Abstract
A collection of Aspergillus fumigatus mutants highly resistant to itraconazole (RIT) at 100 μg ml−1 were selected in vitro (following UV irradiation as a preliminary step) to investigate mechanisms of drug resistance in this clinically important pathogen. Eight of the RIT mutants were found to have a mutation at Gly54 (G54E, -K, or -R) in the azole target gene CYP51A. Primers designed for highly conserved regions of multidrug resistance (MDR) pumps were used in reverse transcriptase PCR amplification reactions to identify novel genes encoding potential MDR efflux pumps in A. fumigatus. Two genes, AfuMDR3 and AfuMDR4, showed prominent changes in expression levels in many RIT mutants and were characterized in more detail. Analysis of the deduced amino acid sequence encoded by AfuMDR3 revealed high similarity to major facilitator superfamily transporters, while AfuMDR4 was a typical member of the ATP-binding cassette superfamily. Real-time quantitative PCR with molecular beacon probes was used to assess expression levels of AfuMDR3 and AfuMDR4. Most RIT mutants showed either constitutive high-level expression of both genes or induction of expression upon exposure to itraconazole. Our results suggest that overexpression of one or both of these newly identified drug efflux pump genes of A. fumigatus and/or selection of drug target site mutations are linked to high-level itraconazole resistance and are mechanistic considerations for the emergence of clinical resistance to itraconazole.
Over the past decade, the incidence of serious infections caused by opportunistic fungal pathogens has increased dramatically due to alterations in the immune status of patients. Unfortunately, the widespread use of triazole antifungal agents to combat these infections has led to the emergence of clinically significant drug resistance that limits therapy and emphasizes the need for a better understanding of the molecular mechanisms conferring drug resistance. Aspergillus fumigatus is the most common species of Aspergillus that causes life-threatening pulmonary disease in severely immunocompromised patients (9). For such patients, the treatment options are largely limited to therapy with the polyene drug amphotericin B, with broad-spectrum triazoles such as itraconazole or voriconazole, and/or with the echinocandin caspofungin (15, 33).
However, amphotericin B therapy can be highly toxic and can result in nephrotoxicity, whereas triazoles are fungistatic and subject to drug resistance (10). The safety profile and high therapeutic index afforded by triazole drugs make them particularly suitable for prophylactic, empirical, and preemptive therapies in bone marrow transplant and other patients with severe immunosuppression. Unfortunately, repeated exposure to triazole drugs is a major risk factor for drug resistance, and itraconazole resistance has been documented in clinical isolates (10) and in spontaneous mutants of A. fumigatus (7, 20).
Fungal azole resistance mechanisms involve both amino acid changes in the target site 14-α-demethylase enzyme encoded by ERG11 (or CYP51) that alter drug-target interactions and those that decrease net azole accumulation. A reduced intracellular level of itraconazole has been identified as a cause of resistance in several posttreatment Candida sp. clinical isolates (31) and in some A. fumigatus isolates (20). Failure to accumulate such antifungal agents has been correlated with overexpression of multidrug resistance (MDR) efflux transporter genes of the ATP-binding cassette (ABC) and the major facilitator superfamily (MFS) classes (39). PCR-based homology cloning has been used to identify AfuMDR1 and AfuMDR2, two ABC-type transporter genes in A. fumigatus, but their role in clinical drug resistance remains unclear (35).
Once an MDR transporter is identified, a quantitative assessment of its expression is critical for deciphering its potential role. Quantitative reverse transcriptase PCR (RT PCR) with reporter probes such as molecular beacons can be used for such quantitative assessments. Molecular beacons are hairpin-shaped self-reporting DNA hybridization probes that are well suited for gene expression studies (21). The molecular beacon loop structure contains a 20-nucleotide (nt) allele-specific target sequence, while the stem structure is capped with a 5′ fluorophore and a 3′ quencher in close proximity. The binding of the loop structure to its specific target induces fluorescence as the fluorophore and quencher physically separate. The fluorescence of the molecular beacon is monitored in real time during PCR, and the level of fluorescence detected indicates the amount of specific product present at that time (3, 36, 37). Molecular beacon technology is similar to that of TaqMan probes, but it is more dynamic and discriminating (34). Furthermore, this approach is preferable to conventional Northern blot analysis because of the narrow linear range associated with visualization of labeled hybridization probes. Molecular beacons have been used successfully to characterize gene expression in sigma factor genes in Mycobacterium tuberculosis (21).
In this study, a collection of mutants conferring high-level resistance to itraconazole were evaluated for multiple resistance mechanisms involving both target site mutations and overexpression of novel multidrug efflux transporters.
MATERIALS AND METHODS
Strains and culture conditions.
A. fumigatus strain H11-20 (19) was the parental wild-type strain used in this study. Mutant strains resistant to itraconazole (RIT) were obtained by mutagenesis of the H11-20 strain. All of the strains were grown and maintained in yeast extract-peptone-dextrose (YEPD) medium (1% [wt/vol] Bacto Yeast Extract, 2% [wt/vol] Bacto Peptone, 2% [wt/vol] dextrose [adjusted to pH 5.7], 2% [wt/vol] Bacto Agar) or Sabouraud dextrose agar (1% [wt/vol] peptone, 4% [wt/vol] dextrose, 1.5% [wt/vol] agar) medium (Becton Dickinson, Franklin Lakes, N.J.). Escherichia coli strain DH10BF′, a host for all plasmid subcloning experiments, was grown in Luria-Bertani broth (Becton Dickinson). E. coli strain LE392 (used as host for propagation of bacteriophages λEMBL3 and λFixII) was grown in NZY medium (0.5% NaCl, 0.2% MgSO4• 7H2O, 0.5% yeast extract, 1% casein hydrolysate, 0.7% agarose, adjusted to pH 7.5). Fungal and bacterial strains were grown at 37°C. The antifungal powders were obtained directly from the drug manufacturers, and the stock solutions were prepared at a concentration of 5 mg ml−1. Amphotericin B (Sigma Chemical Co., St. Louis, Mo.) and ketoconazole (ICN Biomedicals Inc., Costa Mesa, Calif.) were dissolved in dimethyl sulfoxide (Sigma), itraconazole (Janssen Pharmaceutica, Titusville, N.J.) was dissolved in dimethylformamide (Sigma), and flucytosine (Hoffman-La Roche, Inc., Nutley, N.J.) was dissolved in sterile water. The concentrations of itraconazole used for the induction experiments were 10 μg ml−1 for the parental strain and 100 μg ml−1 for the mutant strains.
Mutagenesis and isolation of RIT mutants.
Using UV light as a mutagen at 0.22 J/m2 s−1, conidia from A. fumigatus strain H11-20 were mutagenized at a concentration of 1.0 × 106 ml−1 in sterile distilled water, which reduced spore viability by 90%. The irradiated suspension was washed and suspended in 0.85% saline-0.1% Tween 20. Aliquots of 0.1 ml were spread on YEPD plates containing 10 μg of itraconazole ml−1, and the plates were incubated at 37°C for 3 days. Resistant colonies were selected and retested for growth in the presence of itraconazole (100 μg ml−1).
In vitro antifungal susceptibility testing.
