Abstract
In the mosquito, transamination of 3-HK (3-hydroxykynurenine) to XA (xanthurenic acid) is catalysed by an AGT (alanine glyoxylate aminotransferase) and is the major branch pathway of tryptophan metabolism. Interestingly, malaria parasites hijack this pathway to use XA as a chemical signal for development in the mosquito. Here, we report that the mosquito has two AGT isoenzymes. One is the previously cloned AeHKT [Aedes aegypti HKT (3-HK transaminase)] [Han, Fang and Li (2002) J. Biol. Chem. 277, 15781–15787], similar to hAGT (human AGT), which primarily catalyses 3-HK to XA in mosquitoes, and the other is a typical dipteran insect AGT. We cloned the second AGT from Ae. aegypti mosquitoes [AeAGT (Ae. aegypti AGT)], overexpressed the enzyme in baculovirus/insect cells and determined its biochemical characteristics. We also expressed hAGT for a comparative study. The new cloned AeAGT is highly substrate-specific when compared with hAGT and the previously reported AeHKT and Drosophila AGT, and is translated mainly in pupae and adults, which contrasts with AeHKT that is expressed primarily in larvae. Our results suggest that the physiological requirements of mosquitoes and the interaction between the mosquito and its host appear to be the driving force in mosquito AGT evolution.
Keywords: Aedes aegypti, alanine glyoxylate aminotransferase, Drosophila melanogaster, 3-hydroxykynurenine, 3-hydroxykynurenine transaminase, mosquito
Abbreviations: AGT, alanine glyoxylate aminotransferase; AeAGT, Aedes aegypti AGT; DrAGT, Drosophila melanogaster AGT; hAGT, human AGT; 3-HK, 3-hydroxykynurenine; HKT, 3-HK transaminase; AeHKT, Aedes aegypti HKT; AnHKT, Anopheles gambiae HKT; HTS, high-titre viral stock; LC–MS/MS, liquid chromatography tandem MS; OPT, o-phthaldialdehyde thiol; ORF, open reading frame; PLP, pyridoxal 5-phosphate; RACE, rapid amplification of cDNA ends; XA, xanthurenic acid
INTRODUCTION
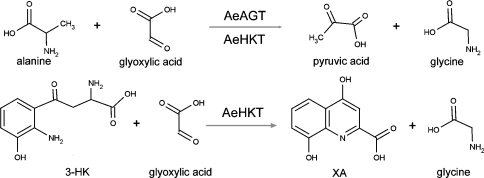
Mosquitoes transmit malaria, dengue fever and West Nile virus, which are major threats to human health and well being in the World. Among them, malaria is considered to be the most prevalent life-threatening disease, with estimates of new cases ranging from 300 millions to 660 millions per year [1]. In mosquitoes, transamination of 3-HK (3-hydroxykynurenine) to XA (xanthurenic acid) is the major branch pathway of tryptophan metabolism [2,3]. XA was identified in 1998 as a natural chemical signal produced by mosquitoes that induces exflagellation of Plasmodium parasites in the midgut [4,5]. Recently, the molecular details of the signalling cascade triggered by XA and resulting in the maturation of Plasmodium gametes have been elucidated [5]. In our previous reports, we detected the presence of a transaminase that catalyses the transamination of 3-HK to XA in Aedes aegypti mosquitoes [2,3]. 3-HK, an intermediate in the tryptophan oxidation pathway, oxidizes easily under physiological conditions, which stimulates radical formation. Therefore it was proposed that this specific mosquito HKT (3-HK transaminase) plays a critical detoxification role in mosquitoes by preventing 3-HK from accumulating. Subsequently, an enzyme with HKT activity was purified and its primary sequence was determined; surprisingly, it shared 45% sequence identity with the hAGT [human AGT (alanine glyoxylate aminotransferase)] [3]. The primary function of the enzyme is transamination of 3-HK to XA and, accordingly, the enzyme was named as AeHKT (Ae. aegypti HKT) [3]. The enzyme-catalysed reaction by HKT or AGT is shown in Scheme 1.
Scheme 1. AeAGT- and AeHKT-catalysed transamination reactions.
AGT plays an important physiological role in living organisms and has been studied in a variety of animals, plants and fungi. It catalyses alanine to pyruvate processing and also converts glyoxylate into glycine. The enzyme is present in peroxisomes or mitochondria in different mammalian species. Recent studies suggest that peroxisomal AGT is responsible for detoxifying glycolate-derived glyoxylate, while the mitochondrial AGT plays a major role in detoxifying hydroxyproline-derived glyoxylate [6–11]. In plants, it has been known to be involved in the photorespiratory glyoxylate cycle within peroxisomes [12,13]. In yeast, disruption of AGT can lead to glycine auxotrophy. For example, a yeast AGT deletion strain was unable to grow when glucose was the only carbon source unless glycine was supplemented [14]. The importance of hepatic AGT in minimizing endogenous oxalate production in some mammals is clearly shown by the autosomal recessive disorder of glyoxylate metabolism primary hyperoxaluria type 1, a potentially lethal condition in which AGT deficiency leads to excessive oxalate synthesis and excretion and the deposition of insoluble calcium oxalate in the kidney [15,16]. AGT also substantially contributes to the metabolism of serine in humans, dogs, cats and rabbits, regardless of its localization in mitochondria or in peroxisomes [17–19]. Therefore it is clear that AGT is essential in living organisms from yeast to humans.
