml. Stock solutions were prepared according to the instructions of the manufacturers and stored at −80°C until use. Working solutions were prepared by dilution of stock solutions in minimum essential medium.
Growth kinetics of T. whipplei Twist, Slow2, and Endo2.
T. whipplei-infected MRC5 cells were grown in a 150-cm2 culture flask (Falcon; Beckman) by incubation at 37°C in a 5% CO2-enriched atmosphere. Minimum essential medium (Life Technologies, GIBCO BRL, Cergy Pontoise, France) supplemented with 10% fetal calf serum and 2 mmol of l-glutamine (Gibco) was used as the incubation medium. Cellular infection was monitored twice a week by scraping infected cells from the culture flasks and microscopic examination of cell smears stained by the Gimenez technique. When heavy infection was detected (usually after ∼1 month of incubation), the cell supernatant was discarded and infected MRC5 cells were detached by using sterile glass beads with 5 ml of fresh medium. The cells were lysed by sonication (three times for 30 s each time on ice at 60 mV), and the resulting bacterial inoculum was diluted 1:100 in culture medium and used to infect confluent MRC5 cell monolayers in 24-well microtiter plates (D. Dutcher, Brumath, France) (0.2 ml of inoculum in 2 ml of medium per well). The plates were incubated at 37°C in 5% CO2. Infected cells in three different wells were harvested every 3 days from day 0 to day 21 postinfection. The growth kinetics of T. whipplei in MRC5 cells were determined by enumeration of the genome copies in each cell suspension by quantitative PCR.
Antibiotic susceptibility testing of T. whipplei by quantitative PCR.
Confluent MRC5 cell monolayers in 24-well microtiter plates were infected with a T. whipplei inoculum as described above. T. whipplei strains Twist, Slow2, and Endo2 were tested for their fluoroquinolone susceptibilities. After incubation of the cultures for 48 h, antibiotics at various concentrations were added to the culture medium (50 μl of 40 times the desired final concentration in 2 ml of medium). Antibiotic-free wells served as growth controls, whereas uninfected MRC5 cells served as negative controls. During antibiotic susceptibility testing, cell cultures were harvested every 3 days (three wells per antibiotic concentration tested) for a total of 12 days, and cell suspensions were frozen at −80°C until DNA extraction for quantitative PCR assays. The lack of toxicity of antibiotics to MRC5 cells was controlled by examination of cell monolayers under an inverted microscope at the time that the cell cultures were harvested. MICs were defined as the minimal antibiotic concentrations that allowed complete inhibition of DNA growth, as measured by quantitative PCR assay.
Escherichia coli ATCC 8739 and Staphylococcus aureus CIP ATCC 49976 were obtained from the Pasteur Institute (Institut Pasteur, Marnes La Coquette, France) and were used as controls for antibiotic susceptibility testing. The activities of ciprofloxacin and levofloxacin were determined on Mueller-Hinton agar (bioMérieux) incubated at 37°C for 18 h. The MICs were in the range of those reported by the Pasteur Institute. The antibiotic activities of the dilutions were checked after 30 days of incubation at 37°C by using E. coli and S. aureus.
Quantitative PCR assay with the LightCycler instrument.
T. whipplei DNA was extracted from MRC5 cells infected in vitro by a previously described protocol (12). Infected MRC5 cell suspensions were centrifuged at 20,000 × g for 15 min at 4°C. After the supernatants were carefully removed, the cell pellets were resuspended in 200 μl of digestion buffer (50 mM Tris-HCl [pH 8.5], 1 mM EDTA, 0.5% sodium dodecyl sulfate, 200 μg of proteinase K per ml) and incubated for 90 min at 55°C. The DNA was than extracted by using QIAamp DNA binding columns (Qiagen, Courtaboeuf, France) and stored at −20°C until it was used for amplification by quantitative PCR assay.