The susceptibility testing of the mutant strains to antifungal agents was performed by assessing the drug MICs according to the NCCLS M38-P microdilution technique proposed by the National Committee for Clinical Laboratory Standards (25). Using approximately 1 ml of sterile 0.85% saline-0.1% Tween 20, conidia from YEPD agar cultures grown for 72 h at 37°C were pooled and the resulting conidial suspensions were transferred to a sterile tube. After spore dispersion with a vortex mixer, the optical densities of the conidial suspensions were read and then the suspensions were diluted in RPMI 1640 broth (Sigma) to approximately 1 × 104 conidia ml−1. Concentrated (2×) antifungal solutions were prepared in RPMI 1640 broth, and a serial dilution series was prepared in a 96-well microtiter plate. Inoculum quantification was performed by plating 0.1 ml of the diluted conidial suspension in each well containing 0.1 ml of drug at a concentration in the following range: amphotericin B, 0.016 to 16 μg ml−1; ketoconazole, 0.016 to 16 μg ml−1; flucytosine, 0.064 to 64 μg ml−1; and itraconazole, 0.1 to 100 μg ml−1. Dimethylformamide at 1% (vol/vol) was used to increase the solubility of itraconazole to the levels indicated. The plates were prepared in duplicate and incubated at 37°C for 48 h. A total of 23 itraconazole-resistant mutant strains were randomly chosen and tested. The MIC was determined as the lowest drug concentration inducing a prominent reduction in growth.
Potentiation of itraconazole-resistant A. fumigatus mutants with the fungal efflux pump inhibitor MC 510027.
Twelve itraconazole-resistant mutants (RIT1 to -6 and RIT8 to -13) and the H11-20 wild-type strain were assayed for restoration of itraconazole susceptibility with the fungal efflux pump inhibitor MC 510027 (Essential Therapeutics, Inc., Mountainside, Calif.). A 10 mg ml−1 stock solution of MC 510027 was prepared in dimethyl sulfoxide. A potentiation assay was performed using a typical checkerboard methodology with the NCCLS method as described above in the presence of 0.0 to 4.0 μg of MC 510027 ml−1. MICs of itraconazole for MC 510027 were determined as the lowest concentration inducing an 80% decrease in growth in comparison to the untreated control.
Isolation of DNA and RNA.
Conidia were inoculated into 15 ml of YEPD medium and incubated with shaking (200 rpm) at 37°C for 12 h. Subsequently, the mycelia were aseptically recovered by filtration through a Whatman number 1 filter and used for DNA extraction. Alternatively, the mycelia were transferred to a flask containing 15 ml of YEPD medium plus 10 or 100 μg of itraconazole ml−1, incubated for 8 h under the same conditions, harvested by filtration, and used for RNA extraction. For DNA extraction, dry mycelia were ground to a fine powder with liquid nitrogen and transferred to 2-ml polypropylene tubes containing 300 μl of extraction buffer (0.25 M NaCl, 25 mM EDTA, 0.5% sodium dodecyl sulfate [SDS], 0.2 M Tris-HCl [pH 8.5]) plus 300 μl of phenol. After 1 min of vortexing, the aqueous phase was recovered by centrifugation in a microcentrifuge at top speed (12,000 × g) for 10 min. DNA was extracted several times with phenol-chloroform and precipitated with isopropanol. Using RNeasy (Qiagen, Valencia, Calif.) according to the manufacturer's instructions, RNA was extracted from the cells. The RNA samples were incubated in the presence of 3 U of DNase I for 30 min at 37°C to eliminate DNA contamination.
PCR and DNA sequencing of CYP51A.
Using Oligo 4.04 primer design software (Molecular Biology Insights, Inc., Cascade, Colo.), PCR and sequencing primers were designed for the N-terminal coding region of CYP51A (GenBank accession number AF338659). The following primers were created: forward primer CYP51AF210 (5′-GTCTCTCATTCGTCCTTGTCCT-3′) and reverse primer CYP51AR709 (5′-CGTTGAGAATAAACTCGTTCCC-3′). The 50-μl amplification mixture included 1× HighFi PCR buffer (Brinkmann, Westbury, New York), 0.5 μM each primer, 0.5 mM deoxynucleoside triphosphate (dNTP) mix (Promega Corp., Madison, Wis.), 2.5 U of Triplemaster DNA polymerase (Brinkmann), and 10 ng of genomic DNA. PCR amplification was carried out in a PTC100 96-well thermal cycler (MJ Research, Waltham, Mass.) at 94°C for 1 min followed by 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s and a final extension step of 72°C for 5 min. Using a Montage PCR96 cleanup kit (Millipore, Bedford, Mass.), PCR products were purified by vacuum filtration. Using a DTCS Quick Start kit (Beckman Coulter, Fullerton, Calif.) and a CEQ 2000 ΧL capillary electrophoresis DNA sequencer (Beckman Coulter), nucleotide sequencing analysis was performed by automated DNA sequencing.
Identification of novel MDR genes by PCR.
PCR amplification with primers matching a consensus in conserved regions of ABCs carrying genes from Saccharomyces cerevisiae (18) was used to identify MDR-like encoding genes from A. fumigatus. The 25-μl amplification reaction mixture consisted of 1× PCR buffer II (Applied Biosystems, Foster City, Calif.), 0.1 mM dNTP mixture, 10 pmol of each primer, 1 U of AmpliTaq Gold DNA polymerase (Applied Biosystems), 10 ng of genomic DNA of the H11-20 strain, and 0.2 mM MgCl2. The following primers were used: ABC1 (5′-GTYGGTTCHTCHGGHTGYGGW-3′) and ABC2 (5′-RTCYAAAGCDGADGTDGCYTCATC-3′) (IDT DNA, Coralville, Iowa). The amplification was carried out at 94°C for 2 min and 94°C for 45 s, followed by a progressive decrease in the annealing temperature from 45 to 40°C for 30 s, an extension of 72°C for 45 s (30 cycles) and a final extension of 72°C for 1 min 30 s. The amplification products were analyzed by gel electrophoresis in a 1% agarose gel. Using a Sure Cloning kit (Amersham Biosciences Corp., Piscataway, N.J.), fragments of about 300 bp were purified and subcloned in the SmaI site of the pUC18 plasmid, creating the pAMN plasmid. Using the BigDye terminator kit on an ABI 377 sequencer (Applied Biosystems), Insert-containing plasmids were subjected to nucleotide sequence analysis by automated DNA sequencing. Sequence homology searches were performed using the BLASTX algorithm (http://www.ncbi.nlm.nih.gov) against the nonredundant database from GenBank/National Center for Biotechnology Information. Sequence alignments were performed using the Bestfit program in a GCG software package.
Screening λFixII and λEMBL3 genomic libraries.
Genomic libraries of A. fumigatus strain NIH 5233 (ATCC 13073) (constructed with λFixII; Stratagene, La Jolla, Calif.) and strain CHUV 192-88 (constructed with λEMBL3 [kindly provided by Michel Monod, CHUV, Lausanne, Switzerland]) were used in this study (5, 16, 23). Recombinant plaques of the genomic libraries were immobilized on Hybond N+ nylon membranes (Amersham Biosciences, Inc.) according to the manufacturer's instructions and probed with PCR-generated fragments radiolabeled with α-32P by using a Random Primed DNA labeling system (Invitrogen, Carlsbad, Calif.). Hybridization was performed overnight in 5× SSC (0.075 M Na3C6H5O7 · 2H2O and 0.75 M NaCl, pH 7.0)-5× Denhardt's solution (0.1% Ficoll type 400, 0.1% polyvinylpyrrolidone, 0.1% bovine serum albumin)-0.5% SDS at 65°C. Filters were washed once in 2× SSC-0.1% SDS for 15 min and once in 1× SSC-0.1% SDS for 15 min at 65°C.