In the present study, we determined that Ae. aegypti mosquitoes have a second AGT coding sequence [AeAGT (Ae. aegypti AGT)] that shares 48% identity with the previously reported AeHKT. A BLAST search of the Anopheles gambiae genomic database revealed that a second AGT is also present in its database. A BLAST search of the genomes of other available model species (including cyanobacteria, archaea, yeast, plants, fruit fly, honeybee, frog, fish, rat, mouse and human) revealed only one AGT in their databases, which raises an essential physiological question as to why two proteins with AGT functions have evolved in the mosquito. Through analysis of the molecular regulation of the two mosquito enzymes and their biochemical functions in comparison with hAGT and Drosophila AGT, we propose the possible functional differentiation and evolution of AeHKT and AeAGT.
EXPERIMENTAL
cDNA cloning and sequencing
The Ae. aegypti EST (expressed sequence tag) (aaest_4508, http://www.tigr.org/tdb/e2k1/aabe/) was used to design a forward primer and a reverse primer. PCR amplification was then performed using a template of first-strand cDNA synthesized from total RNA of Ae. aegypti larvae and pupae (Clontech) and a 730 bp fragment was obtained and sequenced. To clone the full-length AeAGT cDNA, a 5′-RACE (5′-rapid amplification of cDNA ends) reaction was performed using an available kit (Invitrogen), which resulted in the cloning of a full-length cDNA.
Preparation of polyclonal antibodies and Western-blot analysis
To produce antibodies against AeAGT, its recombinant protein was expressed in a bacterial expression system, pET21b (Novagen). A pair of gene-specific primers (forward: 5′-CATATG-TGTCTCAGTGCGTCCGGT-3′; reverse: 5′-AAGCTTCTGGACGTCCGGTTTCAC-3′) containing specific restriction sites (underlined nucleotides) was used to amplify a full-length AeAGT cDNA that was subsequently ligated into the pET21b vector. The recombinant pET21b was transformed into Escherichia coli BL21 (DE3) cells and the recombinant protein that was produced in inclusion bodies was solubilized with a denaturing buffer [20 mM phosphate containing 8 M urea, 500 mM NaCl, 20 mM imidazole, pH 7.4, and TBST (25 mM Tris/HCl, 125 mM NaCl and 0.1% Tween 20, pH 8.0)] and purified using an Ni2+-charged His-Bind resin column (1.5 cm×2.5 cm; Novagen) according to the manufacturer's instructions. The affinity-purified protein was further separated by SDS/PAGE and the recombinant protein was excised, eluted from the gel slices, dialysed against 20 mM Tris/HCl and used as antigen to generate anti-AeAGT polyclonal antibodies in a rabbit (Immunological Resource Center, University of Illinois).
For Western-blot analysis, crude proteins from larvae at 1–5 days after hatching, pupae at 0.5 and 12 h after pupation, and adults at 3 days after emergence, were extracted by homogenization in 20 mM potassium phosphate buffer (pH 7.0) containing 1 μM PMSF, 1 mM 1-phenyl-2-thiourea and 5 mM EDTA using a Polytron homogenizer. The homogenates were centrifuged at 13000 g for 10 min at 4 °C to obtain soluble proteins that were analysed using SDS/PAGE. Protein concentration was determined by a Bio-Rad protein assay kit using BSA as a standard, and equal amounts of crude proteins were separated on the same gel. After electrophoresis, the proteins were transferred on to a nitrocellulose membrane with a transfer buffer [25 mM Tris and 192 mM glycine in 20% (v/v) methanol]. The membrane then was blocked by incubation in 1% BSA solution for 2 h at room temperature (22 °C) and then incubated with rabbit anti-recombinant AeAGT polyclonal antibodies (a dilution of 1:1000 in TBST) for 1 h at room temperature. After washing in TBST, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody (a dilution of 1:20000 in TBST; Sigma) for 1 h at room temperature. After extensive washing, SuperSignal West Pico chemiluminescent substrate (Pierce) was used to detect AeAGT. The blots were visualized on an X-ray film (Kodak).