DNA extracts were amplified by the LightCycler PCR assay (Roche Diagnostics) in glass capillaries (volume 20 μl) with primers that allowed amplification of the T. whipplei intergenic spacer (ITS) region (7). These primers were chosen to allow amplification of one copy of target gene per bacterium. The PCR mixture (20 μl) contained 2 μl of extracted DNA, 13.2 μl of H2O, 1.6 μl of MgCl2 (25 mmol), 1 μl (i.e., 10 pmol) of forward primer TwITSF (Table 2), 1 μl (i.e., 10 pmol) of reverse primer TwITSR (Table 2), and 2 μl of DNA-Master Hybridization Probes (Roche Diagnostics) containing Taq DNA polymerase, reaction buffer, a deoxynucleoside triphosphate mixture, and 10 mmol MgCl2 (concentrated 10 times). Cycling conditions consisted of an initial denaturation at 95°C for 8 min, followed by 40 cycles with denaturation at 95°C for 15 s, annealing at 56°C for 5 s, and extension at 72°C for 8 s, with a ramping time of 20°C/s. A calibration curve for DNA quantification was determined by amplifying 10-fold serial dilutions of DNA extracted from the primary T. whipplei inoculum used to infect MRC5 cells.
TABLE 2.
Oligonucleotide primers used for PCR amplification and sequencing
| Gene | Primers | Nucleotide sequence |
|---|---|---|
| its | TwITSF | 5′-CCGAGGCTTATCGCAGATTG-3′ |
| TwITSR | 5′-GGTGACTTAACCTTTTTGGAG-3′ | |
| gyrA QRDR | gyrAF1 | 5′-GATGGCTTAAAGCCCGTACA-3′ |
| gyrAR1 | 5′-CGACGGCTATTCCGC-3′ | |
| parC QRDR | parCF1 | 5′-GATGGCTTAAAGCCCGTACA-3′ |
| parCR1 | 5′-CGACGGCTATTCCGC-3′ | |
| gyrA QRDR | GyrAF2 | 5′-CGGGCATTGCCAGACGC-3′ |
| GyrAR2 | 5′-CGGGGGTATACTGGTTGCC-3′ | |
| parC QRDR | ParCF2 | 5′-CGGGCATTGCCAGACGC-3′ |
| parCR2 | 5′-CGGGGGTATACTGGTTGCC-3′ |
Identification of gyrA and parC in the T. whipplei Twist genome.
Two gyrA-like sequences were identified in the T. whipplei Twist genome by comparison (1) with known gyrA sequences deposited in GenBank with the BLAST program. Their open reading frames (ORFs) were determined by using the ORF finder program (28). Their G+C contents were determined by using the FREQBNK program (INFOBIOGEN, Evry, France). These ORFs were defined as putative gyrA and parC genes, which encode the A subunits of DNA gyrase and topoisomerase IV, respectively, by determination of the best sequence homologies of their protein counterparts (GyrA and ParC) with the same protein sequences from Mycoplasma pneumoniae and M. tuberculosis. T. whipplei GyrA was also differentiated from ParC by constructing a phylogenetic tree with known GyrA and ParC sequences by using MEGA (Molecular Evolutionary Genetics Analysis) software (version 2.1) and the neighbor-joining method (amino acid proportion distance, with bootstrap testing). The GyrA sequences used were those of E. coli K-12 (16130166), Haemophilus influenzae (16271976), Pseudomonas aeruginosa (15595198), Rickettsia conorii (15892196), Rickettsia prowazekii (3860770), Neisseria gonorrhoeae (2120873), Neisseria meningitidis (15794492), Bartonella bacilliformis (18920720), Agrobacterium tumefaciens (17933925), Mycobacterium leprae (15826871), Mycobacterium tuberculosis (13879047), Mycobacterium smegmatis (1346234), Streptomyces coelicolor (1093585), Treponema pallidum (15639000), Enterococcus faecalis (15982568), Streptococcus pneumoniae (15903142), Listeria monocytogenes (16802048), S. aureus (21203170), Clostridium perfringens (18143657), Bacillus subtilis (16077075), Corynebacterium urealyticum (21518737), M. pneumoniae (13507743), Ureaplasma urealyticum (13357558), Chlamydia trachomatis (13627049), and Chlamydia pneumoniae (12644500). The ParC sequences used were those of H. influenzae (16271976), E. coli K-12 (16130915), P. aeruginosa (4176378), R. prowazekii (3860637), R. conorii (15891923), N. gonorrhoeae (2120882), N. meningitidis (15794693), A. tumefaciens (17739557), S. pneumoniae (20530728), L. monocytogenes (21328243), E. faecalis (15982571), S. aureus (21204410), C. perfringens (18311050), B. subtilis (3914271), C. urealyticum (21518734), M. pneumoniae (13507739), and U. urealyticum (13357558).