Primers and molecular beacon design.
Primer pairs were designed based on unique regions of the AfuMDR3, AfuMDR4, and β-tubulin genes of A. fumigatus. The β-tubulin-encoding gene of A. fumigatus (GenBank accession number AF057315) (13) was used as an internal reference. Using an Oligo 4.04 program (Molecular Biology Insights, Inc.), all primer sets were designed to anneal to their respective target at the same temperature (50°C). The expected sizes of the PCR products and the primer sequences used were as follows: for MDR3 (131 bp), the forward primer sequence was 5′-CTATATCGGGTCAGTCCTGG-3′ and the reverse primer sequence was 5′-GACCCAGAACAAGGAATCCGAC-3′; for MDR4 (158 bp), the forward primer sequence was 5′-TTCTACGATCCCGATTCAGG-3′ and the reverse primer sequence was 5′-GACGACACTAAGCCATATGC-3′; and for β-tubulin (115 bp), the forward primer sequence was 5′-CAATGGCTCCTCCGATCTCC-3′ and the reverse primer sequence was 5′-TCCATGGTACCAGGCTCG-3′.
Three molecular beacons were designed and synthesized specifically for the AfuMDR3, AfuMDR4, and β-tubulin genes of A. fumigatus, and each hybridized to its specific PCR amplicon. We chose the molecular beacon sequences by entering the sequences of the internal PCR product into Oligo 4.04 primer analysis software. A DNA folding program was used to estimate the stability of the designed hairpin stem (available at http://www.bioinfo.rpi.edu/applications/mfold/old/dna.) Using a controlled-pore glass column (Glen Research, Sterling, Va.) to introduce a 4-dimethylaminoazobenzene-4′-sulfonyl group (DABCYL) at the 3′ end of the oligodeoxyribonucleotide and a tetrachloro-6-carboxyfluorescein phosphoramidite (TET) to introduce the fluorophore at the 5′ end of the molecule, the molecular beacons were synthesized completely on a DNA synthesizer. Each molecular beacon was purified by high-pressure liquid chromatography. A detailed protocol for synthesizing molecular beacons is available at http://www.molecular-beacons.org. The hairpin stems were designed to dissociate at a temperature 7 to 8°C higher than the 50°C detection temperature. The molecular beacon sequences and their fluorophores used were as follows: for MB-MDR3, TET-5′-CGCACGGCTCAATATCTCATG GAACCAAGCGTGCG-3′-DABCYL; for MB-MDR4, TET-5′-CCGTCGACCTGCGCGGGAATATCGGCTATGCGACGG-3′-DABCYL; and for MB-BTUB, TET-5′-CGTGCGGTGCCGTTCTGGTCGATCTCGCGCACG-3′-DABCYL.
Itraconazole induction of gene expression.
Conidia were inoculated into 15 ml of YEPD medium and incubated with shaking (200 rpm) at 37°C for 12 h. Mycelia were recovered by vacuum filtration through Whatman number 1 filter paper and transferred to a flask containing 15 ml of fresh YEPD medium containing 1% dimethylformamide and either 10 or 100 μg of itraconazole ml−1 for the parental or mutant strain, respectively. The mycelia were incubated for 8 h with shaking (200 rpm) at 37°C. Control cells in the absence of itraconazole were incubated in YEPD medium for 8 h with shaking (200 rpm) at 37°C.
RT assay followed by PCR.
First-strand cDNA was synthesized from 1 μg of total RNA by incubation with 10 pmol of each antisense primer (three different primers were used in the same annealing mixture) and 1× Omniscript RT PCR buffer (Qiagen) in a volume of 10 μl at 37°C for 5 min, followed by chilling on ice for 10 min. Subsequently, a 0.5 mM dNTP mixture, 1× Omniscript RT PCR buffer, and 4 U of reverse transcriptase were added to the annealing mixture to achieve a final volume of 20 μl. The reaction mixture was incubated at 37°C for 1 h and heated at 94°C for 6 min. Samples incubated in the absence of reverse transcriptase were used as controls. Samples were diluted (1:15) in water, and 5-μl aliquots of first-strand cDNA were used for PCR. The final concentrations for the 50-μl reaction mixtures were 0.2 μM each molecular beacon, 10 pmol of each primer (reverse and forward), 2 U of AmpliTaq Gold DNA polymerase (Applied Biosystems), 0.25 mM dNTPs, 4 mM MgCl2, 5 μl of 10× PCR buffer II (Applied Biosystems), and 5 μl of 250-ng cDNA. The thermal cycling program consisted of 10 min on a spectrofluorometric thermal cycler (Applied Biosystems 7700 Prism) at 95°C followed by 10 cycles of 20 s at 95°C, 30 s at 60°C, and 30 s at 72°C and 40 cycles of 20 s at 95°C, 30 s at 50°C, and 30 s at 72°C. Real-time fluorescence was monitored during the reaction and plotted against the number of thermal cycles.
Quantitative analysis of the data.
In real-time PCR analysis, quantification is based on the threshold cycle value, which is inversely proportional to the logarithm of the initial copy number (37). Using specific amounts of target obtained by serial dilution of known genomic DNA as templates over several orders of magnitude, real-time PCR was performed for each primer pair and its molecular beacon. Using an A. fumigatus genome size value of 30 Mb (as determined by Brookman and Denning [4]), the initial template amount used in the parallel reactions was estimated. A linear relationship was obtained by plotting the threshold cycle against the logarithm of a known amount of an initial template. The equation of the line that best fits the data was determined by regression analysis. With molecular beacons used for the quantification method, the R2 value was calculated for each data set to estimate the accuracy of the real-time PCR. The amount of target copies contained in an unknown sample was determined by extrapolating from the linear regression of the standard curve obtained for each primer-molecular beacon set (12).
Virulence in murine aspergillosis model.
A systemic aspergillosis model was used in this study (40). Female BALB/c mice (20 to 22 g) were rendered susceptible to A. fumigatus infection by treatment with cyclophosphamide at 200 mg kg−1 via intravenous injection through the lateral tail vein 3 days prior to infection. Infection of 8 to 10 animals per group commenced with injection through the lateral tail vein of 0.1 ml of saline containing 105, 106, or 107 spores of A. fumigatus. Survival after 10 to 12 days was used to determine fungal virulence. Surviving mice were euthanized with CO2.
Nucleotide sequence accession numbers.
Primary nucleotide and deduced amino acid sequences for AfuMDR3 and AfuMDR4 from the cloned fragments are available from GenBank under accession numbers AF503774 and AF503773, respectively.
RESULTS
Isolation and characterization of RIT mutants.
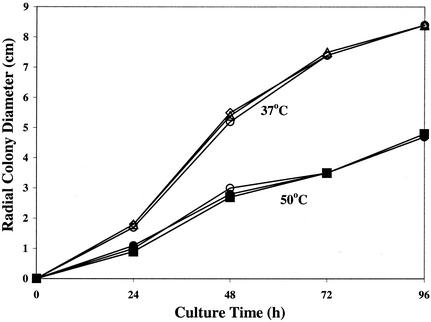
Following UV irradiation, a collection of fifty-five RIT colonies of the H11-20 clinical strain was isolated on YEPD agar plates containing 10 μg of itraconazole ml−1. Of these mutants, 23 were selected for further analysis and were found to grow in the presence of 100 μg of itraconazole ml−1, which is at least 62.5-fold higher than the MIC of the drug for the parental strain (0.8 to 1.6 μg ml−1). Four mutants, RIT4, RIT5, RIT8, and RIT11, showed cross-resistance to ketoconazole (MIC, 8 to 16 μg ml−1), while the remaining strains showed the same (2.0 to 4.0 μg of ketoconazole ml−1) or enhanced (<2.0 μg ml−1) susceptibility when compared to the parental H11-20 strain. All RIT mutants were fully susceptible to amphotericin B (0.125 to 0.5 μg ml−1) (data not shown). There was no significant difference in cell growth rate between the RIT mutants and the parental wild type, as determined by radial colony growth on solid Sabouraud dextrose medium at different temperatures (Fig. 1). Six mutant strains, RIT1, RIT4, and RIT10 to -13, were evaluated for virulence in a murine model of systemic aspergillosis (40). All strains (except RIT1) produced more than 90% mortality within 5 to 6 days postinfection with 106 spores compared with the parental strain, which showed equivalent mortality after 4 to 5 days. Although the difference in mortality between the RIT mutants and the wild-type strain was significant (P < 0.05; Kaplan-Meier test), it was apparent that the RIT mutants were still highly virulent.
FIG. 1.
Radial cell growth of RIT mutants. Representative colony growth of RIT mutants was determined by measuring the average diameter of three colonies for strains RIT1 (squares) and RIT4 (triangles) and for parental strain H11-20 (circles) at 37°C (open symbols) and 50°C (filled symbols) over 96 h. Colony formation was initiated by spotting medium with 1,000 spores of each strain.
Target site mutations.
Naturally occurring ERG11 (CYP51A) mutations identified in Candida albicans azole-resistant isolates are clustered in three hot-spot regions in the primary sequence from amino acid residues 72 to 467 (27, 28). DNA sequence analysis of the RIT mutants in these regions failed to identify any mutations. However, it was recently reported that mutation of codon 54 in the A. fumigatus CYP51A gene conferred resistance to itraconazole (22). When the N-terminal region of the RIT mutants was examined, eight mutant strains were found to have glycine substitutions in codon 54 (Table 1). These strains were RIT1 to -3, RIT12, and RIT13 (G54E); RIT15 and RIT18 (G54K); and RIT19 (G54R).
TABLE 1.
CYP51A mutations and expression levels of AfuMDR3 and AfuMDR4 in RIT mutant strains
| Strain |
AfuMDR3 cDNA level
|
AfuMDR4 cDNA level
|
CYP51A mutation | ||
|---|---|---|---|---|---|
| Constitutively expressed straina | Itraconazole-treated strainb | Constitutively expressed straina | Itraconazole-treated strainb | ||
| H11-20 | 1.0 (0.8-1.2) | 1.3 (0.6-2.7) | 1.0 (0.8-1.2) | 2.2 (1.9-2.6) | |
| RIT1 | 2.4 (0.8-7.1) | 0.5 (0.2-1.4) | 1.4 (0.8-2.5) | 1.4 (0.8-2.4) | G54E |
| RIT2 | 29.9 (27.7-32.2) | 3.2 (3.2-3.3) | 5.2 (5.0-5.3) | 2.8 (2.6-3.0) | G54E |
| RIT3 | 30.9 (26.6-35.9) | 1.0 (0.9-1.1) | 6.8 (5.8-8.1) | 1.2 (0.5-3.2) | G54E |
| RIT4 | 1.9 (0.4-8.9) | 0.8 (0.3-1.9) | 1.7 (0.8-3.6) | 0.7 (0.4-1.1) | |
| RIT5 | 121.5 (84.7-174.4) | 1.1 (0.8-1.4) | 14.7 (10.3-21.0) | 1.1 (1.0-1.2) | |
| RIT6 | 0.6 (0.1-2.5) | 2.1 (0.9-5.0) | 1.5 (1.0-2.3) | 1.2 (0.7-2.0) | |
| RIT8 | 17.7 (6.3-50.0) | 0.6 (0.3-1.5) | 2.1 (1.1-3.9) | 0.8 (0.6-1.2) | |
| RIT9 | 0.7 (0.3-2.0) | 0.8 (0.2-2.6) | 1.3 (0.6-3.2) | 0.3 (0.1-1.4) | |
| RIT10 | 36.3 (9.8-134.6) | 0.6 (0.6-0.7) | 9.4 (2.9-31.3) | 0.4 (0.4-0.4) | |
| RIT11 | 20.7 (13.6-31.5) | 4.8 (3.4-6.8) | 1.6 (1.0-2.4) | 11.2 (7.6-16.5) | |
| RIT12 | 2.7 (2.0-3.8) | 2.4 (0.6-8.7) | 4.1 (3.0-5.7) | 0.6 (0.3-1.0) | G54E |
| RIT13 | 1.5 (1.2-1.8) | 139.6 (83.0-234.7) | 1.0 (0.9-1.2) | 60.8 (37.4-98.8) | G54E |
| RIT14 | 9.9 (2.0-48.3) | 0.6 (0.2-1.9) | 1.2 (0.6-2.8) | 2.4 (1.7-3.6) | |
| RIT15 | 2.4 (0.6-10.0) | 1.9 (0.7-4.7) | 1.1 (0.5-2.5) | 0.5 (0.3-1.0) | G54K |
| RIT16 | 6.4 (3.6-11.2) | 0.5 (0.2-1.5) | 2.6 (1.3-5.0) | 0.3 (0.2-0.6) | |
| RIT18 | 4.7 (1.7-12.9) | 2.3 (0.5-10.2) | 3.1 (1.9-5.1) | 2.9 (2.2-4.0) | G54K |
| RIT19 | 1.2 (0.5-3.2) | 2.6 (1.0-6.9) | 2.4 (4.9-1.1) | 1.1 (0.4-2.7) | G54R |
| RIT20 | 0.1 (0.0-0.4) | 3.2 (0.4-23.7) | 1.2 (0.3-4.5) | 1.3 (0.5-3.2) | |
| RIT21 | 0.8 (0.1-6.3) | 3.6 (1.2-11.0) | 0.4 (0.1-2.7) | 1.2 (0.2-7.6) | |
| RIT22 | 0.2 (0.0-0.6) | 1.9 (0.9-4.2) | 0.3 (0.1-1.3) | 1.1 (0.4-3.4) | |
| RIT23 | 2.6 (0.3-19.2) | 2.8 (0.3-24.8) | 0.5 (0.1-2.7) | 1.4 (0.2-8.5) | |
| RIT24 | 0.04 (0.002-0.9) | 29.3 (8.6-100.2) | 0.3 (0.1-1.0) | 2.0 (0.8-4.9) | |
| RIT26 | 2.8 (0.8-9.9) | 1.4 (0.4-5.3) | 0.4 (0.1-0.9) | 0.3 (0.04-2.6) | |
cDNA levels were calculated relative to that of the untreated parental strain H11-20. The numbers represent the averages of 3 to 5 replicates, with the ranges shown in parentheses.