Determination of gyrA and parC QRDRs in T. whipplei Twist.
The T. whipplei Twist gyrA and parC QRDRs were defined by best alignment with known homologous QRDR sequences from other species, including E. coli K-12 (16127994), M. tuberculosis (13879047), M. leprae (1041443), Mycobacterium avium (1041431), Mycobacterium intracellulare (2815375), M. smegmatis (1346234), and Mycobacterium fortuitum (2815371).
Determination of gyrA and parC QRDR sequences in an additional 11 samples and comparison of their protein counterparts with the BLAST program.
T. whipplei DNA was extracted as described above from MRC5 cells infected with strain Slow2 or Endo2 and from eight duodenal biopsy specimens and one cardiac valve specimen obtained from patients with Whipple's disease (Table 1). Tissue samples were preincubated in 200 μl of digestion buffer at 55°C for 3 h. T. whipplei DNA in biopsy samples was previously demonstrated by amplification and sequencing of the T. whipplei rpoB and its genes (5).
The extracted DNA was used to amplify the T. whipplei gyrA QRDR with primers gyrAF1 and gyrAR1 (Table 2) or the T. whipplei parC QRDR with primers parCF1 and parCR1 (Table 2). The sequences of these primers were defined from the T. whipplei Twist gyrA and parC sequences. PCRs were performed with a Perkin-Elmer 9600 thermocycler under the following conditions: a first denaturation step at 95°C for 2 min; 40 three-step cycles of 94°C for 30 s, 48°C for 30 s for primers gyrAF1 and gyrAR1 or 55°C for 30 s for primers parCF1 and parCR1, and 68°C for 1 min; and a final 6-min extension step at 68°C.
DNA sequencing was performed with an internal dRhodamine terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase (FS; Perkin-Elmer Applied Biosystems, Warrington, United Kingdom), according to the instructions of the manufacturer. The 3′ and 5′ ends of the amplified fragments obtained in the different PCR assays were sequenced after precipitation and purification with 70% ethanol and 0.5 mmol MgCl2. The primers used were gyrAF2 and gyrAR2 for the gyrA QRDR sequences and parCF2 and parCR2 for the parC QRDR sequences (Table 2). The cycle sequencing reaction mixtures comprised 4 μl of ready reaction mixture, 1 μl (i.e., 10 pmol) of forward primer for direct DNA strand sequencing or 1 μl (i.e., 10 pmol) of reverse primer for cDNA strand sequencing, and 4 μl (i.e., 200 ng) of template DNA. The mixture was brought to 10 μl with deionized water. Amplification was performed by use of 30 cycles of 95°C for 20 s, 50°C for 10 s, and 60°C for 2 min. Electrophoresis was performed with an ABI PRISM 310 genetic analyzer (Perkin-Elmer).
The DNA and amino acid sequences of the QRDRs obtained for the 12 T. whipplei strains studied were aligned and compared by using the CLUSTAL multialignment package (11).
RESULTS
T. whipplei growth kinetics.
Growth of T. whipplei isolates was exponential, with a 2.7 ± 1.9 log increase in bacterial concentration over 12 days of incubation for strain Twist, a 2.25 ± 0.98 log increase for strain Endo2, and a 1.96 ± 1.16 log increase for strain Slow2. During this period, the doubling time of T. whipplei Twist was found to be between 32 and 36 h, whereas that of Endo2 was between 34 and 38 h and that of strain Slow2 was between 41 and 48 h. Bacterial growth then reached a plateau.