Values were obtained by using the cDNA copy number of each respective untreated strain as the baseline for strains in the presence of drug following normalization based on β-tubulin cDNA copy number. Cells were induced with 10 (H11-20) or 100 μg of itraconazole ml−1 for 8 h. Standard deviations are in parentheses.
RIT mutants are sensitized by a drug efflux pump inhibitor.
More than 65% of the RIT mutants did not show target site mutations, suggesting that other resistance mechanisms were operational. To help identify the contribution of drug efflux pumps in itraconazole resistance, the compound MC 510027 (Essential Therapeutics, Inc.), a milbemycin derivative that blocks ABC-class drug efflux (M. Warren, A. Mistry, J. Blais, A. L. Staley, J. L. Galazzo, H. Fuernkranz, S. Chamberland, M. D. Lee, W. J. Watkins, O. Lomovskaya, G. H. Miller, and D. Sanglard, Abstr. 40th Intersci. Conf. Antimicrob. Agents and Chemother., abstr. 114, 2000), was used to evaluate whether it could restore itraconazole sensitivity to the RIT mutants. Twelve tester strains were evaluated for itraconazole susceptibility for drug concentrations from 0.016 to 16.0 μg ml−1 in the presence of MC 510027 at concentrations up to 4 μg ml−1. A total of 10 of the strains (RIT1 to -3, RIT5, and RIT8 to -13) showed a restoration of drug susceptibility, with drug MICs of ≤2 μg ml−1 in the presence of MC 510027 at 4 μg ml−1. RIT4 was weakly affected by the pump inhibitor (MIC, 4 to 8 μg ml−1), and RIT6 was unaffected (MIC ≥ 16 μg ml−1). These data suggested that drug efflux transporters are likely to play a role in resistance.
Identification of ABC genes of A. fumigatus.
In the absence of a genome sequence for A. fumigatus, an attempt was made to identify potential drug efflux transporters. Degenerate oligonucleotide primers ABC1 and ABC2, corresponding to consensus ATP-binding and ABC signature motifs, were used for PCR amplification of potential ABC transporter genes in A. fumigatus. Agarose gel electrophoresis of PCR products revealed a strong broad band at the expected size of ∼300 bp. The DNA fragments were isolated and subcloned, and examination of the plasmids from about 200 transformant colonies produced six different sequences that contained putative transporter genes that were distinct from transporters AfuMDR1, AfuMDR2, and atrF (32, 35). Using the BLASTX algorithm, the partial sequences of these DNA fragments were compared against a nonredundant data bank and were found to correspond to novel sequences.
Cloning and characterization of AfuMDR3 and AfuMDR4.
Using primers specific to each sequence, RT PCR was used to probe mutants RIT1 to -12 for overexpression of the six novel partial gene sequences as well as of AfuMDR1 and AfuMDR2. Following cell growth in the presence of 10 μg of itraconazole ml−1, it was found that two of the six novel gene sequences were consistently overexpressed in many of the RIT mutants. Under these conditions, AfuMDR1 and AfuMDR2 showed minimal differential expression relative to the parent strain (data not shown). The two putative genes showing prominent changes, designated AfuMDR3 and AfuMDR4, were then cloned and analyzed in more detail. A 131-bp PCR fragment of AfuMDR3 was used as a probe to screen a genomic library of A. fumigatus. Four positive clones were identified and purified from ∼60,000 recombinant phage plaques. Restriction analysis of these clones with endonucleases EcoRI, HindIII, NotI, and KpnI followed by Southern blotting of purified bacteriophage DNA showed that all of the clones had fragments in common that hybridized with the 131-bp probe (data not shown). The purified bacteriophage DNA was used for sequence analysis by primer walking. A 4,596-nt contig (obtained by the overlapping of results of the sequence analysis) revealed the presence of an open reading frame of 1,741 bp (GenBank accession number AF503774). A predicted intron of 61 nt presented a GT dinucleotide-splicing donation site sequence at the 5′ end and an AG-splicing acceptor sequence site at the 3′ end that are identical to those observed in genes of fungi and other organisms (6, 11). The expected translation product of AfuMDR3 was a 515-amino- acid polypeptide with a predicted molecular mass of 60,242 Da and a pI value of 7,961. Comparison of the deduced amino acid sequence of AfuMDR3 with those of proteins in the available databases revealed significant similarity to several members of MFS transporters of yeast. These include the probable membrane protein Yor378wp of S. cerevisiae (40%), the probable membrane protein YMR279cp in S. cerevisiae (35%), a MFS transporter of unknown specificity of Schizosaccharomyces pombe (42%), the protein encoded by ATR1 that confers resistance to aminotriazole in S. cerevisiae (33%) and a membrane protein involved in the drug efflux of Streptomyces coelicolor (24%). The similarities of the AfuMDR3 gene product and related proteins extend over the entirety of the sequences. The hydropathy profiles and homology analysis suggested the presence of 14 transmembrane domains as observed for members of cluster II or III, according to the classification proposed by Goffeau et al. (14) for MFS transporters of S. cerevisiae.
A 158-bp PCR fragment of AfuMDR4 was used as a probe to screen the same genomic library of A. fumigatus as described above. Two positive clones were identified, and purified bacteriophage DNA was digested with the restriction endonucleases EcoRI, HindIII, NotI, and KpnI. Southern blot analysis showed fragments in common that hybridized with the probe used (data not shown). Fragments of about 4.5 and 7 kb (which were obtained by digestion with EcoRI and KpnI, respectively) were subcloned into pUC19. DNA sequence analysis showed a contig of 5,300 nt that revealed the presence of an open reading frame of 4,032 bp corresponding to AfuMDR4 (GenBank accession number AF503773). A BLAST search for AfuMDR4 in the entire GenBank database at the amino acid level revealed strong identity to several ABC transporters: 35% identity with the ABC transporter of Aureobasidium pullulans, 33% identity with the ABCD transporter of Aspergillus nidulans, 33% and 20% identities with the proteins encoded by AfuMDR1 and atrF of A. fumigatus, respectively, and 32% identity with the ABC protein of Gallus gallus. The deduced product of AfuMDR4 gene is composed of two homologous halves, each comprising typical hydrophilic and hydrophobic domains. Hydropathy analysis suggested the presence of six putative transmembrane domains for each of the two hydrophobic domains. In the N-terminal ABC domain, Walker A (GPSGGGKST) and Walker B (LLILDEATAALD) motifs and the ABC signature (LSGGQKQRIALA) were observed. In the C-terminal ABC domain, Walker A (GPSGSGKST) and Walker B (LLLLDESTSALD) motifs and the ABC signature (SGGQRQRLSIA) were also recognized. The predicted molecular mass of the deduced protein encoded by AfuMDR4 was 146,456 Da, and the pI was 7.94.
AfuMDR3 and AfuMDR4 gene expression in RIT mutants.
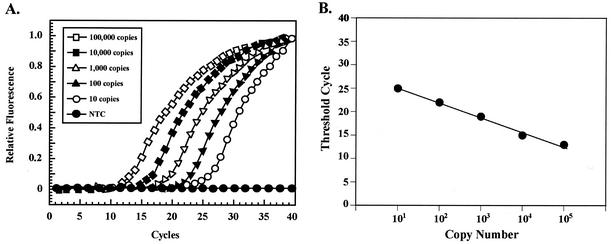
Molecular beacons MB-MDR3, MB-MDR4, and MB-BTUB were designed for specific regions of AfuMDR3 and AfuMDR4 and the A. fumigatus β-tubulin gene, respectively (see Materials and Methods). The A. fumigatus β-tubulin gene was used as an internal standard, because its mRNA levels are constitutively expressed at a high level under various environmental conditions. A linear relationship was observed between the cycle threshold value (i.e., that of the cycle producing the initial linear fluorescence response) and the logarithm of a known amount of initial template molecules (Fig. 2). Total RNA samples isolated from the parental and the mutant strains (grown in the absence and the presence of itraconazole) were used as templates to synthesize cDNA specific for AfuMDR3, AfuMDR4, and β-tubulin transcripts. The amount of cDNA produced was proportional to the amounts of specific transcripts present in the original RNA. Variations in the initial amount of mRNA among the different samples were normalized to the level of β-tubulin mRNA, yielding a ratio between the specific gene cDNA copy number and the β-tubulin cDNA copy number. Table 1 shows that the highest levels of constitutively expressed AfuMDR3 mRNA were observed with the RIT2, RIT3, RIT5, RIT8, RIT10, RIT11, and RIT14 mutant strains, resulting in 29.9-, 30.9-, 121.5-, 17.7-, 36.3-, 20.7- and 9.9-fold more copies of mRNA, respectively, than those of the H11-20 parental strain. The highest levels of AfuMDR4 mRNA were obtained for the RIT5 and RIT10 strains, which displayed 14.7- and 9.4-fold more mRNA than the parental strain (Table 1). Constitutive overexpression of AfuMDR3 and AfuMDR4 was observed to some extent in many of the RIT mutants. The most prominent cases were those of RIT5 and RIT10, which showed high-level expression of both genes. Only two mutants, RIT8 and RIT14, showed prominent overexpression of a single gene.
FIG. 2.
Quantitative relationship between the levels of input mRNA and cDNA as determined by molecular beacon analysis. (A) The quantitative properties of molecular beacons were assessed by amplifying known amounts of AfuMDR3 cDNA by real-time PCR with an AfuMDR3-specific molecular beacon. NTC, no copies. (B) A linear relationship was obtained by plotting the resulting threshold cycle values against the logarithm of the initial template amount. The best-fit line was determined through linear regression analysis. The resulting relationships for β-tubulin, AfuMDR3, and AfuMDR4 genes were used to quantify the amounts of their respective transcripts present in unknown samples.
To explore whether gene expression responded to the presence of the itraconazole, cells were treated with the drug at 10 (parent strain) or 100 (RIT mutants) μg/ml for 8 h. Differences in the expression levels of AfuMDR3 and AfuMDR4 after exposure to itraconazole are shown in Table 1. The AfuMDR3 gene in the parent strain showed little induction (1.3-fold) by itraconazole, while the AfuMDR4 gene was induced about 2.2-fold (Table 1). Following exposure to itraconazole, the levels of AfuMDR3 transcripts increased 4.8-, 139.6-, and 29.3-fold in RIT11, RIT13, and RIT24, while the levels of AfuMDR4 gene transcripts increased 11.2- and 60.8-fold after exposure of the RIT11 and RIT13 strains, respectively, to itraconazole. In the remaining RIT strains, expression of AfuMDR3 and AfuMDR4 either remained constant or was slightly reduced (from 1.3- to 3.1-fold) after exposure to itraconazole (Table 1).
Collectively, the constitutive and drug-induced gene expression profiles suggest that both AfuMDR3 and AfuMDR4 are overexpressed in many of the mutants, including some with target site mutations (RIT2, RIT3, and RIT13).
DISCUSSION
In this study, RIT mutants of A. fumigatus with high-level resistance to itraconazole (MIC > 100 μg ml−1) were isolated following mutagenesis (using UV light as a genotoxic agent) of a susceptible clinical isolate. Four strains were cross-resistant to ketoconazole (MIC > 16 μg ml−1), although all of the mutant strains were susceptible to amphotericin B (MIC < 1 μg ml−1), indicating that resistance was likely to be mechanism specific. A. fumigatus is inherently resistant to limited-spectrum azole antifungal drugs such as fluconazole, suggesting that baseline resistance mechanisms to these agents exist in wild-type strains. Importantly, the RIT strains grew normally (Fig. 1) and the stability of the itraconazole-resistant phenotype was unaltered even after 50 consecutive transfers in rich medium without drug selection (data not shown). Most RIT mutants retained virulence in a murine aspergillosis model, indicating that the drug resistance phenotype did not render A. fumigatus nonvirulent (as has been suggested on the basis of the fact that clinical resistance has been limited).
UV-induced mutations have the potential to promote different types of resistance mechanisms, including changes in the target lanosterol-14-α-demethylase (30) and induction of high-capacity drug efflux pumps (1, 26, 29). Several of the mutants screened had an amino acid change at position 54 in the target gene CYP51A. Substitutions at this position have recently been shown to confer itraconazole resistance to A. fumigatus (22). Naturally occurring ERG11 (CYP51A) mutations in C. albicans azole-resistant clinical isolates can be divided into four hot spots on the basis of their association with different structural regions observed in the recently described MTCYP51 structure (28). The first hot spot (comprising G464S, G465S, and R467K) associates with the N-terminal part of the cysteine pocket. A second hot spot is mapped to the C terminus of the G helix and H helix, and a third hot spot (comprising F72L, F105L, S405F, and T229A) associates with the domain interface. The fourth hot spot (comprising D116E, F126L, K128T, G129A, Y132H, K143R, F145L, K147R, A149V, and D153E) is located in the region between the B′ and C helices that have been postulated to be involved in inhibitor- or substrate-induced structural changes. The presence of mapping mutations identified in C. albicans azole-resistant isolates indicates that azole resistance in fungi develops in protein regions involved in orchestrating the passage of CYP51p through different conformational stages along the catalytic cycle rather than in residues directly contacting the triazole. The position of Gly54 at the fringe of the fourth hot spot suggests that it performs a similar function.
Decreased intracellular accumulation of itraconazole has been reported for drug-resistant clinical and in vitro-generated isolates of A. fumigatus (10, 20), suggesting that drug efflux pumps can contribute significantly to resistance phenotypes. The ABC and the MFS comprise the two major classes of efflux pumps known to contribute to drug resistance (8, 27, 38). For this report, two novel genes encoding putative efflux transporters in A. fumigatus, AfuMDR3 and AfuMDR4, were identified (utilizing degenerate primers designed for conserved ABC transporter motifs) by PCR amplification of genomic DNA. AfuMDR3 encodes a transporter that shows high similarity (30 to 42%) to transporters from the fungal MFS class. This result was somewhat unexpected, given the fact that the primers used for PCR amplification were designed for ABC transporters. A reasonable explanation for this finding is that the degenerate primers, in combination with the low temperatures used for annealing during PCR amplification, amplified a DNA fragment with relatively low identity to the ABC region. This explanation was corroborated by the fact that AfuMDR3 displays about 25% identity with the primers used for PCR amplification, ABC1 and ABC2. This is the first filamentous fungal MFS transporter gene identified as possibly involved in drug resistance. The second gene, AfuMDR4, is the fourth ABC transporter gene described for A. fumigatus, following the previously reported descriptions of AfuMDR1, AfuMDR2, and atrF (32, 35).