Fluoroquinolone susceptibility testing of T. whipplei Twist, Slow2, and Endo2 strains.
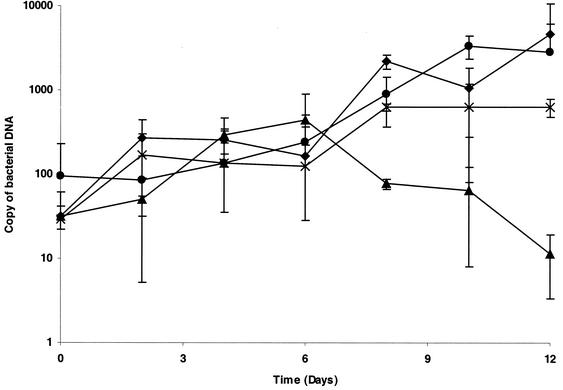
As determined by quantitative PCR assay, increases in the number of DNA copies of T. whipplei Twist, Slow2, and Endo2 were completely abolished with ciprofloxacin at 4 or 8 μg/ml (Fig. 1) but not with ciprofloxacin at 1 or 2 μg/ml. Thus, the MIC of ciprofloxacin for T. whipplei was evaluated to be 4 μg/ml. Levofloxacin was more effective, with MICs ranging from 0.25 to 0.5 μg/ml. The antibiotic dilution was stable for 30 days and had the same activity against control strains.
FIG. 1.
Growth kinetics and susceptibilities to fluoroquinolones of T. whipplei Endo2. Bacterial loads were determined by enumeration of the bacterial DNA copies by quantitative PCR. Comparable results were found for the T. whipplei Twist and Slow2 strains. ⧫, growth control; •, ciprofloxacin at 1 μg/ml; ∗, ciprofloxacin at 4 μg/ml; ▴, ciprofloxacin at 8 μg/ml.
T. whipplei Twist gyrA and parC.
The putative T. whipplei gyrA sequence is a 2,460-bp ORF (AE014184). The start codon, TTG, was determined with the ORF finder (28). The ORF extends to a stop codon, TAG, at nucleotide 2,460. The ITS G+C content is 49.3%. Translation of the ORF corresponds to a putative 819-amino-acid (aa) protein, with a calculated molecular mass of 90,668 kDa, comparable to that of S. coelicolor GyrA (i.e., 818 aa with a calculated molecular mass of 88.586 kDa) (3).
The putative T. whipplei parC ORF is 2,475 bp (GenBank accession number AE014184). The start codon, TTG, was determined with the ORF finder, as described above (28). The ORF extends to a stop codon, TAG, at nucleotide 2475. The ITS G+C content is 43.92%. Translation of the ORF corresponds to an 824-aa protein, with a calculated molecular mass of 91.190 kDa.
For the T. whipplei GyrA sequence, we found identities with the M. pneumoniae GyrA and ParC sequences of 40 and 29%, respectively, and identity with the M. tuberculosis GyrA sequence of 54%. For the T. whipplei ParC sequence, we found identities with the M. pneumoniae GyrA and ParC sequences of 33 and 38%, respectively, and identity with the M. tuberculosis GyrA sequence of 34%.
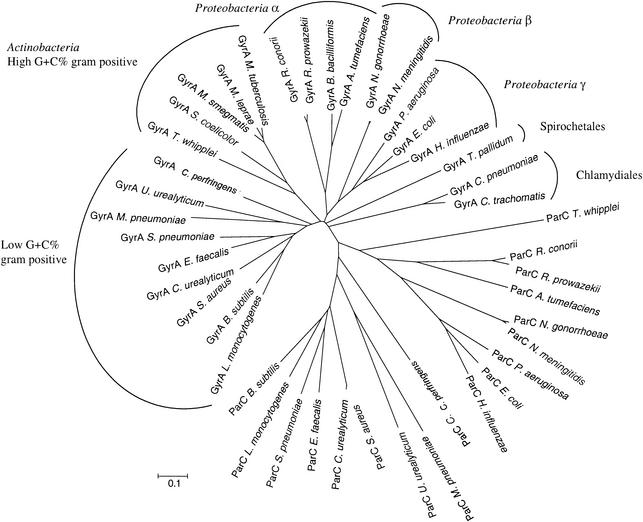
Also, a phylogenetic tree constructed with known GyrA and ParC protein sequences clearly distinguished the two types of protein sequences and allowed differentiation of putative GyrA and ParC sequences in T. whipplei (Fig. 2).