To explore the involvement of AfuMDR3 and AfuMDR4 in the itraconazole resistance phenotypes displayed by the RIT mutants, real-time RT PCR with molecular beacons was used to assess gene expression levels. Quantitative PCR with molecular beacons was used to evaluate expression levels in place of classical Northern blot analysis, because this approach provides a more quantitative evaluation of mRNA levels (21). AfuMDR3 and AfuMDR4 were constitutively expressed in all of the strains tested. The cDNA levels of AfuMDR3 in the RIT2, RIT5, RIT8, RIT10, RIT14, and RIT16 mutant strains were from 6.4- to 121.5-fold higher than that in the parental strain (Table 1). Similar results were obtained in analyses of constitutive AfuMDR4 cDNA levels, where RIT2, RIT3, RIT5, RIT10, and RIT12 displayed higher constitutive levels than the H11-20 parental strain (Table 1). Importantly, in the presence of itraconazole, significant overexpression of these genes was observed. Overexpression levels of 139.6-, 29.3-, and 4.8-fold were observed for AfuMDR3 in RIT13, RIT24, and RIT11 mutants, respectively, while increases of about 11.2- and 60.8-fold in the expression of AfuMDR4 were observed for the RIT11 and RIT13 mutant strains. A concomitant increase in levels of AfuMDR3 and AfuMDR4 cDNAs in RIT11 and RIT13 mutant strains after exposure to itraconazole suggests that they are under the control of the same activator or that AfuMDR3 controls AfuMDR4 expression or that AfuMDR4 controls AfuMDR3 expression.
These results suggest that mutations induced as a consequence of UV irradiation resulted in either gain of function of alleles for AfuMDR3 and AfuMDR4 or their common activator (or loss of function of a repressor of a common activator). It has been shown that pleiotropic drug resistance in yeast can be brought about by the alteration or amplification of several different genes whose products can act either as membrane transporters or as transcriptional regulators (2). Such behavior is well known for the S. cerevisiae transcriptional activators PDR1 and PDR3, which positively influence expression of drug transporter genes such as PDR5 (17). The results obtained in this study suggest that AfuMDR3 and AfuMDR4, and possibly other unidentified drug efflux pumps, are likely to contribute to itraconazole resistance in the RIT mutants. This notion is supported by the finding that itraconazole susceptibility is restored in RIT1 to -3, RIT5, and RIT8 to -13 in the presence of MC 510027, a milbemycin derivative that blocks drug efflux pumps. Susceptibility was not restored in RIT4 or RIT6, which did not overexpress AfuMDR3 and AfuMDR4. One mutant, RIT9, showed a shift in susceptibility in response to the presence of the pump inhibitor but did not differentially express either AfuMDR3 or AfuMDR4 and did not have a target site mutation. This strain appears to have a yet-undefined resistance mechanism. Interestingly, all mutants with Gly54 mutations conferring target-based resistance were sensitized by the presence of the pump inhibitor, suggesting that high-level resistance involves multiple mechanisms, including target-based changes and drug efflux.
Ideally, gene disruption experiments can be used to confirm the phenotypic role ascribed to a certain gene product. However, it is apparent that both AfuMDR3 and AfuMDR4 are being overexpressed in many of the mutants and that resistance is likely to reflect a complex expression of multiple genes. In fact, it is likely that other transport genes are coregulated in these mutants. But such a determination will have to await completion of the A. fumigatus genome for the identification of candidate genes. Given the nature of the RIT mutants, disruption of a single gene is not likely to be instructive, since multiple genes must be disrupted. The paucity of dominant markers in A. fumigatus and the failure to successively integrate different markers in the same strain makes the engineering of multiple deletion or disruption strains extremely impractical. It should be noted that the role of multiple efflux systems in triazole resistance is supported by the recent study of Mosquera and Denning (24), who tested for cross-resistance to posaconazole, ravuconazole, and voriconazole in 17 clinical isolates of A. fumigatus for which itraconazole MICs were elevated. A complex pattern of cross-resistance and hypersusceptibility was seen with voriconazole and ravuconazole, suggesting marked differences in activity and mechanisms of resistance.
In summary, high-level itraconazole resistance in in vitro-induced mutants of A. fumigatus is linked to mutation of Gly54 and to the overexpression of the previously undefined drug efflux transporters AfuMDR3 and AfuMDR4. These genes were constitutively overexpressed in some mutants, while in others they were induced by the presence of drug. High-level itraconazole resistance is closely linked with target site mutations and/or pleiotropic overexpression of MDR pumps. The presence of multiple resistance mechanisms might be an important consideration for the emergence of clinical resistance to itraconazole.
Acknowledgments
We acknowledge Renata C. Pascon, Marcelo A. Vallim (Universidade de São Paulo), and Paul M. McNicholas (SPRI) for their support and valuable discussion and Ryota Kashiwazaki (PHRI) for technical assistance.
A.M.N. was supported by a fellowship from CAPES (Brazil).