FIG. 2.
Phylogenetic tree of GyrA and ParC sequences from T. whipplei and various representative species of the domain Bacteria. Protein sequences were aligned by using the ClustalW program from the Pôle BioInformatique Lyonnais (Lyon, France). The tree was constructed with MEGA software (version 2.1) by the neighbor-joining methodology (amino acid p-distance, including site, complete deletion, and bootstrap testing).
Determination of gyrA and parC QRDRs in T. whipplei Twist.
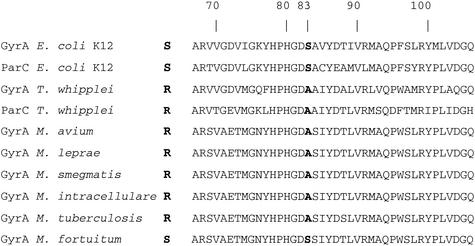
The T. whipplei GyrA and ParC QRDRs are presented in Fig. 3. The T. whipplei GyrA QRDR extends from the alanine at position 65 to the glutamine at position 104. Its ParC QRDR extends from the alanine at position 80 to the histidine at position 119. An alanine residue was found at position 81 of the T. whipplei GyrA QRDR, corresponding to the serine at position 83 in the E. coli GyrA sequence. An alanine residue was also found at position 96 of the T. whipplei ParC QRDR, corresponding to the serine at position 80 in the E. coli ParC sequence (19).
FIG. 3.
T. whipplei GyrA and ParC QRDR alignment with homologous sequences from E. coli K-12 (16127994), M. tuberculosis (13879047), M. leprae (1041443), M. avium (1041431), M. intracellulare (2815375), M. smegmatis (1346234), and M. fortuitum (2815371).
Determination of gyrA and parC QRDRs in an additional 11 T. whipplei strains.
The primers whose sequences were defined from the T. whipplei Twist gyrA and parC sequences allowed us to amplify and sequence the gyrA and parC QRDRs from two other T. whipplei strains cultured (i.e., strains Endo2 and Slow2), as well as from the T. whipplei DNA contained in uncultured digestive or cardiac valve biopsy specimens from nine additional patients with Whipple's disease.
For the gyrA QRDR, a 460-bp DNA fragment was amplified from the 11 additional specimens tested. When these DNA fragments were aligned with the T. whipplei Twist gyrA QRDR, nucleotide sequence variations were found only at positions 120 (A versus G) and 354 (T versus C), which corresponded to silent mutations. An alanine residue at position 81was found in the sequences of all strains. For the parC QRDR, 400-bp DNA fragments were amplified and aligned with the T. whipplei Twist parC QRDR, as described above. Variations in nucleotide sequences were found at positions 432 (T versus G), 489 (C versus T), and 531 (C versus T), which corresponded to silent mutations. An alanine residue was found at position 96 in the sequences of all strains tested.
DISCUSSION
T. whipplei, the agent of Whipple's disease, is extremely fastidious. Its recent in vitro propagation in our laboratory provided us the opportunity to test its antibiotic susceptibility for the first time. As phenotypic methods were not applicable, we determined the in vitro growth kinetics of T. whipplei using quantitative PCR technology, allowing enumeration of specific target its DNA copies. We found a doubling time of 32 to 36 h for the T. whipplei Twist strain, much lower than that previously reported (i.e., 18 days) for the same strain at the time of primary isolation (18) (Fig. 1). This suggests in vitro selection of bacteria most adapted to the MRC5 cell culture system after multiple passages. This doubling time, however, remains higher than that of the slowly growing bacterium M. tuberculosis (14.3 h minimum to 24 h) (15).
Molecular evaluation of fluoroquinolone MICs, defined as the minimum concentration of antibiotic that allowed complete inhibition of an increase in its copy numbers, revealed the relative resistance of three T. whipplei strains to ciprofloxacin, with the MICs being comparable to those previously reported for M. tuberculosis (26).
Comparison of the whole T. whipplei Twist genome with known gyrA sequences with the BLAST program allowed identification of two gyrA-type sequences, which were differentiated into gyrA and parC genes on the basis of the sequence homologies of their protein counterparts with known GyrA and ParC sequences. Both genes are considered paralogs and are found in many of the complete bacterial genomes that are available.
In both the T. whipplei GyrA and the T. whipplei ParC sequences, alanines were found at positions 81 and 96, respectively, corresponding to the serine at position 83 in E. coli GyrA (10) and the serine at position 80 in E. coli ParC (29), respectively. We sequenced the same QRDRs of both genes from two other T. whipplei isolates as well as from nine uncultured strains whose DNA was directly extracted from tissue specimens from Whipple's disease patients.
GyrA-mediated resistance to fluoroquinolones has been well characterized in many bacteria. Many examples exist to demonstrate that species naturally bearing a serine residue at position 83 (E. coli numbering) are usually susceptible to fluoroquinolones (14), whereas the presence of an alanine at this critical position usually corresponds to natural or acquired resistance to these antibiotics (4, 13, 13, 14, 21, 31, 34). GyrA-mediated natural resistance to fluoroquinolones has been described in Mycobacterium species, which are phylogenetically closely related to T. whipplei. Poor susceptibilities to fluoroquinolones are found in M. tuberculosis, M. avium, M. intracellulare, Mycobacterium marinum, Mycobacterium chelonae, Mycobacterium abcessus, and M. smegmatis, species that bear an alanine residue at position 83 (E. coli numbering) in the GyrA QRDR (8, 9, 26). In contrast, a serine residue is found in M. fortuitum, Mycobacterium peregrinum, and Mycobacterium aurum, which are naturally more susceptible to these antibiotics (8, 9, 26). Other critical positions in the GyrA QRDR have been described as positions 84 and 87 in E. coli (31) and corresponding positions in other species (23, 31, 32). High-level resistance to fluoroquinolones has been described in M. tuberculosis due to amino acid substitution in the wild-type GyrA sequence at the critical positions 83, 84, and 87, with MICs being more than 100-fold those for wild-type strains when these mutations accumulate (16). ParC-mediated resistance has been described in gram-negative bacteria such as E. coli (17, 29). Critical positions in E. coli ParC correspond to positions 80 and 84, and increased fluoroquinolone resistance has been specifically associated with replacement of the serine at position 80 by an alanine. In gram-negative bacteria, these mutations are usually combined with amino acid sequence alterations in GyrA. In contrast, topoisomerase IV is considered the primary target of fluoroquinolones in the gram-positive species of the genera Staphylococcus and Streptococcus (22, 30). Surprisingly, levofloxacin was efficient, and this may suggest another resistance mechanism.
In conclusion, we have presented the first in vitro evaluation of the antibiotic susceptibilities of T. whipplei. Because of the fastidious nature of this bacterium, only a molecular method based upon quantitative PCR technology allowed determination of the MICs. Relative resistance to ciprofloxacin was found in three human strains of T. whipplei. Identification of gyrA and parC in the T. whipplei genome allowed us to correlate fluoroquinolone resistance with specific QRDR sequences. These sequences were then demonstrated to be the wild types by amplification and sequencing of homologous sequences in two additional isolates as well nine uncultured strains. We speculate that T. whipplei has a topoisomerase-mediated natural low-level resistance. The T. whipplei model can be considered a paradigm for an obligate molecular approach to the testing of antibiotic susceptibility in a fastidiously growing microorganism.