REFERENCES
- 1.Balzi, E., and A. Goffeau. 1994. Genetics and biochemistry of yeast multidrug resistance. Biochim. Biophys. Acta 1187:152-162. [DOI] [PubMed] [Google Scholar]
- 2.Balzi, E., and A. Goffeau. 1995. Yeast multidrug resistance: the PDR network. J. Bioenerg. Biomembr. 27:71-76. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet, G., S. Tyagi, A. Libchaber, and F. R. Kramer. 1999. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl. Acad. Sci. USA 96:6171-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brookman, J. L., and D. W. Denning. 2000. Molecular genetics in Aspergillus fumigatus. Curr. Opin. Microbiol. 3:468-474. [DOI] [PubMed] [Google Scholar]
- 5.Burghoorn, H. P., P. Soteropoulos, P. Paderu, R. Kashiwazaki, and D. S. Perlin. 2002. Molecular evaluation of the plasma membrane proton pump from Aspergillus fumigatus. Antimicrob. Agents Chemother. 46:615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, T., and M. Q. Zhang. 1998. Pombe: a gene-finding and exon-intron structure prediction system for fission yeast. Yeast 14:701-710. [DOI] [PubMed] [Google Scholar]
- 7.Dannaoui, E., E. Borel, M. F. Monier, M. A. Piens, S. Picot, and F. Persat. 2001. Acquired itraconazole resistance in Aspergillus fumigatus. J. Antimicrob. Chemother. 47:333-340. [DOI] [PubMed] [Google Scholar]
- 8.Del Sorbo, G., H. Schoonbeek, and M. A. De Waard. 2000. Fungal transporters involved in efflux of natural toxic compounds and fungicides. Fungal Genet. Biol. 30:1-15. [DOI] [PubMed] [Google Scholar]
- 9.Denning, D. W. 1996. Diagnosis and management of invasive aspergillosis. Curr. Clin. Top. Infect. Dis. 16:277-299. [PubMed] [Google Scholar]
- 10.Denning, D. W., K. Venkateswarlu, K. L. Oakley, M. J. Anderson, N. J. Manning, D. A. Stevens, D. W. Warnock, and S. L. Kelly. 1997. Itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 41:1364-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deutsch, M., and M. Long. 1999. Intron-exon structures of eukaryotic model organisms. Nucleic Acids Res. 27:3219-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freeman, W. M., S. J. Walker, and K. E. Vrana. 1999. Quantitative RT-PCR: pitfalls and potential. BioTechniques 26:112-125. [DOI] [PubMed] [Google Scholar]
- 13.Geiser, D. M., J. C. Frisvad, and J. W. Taylor. 1998. Evolutionary relationships in Aspergillus section Fumigati inferred from partial β-tubulin and hydrophobin DNA sequences. Mycologia 90:831-845. [Google Scholar]
- 14.Goffeau, A., J. Park, I. T. Paulsen, J. L. Jonniaux, T. Dinh, P. Mordant, and M. H. Saier, Jr. 1997. Multidrug-resistant transport proteins in yeast: complete inventory and phylogenetic characterization of yeast open reading frames with the major facilitator superfamily. Yeast 13:43-54. [DOI] [PubMed] [Google Scholar]
- 15.Herbrecht, R., D. W. Denning, T. F. Patterson, J. E. Bennett, R. E. Greene, J. W. Oestmann, W. V. Kern, K. A. Marr, P. Ribaud, O. Lortholary, R. Sylvester, R. H. Rubin, J. R. Wingard, P. Stark, C. Durand, D. Caillot, E. Thiel, P. H. Chandrasekar, M. R. Hodges, H. T. Schlamm, P. F. Troke, and B. de Pauw. 2002. Voriconazole versus amphotericin B for primary therapy of invasive aspergillosis. N. Engl. J. Med. 347:408-415. [DOI] [PubMed] [Google Scholar]
- 16.Jaton-Ogay, K., M. Suter, R. Crameri, R. Falchetto, A. Fatih, and M. Monod. 1992. Nucleotide sequence of a genomic and a cDNA clone encoding an extracellular alkaline protease of Aspergillus fumigatus. FEMS Microbiol. Lett. 71:163-168. [DOI] [PubMed] [Google Scholar]
- 17.Kolaczkowski, M., A. Kolaczowska, J. Luczynski, S. Witek, and A. Goffeau. 1998. In vivo characterization of the drug resistance profile of the major ABC transporters and other components of the yeast pleiotropic drug resistance network. Microb. Drug. Resist. 4:143-158. [DOI] [PubMed] [Google Scholar]
- 18.Kuchler, K., H. M. Goransson, M. N. Viswanathan, and J. Thorner. 1992. Dedicated transporters for peptide export and intercompartmental traffic in the yeast Saccharomyces cerevisiae. Cold Spring Harbor Symp. Quant. Biol. 57:579-592. [DOI] [PubMed] [Google Scholar]
- 19.Kurtz, M. B., E. M. Bernard, F. F. Edwards, J. A. Marrinan, J. Dropinski, C. M. Douglas, and D. Armstrong. 1995. Aerosol and parenteral pneumocandins are effective in a rat model of pulmonary aspergillosis. Antimicrob. Agents Chemother. 39:1784-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manavathu, E. K., J. A. Vazquez, and P. H. Chandrasekar. 1999. Reduced susceptibility in laboratory-selected mutants of Aspergillus fumigatus to itraconazole due to decreased intracellular accumulation of the antifungal agent. Int. J. Antimicrob. Agents 12:213-219. [DOI] [PubMed] [Google Scholar]
- 21.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 22.Mann, P. A., R. M. Parmegiani, S.-Q. Wei, C. A. Mendrick, X. Li, D. Loebenberg, B. DiDomenico, R. S. Hare, S. S. Walker, and P. M. McNicholas. 2003. Mutations in Aspergillus fumigatus resulting in reduced susceptibility to posaconazole appear to be restricted to a single amino acid in the cytochrome P450 14α-demethylase. Antimicrob. Agents Chemother. 47:577-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monod, M. 1994. Construction of a genomic library of Aspergillus fumigatus, p. 29-32. In B. Maresca and G. Kobayashi (ed.), Molecular biology of pathogenic fungi. Telos, New York, N.Y.
- 24.Mosquera, J., and D. W. Denning. 2002. Azole cross-resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 46:556-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. 1998. Reference method for broth dilution antifungal susceptibility testing of conidium-forming filamentous fungi. Proposed standard M38-P. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 26.Osherov, N., D. P. Kontoyiannis, A. Romans, and G. S. May. 2001. Resistance to itraconazole in Aspergillus nidulans and Aspergillus fumigatus is conferred by extra copies of the A. nidulans P-450 14α-demethylase gene, pdmA. J. Antimicrob. Chemother. 48:75-81. [DOI] [PubMed] [Google Scholar]
- 27.Perea, S., J. L. López-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillán, M. Martínez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Podust, L. M., T. L. Poulos, and M. R. Waterman. 2001. Crystal structure of cytochrome P450 14α-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc. Natl. Acad. Sci. USA 98:3068-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanglard, D., F. Ischer, D. Calabrese, P. A. Majcherczyk, and J. Bille. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43:2753-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slaven, J. W., M. J. Anderson, D. Sanglard, G. K. Dixon, J. Bille, I. S. Roberts, and D. W. Denning. 2002. Increased expression of a novel Aspergillus fumigatus ABC transporter gene, atrF, in the presence of itraconazole in an itraconazole resistant clinical isolate. Fungal Genet. Biol. 36:199-206. [DOI] [PubMed] [Google Scholar]
- 33.Stone, E. A., H. B. Fung, and H. L. Kirschenbaum. 2002. Caspofungin: an echinocandin antifungal agent. Clin. Ther. 24:329, 351-377. [DOI] [PubMed]
- 34.Tapp, I., L. Malmberg, E. Rennel, M. Wik, and A. C. Syvanen. 2000. Homogeneous scoring of single-nucleotide polymorphisms: comparison of the 5′-nuclease TaqMan assay and molecular beacon probes. BioTechniques 28:732-738. [DOI] [PubMed] [Google Scholar]
- 35.Tobin, M. B., R. B. Peery, and P. L. Skatrud. 1997. Genes encoding multiple drug resistance-like proteins in Aspergillus fumigatus and Aspergillus flavus. Gene 200:11-23. [DOI] [PubMed] [Google Scholar]
- 36.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 37.Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49-53. [DOI] [PubMed] [Google Scholar]
- 38.Van Bambeke, F., E. Balzi, and P. M. Tulkens. 2000. Antibiotic efflux pumps. Biochem. Pharmacol. 60:457-470. [DOI] [PubMed] [Google Scholar]
- 39.Vanden Bossche, H., F. Dromer, I. Improvisi, M. Lozano-Chiu, J. H. Rex, and D. Sanglard. 1998. Antifungal drug resistance in pathogenic fungi. Med. Mycol. 36:119-128. [PubMed] [Google Scholar]
- 40.Verweij, P. E., K. L. Oakley, J. Morrissey, G. Morrissey, and D. W. Denning. 1998. Efficacy of LY303366 against amphotericin B-susceptible and -resistant Aspergillus fumigatus in a murine model of invasive aspergillosis. Antimicrob. Agents Chemother. 42:873-878. [DOI] [PMC free article] [PubMed] [Google Scholar]