Acknowledgments
We thank P. Kelly for correction of the English in the manuscript.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badak, F. Z., S. Goksel, R. Sertoz, A. Guzelant, A. Kizirgil, and A. Bilgic. 1999. Cord formation in MB/BacT medium is a reliable criterion for presumptive identification of Mycobacterium tuberculosis complex in laboratories with high prevalence of M. tuberculosis. J. Clin. Microbiol. 37:4189-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calcutt, M. J. 1994. Gene organization in the dnaA-gyrA region of the Streptomyces coelicolor chromosome. Gene 151:23-28. [DOI] [PubMed] [Google Scholar]
- 4.Cullen, M., A. Wyke, R. Kuroda, and L. Fisher. 1989. Cloning and characterization of a DNA gyrase A gene from Escherichia coli that confers clinical resistance to 4-quinolones. Antimicrob. Agents Chemother. 33:886-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drancourt, M., A. Carlioz, and D. Raoult. 2001. rpoB sequence analysis of cultured Tropheryma whippelii. J. Clin. Microbiol. 39:2425-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drlica, K. 1999. Mechanism of fluoroquinolone action. Curr. Opin. Microbiol. 2:504-508. [DOI] [PubMed] [Google Scholar]
- 7.Fenollar, F., and D. Raoult. 2001. Molecular techniques in Whipple's disease. Expert Rev. Mol. Diagn. 1:299-309. [DOI] [PubMed] [Google Scholar]
- 8.Guillemin, I., V. Jarlier, and E. Cambau. 1998. Correlation between quinolone susceptibility patterns and sequences in the A and B subunits of DNA gyrase in mycobacteria. Antimicrob. Agents Chemother. 42:2084-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guillemin, I., W. Sougakoff, E. Cambau, V. Revel-Viravau, N. Moreau, and V. Jarlier. 1999. Purification and inhibition by quinolones of DNA gyrases from Mycobacterium avium, Mycobacterium smegmatis and Mycobacterium fortuitum bv. peregrinum. Microbiology 145:2527-2532. [DOI] [PubMed] [Google Scholar]
- 10.Herrera, G., V. Aleixandra, A. Urios, and M. Blanco. 1993. Quinolone action in Escherichia coli cells carrying gyrA and gyrB mutations. FEMS Microbiol. Lett. 106:187-191. [DOI] [PubMed] [Google Scholar]
- 11.Higgins, D. G., and P. M. Sharp. 1988. Clustal: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 12.Hinrikson, H. P., F. Dutly, and M. Altwegg. 2000. Evaluation of a specific nested PCR targeting domain III of the 23S rRNA gene of “Tropheryma whippelii” and proposal of a classification system for its molecular variants. J. Clin. Microbiol. 38:595-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houssaye, S., L. Gutmann, and E. Varon. 2002. Topoisomerase mutations associated with in vitro selection of resistance to moxifloxacin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:2712-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, W. M. 1996. Bacterial diversity based on type II DNA topoisomerase genes. Annu. Rev. Genet. 30:79-107. [DOI] [PubMed] [Google Scholar]
- 15.James, B. W., A. Williams, and P. D. Marsh. 2000. The physiology and pathogenicity of Mycobacterium tuberculosis grown under controlled conditions in a defined medium. J. Appl. Microbiol. 88:669-677. [DOI] [PubMed] [Google Scholar]
- 16.Kocagoz, T., C. J. Hackbarth, I. Unsal, E. Y. Rosenberg, H. Nikaido, and H. F. Chambers. 1996. Gyrase mutations in laboratory-selected, fluoroquinolone-resistant mutants of Mycobacterium tuberculosis H37Ra. Antimicrob. Agents Chemother. 40:1768-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumagai, Y., J. I. Kato, K. Hoshino, T. Akasaka, K. Sato, and H. Ikeda. 1996. Quinolone-resistant mutants of Escherichia coli DNA topoisomerase IV parC gene. Antimicrob. Agents Chemother. 40:710-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.La Scola, B., F. Fenollar, P. E. Fournier, M. Altwegg, M. N. Mallet, and D. Raoult. 2001. Description of Tropheryma whipplei gen. nov., sp. nov., the Whipple's disease bacillus. Int. J. Syst. E vol. Microbiol. 51:1471-1479. [DOI] [PubMed] [Google Scholar]
- 19.Mateos-Colino, A., C. Falo Zamora, M. Maner, and P. Fernandez-Viladrich. 1996. Sindrome febril y derrame pleural aislado como presentacion de fiebre Q. Rev. Clin. Espanola 196:78-79. (In Spanish.) [PubMed]
- 20.Musser, J. M. 1995. Antimicrobial agents resistance in mycobacteria: molecular genetic insights. Clin. Microbiol. Rev. 8:496-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oram, M., and L. Fisher. 1991. 4-Quinolone resistance mutations in the DNA gyrase of Escherichia coli clinical isolates identified by using the polymerase chain reaction. Antimicrob. Agents Chemother. 35:387-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan, X. S., and L. M. Fisher. 1996. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J. Bacteriol. 178:4060-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan, X. S., P. J. Hamlyn, R. Talens-Visconti, F. L. Alovero, R. H. Manzo, and L. M. Fisher. 2002. Small-colony mutants of Staphylococcus aureus allow selection of gyrase-mediated resistance to dual-target fluoroquinolones. Antimicrob. Agents Chemother. 46:2498-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piddock, L. J. 1999. Mechanisms of fluoroquinolone resistance: an update 1994-1998. Drugs 58(Suppl. 2):11-18. [DOI] [PubMed] [Google Scholar]
- 25.Raoult, D., M. L. Birg, B. La Scola, P. E. Fournier, M. Enea, H. Lepidi, V. Roux, J. C. Piette, F. Vandenesch, D. Vital Durand, and T. J. Marrie. 2000. Cultivation of the bacillus of Whipple's disease. N. Engl. J. Med. 342:620-625. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez, J. C., M. Ruiz, A. Climent, and G. Royo. 2001. In vitro activity of four fluoroquinolones against Mycobacterium tuberculosis. Int. J. Antimicrob. Agents 17:229-231. [DOI] [PubMed] [Google Scholar]
- 27.Rolain, J. M., L. Stuhl, M. Maurin, and D. Raoult. 2002. Evaluation of antibiotic susceptibilities of three rickettsial species including Rickettsia felis by a quantitative PCR DNA assay. Antimicrob. Agents Chemother. 46:2747-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rombel, I. T., K. F. Sykes, S. Rayner, and S. A. Johnston. 2002. ORF-FINDER: a vector for high-throughput gene identification. Gene 282:33-41. [DOI] [PubMed] [Google Scholar]
- 29.Vila, J., J. Ruiz, P. Goni, and M. T. De Anta. 1996. Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 40:491-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, T., M. Tanaka, and K. Sato. 1998. Detection of grlA and gyrA mutations in 344 Staphylococcus aureus strains. Antimicrob. Agents Chemother. 42:236-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waters, B., and J. Davies. 1997. Amino acid variation in the GyrA subunit of bacteria potentially associated with natural resistance to fluoroquinolone antibiotics. Antimicrob. Agents Chemother. 41:2766-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weigel, L. M., G. J. Anderson, and F. C. Tenover. 2002. DNA gyrase and topoisomerase IV mutations associated with fluoroquinolone resistance in Proteus mirabilis. Antimicrob. Agents Chemother. 46:2582-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yagupsky, P. V., D. A. Kaminski, K. M. Palmer, and F. S. Nolte. 1990. Cord formation in BACTEC 7H12 medium for rapid, presumptive identification of Mycobacterium tuberculosis complex. J. Clin. Microbiol. 28:1451-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida, H., T. Kojima, J.-I. Yamagishi, and S. Nakamura. 1988. Quinolone-resistant mutations of the gyrA gene of Escherichia coli. Mol. Gen. Genet. 211:1-7. [DOI] [PubMed] [Google Scholar]