Production of AeAGT and hAGT recombinant baculovirus
The ORF (open reading frame) of AeAGT was PCR-amplified from a cDNA template from Ae. aegypti pupae using a forward primer (5′-CTGCAGATGGAATACAAAGTGACACCA-3′) and a reverse primer (5′-AAGCTTACATTTTGCCCTTAACCTGGA-3′) containing PstI and HindIII restriction sites (underlined nucleotides) respectively. The hAGT was amplified from a first-strand human liver cDNA (Clontech) using a forward primer (5′-CTCGAGATGGCCTCTCACAAGCTGC-3′) and a reverse primer (5′-AAGCTTTCACAGCTTCTTCTTGGGGC-3′) containing XhoI and HindIII restriction sites (underlined nucleotides) respectively. The PCR product was inserted into a pGEM-T Easy Vector (Promega) and then subcloned into a baculovirus transfer vector, pBlueBac4.5 (Invitrogen). The recombinant AeAGT pBlueBac4.5 transfer vector was co-transfected with linearized Bac-N-Blue™ [AcMNPV (Autographa californica multiple nuclear polyhedrosis virus)] viral DNA in the presence of InsectinPlus™ insect cell-specific liposomes into Spodoptera frugiperda (Sf9) insect cells (Invitrogen). The recombinant baculoviruses were purified by a plaque assay. Blue putative recombinant plaques were transferred on to 12-well microtitre plates and amplified in Sf9 cells. Viral DNA was isolated for PCR analysis to determine the purity of recombinant viruses. An HTS (high-titre viral stock) of pure recombinant virus was generated through amplification in a suspension of cultured Sf9 cells. The titre of HTS and a time course at an MOI (multiplicity of infection) of 6 was established according to the manufacturer's instructions.
Recombinant AeAGT and hAGT protein expression and purification
Sf9 cells were cultured in spinner flasks in TNM-FH medium (MP Biomedicals, Solon, OH, U.S.A.) containing 10%(v/v) fetal bovine serum (Gibco BRL). An HTS of pure recombinant virus was inoculated into the culture at a cell density of 2.5×106 cells/ml and was harvested on the fourth day after AeAGT or hAGT recombinant virus inoculation by centrifugation at 800 g for 15 min at 4 °C. The cell pellet then was dissolved in a lysis buffer (0.1% Triton X-100 in PBS containing 1 μM PMSF). After sonication in cold water, the cell lysate was centrifuged at 20000 g for 20 min at 4 °C and the supernatant was collected. Recombinant AeAGT or hAGT was sequentially purified using DEAE-Sepharose (Amersham Biosciences), phenyl-Sepharose (Amersham Biosciences), hydroxyapatite (Bio-Rad) and gel filtration (Amersham Biosciences) chromatographies. The molecular mass and the purity of the proteins were assessed by SDS/PAGE analysis.
Biochemical characterization
Kynurenine aminotransferase and HKT activity assays were based on previously described methods [3]. Briefly, a reaction mixture of 50 μl, containing either 10 mM L-kynurenine or 5 mM 3-HK, 5 mM glyoxylate, 40 μM PLP (pyridoxal 5-phosphate) and 1 μg of protein sample, was prepared using 100 mM potassium phosphate buffer (pH 7.5). The mixture was incubated for 5 min at 45 °C and the reaction was stopped by adding an equal volume of 0.8 M formic acid. Supernatant of the reaction mixture, obtained by centrifugation at 15000 g for 10 min at 4 °C, was analysed by HPLC with UV detection at 330 and 340 nm for kynurenic acid and XA respectively.
For other aminotransferase activity assays, an assay mixture, containing 10 mM specific amino acids, 5 mM glyoxylate or pyruvate (to test glycine), 40 μM PLP, 100 mM potassium phosphate buffer (pH 7.5), and 1 μg of enzyme in a total volume of 50 μl, was made and it was incubated for 5 min at 45 °C. Using HPLC with electrochemical detection, the product was quantified based on the detection of OPT (o-phthaldialdehyde thiol)–amino acid product conjugate after the corresponding reaction mixtures were derivatized by OPT reagent [20]. The kinetic parameters of the recombinant enzymes towards the amino acid or α-oxo acid were calculated by fitting the Michaelis–Menten equation to the experimental data using the Enzyme Kinetics Module in SigmaPlot 8.2 (SPSS Science). Protein concentration was determined using a Bio-Rad protein assay kit with BSA as a standard.
To determine the effect of pH and temperature on AeAGT and hAGT activity, a buffer mixture consisting of 50 mM phosphate and 50 mM boric acid was prepared and the pH of the buffer was adjusted to 6.0, 7.0, 8.0, 9.0 and 10.0 respectively. The buffer mixture was selected to maintain a consistent buffer composition and ionic strength, yet have sufficient buffering capacity for a relatively broad pH range. To test the enzyme activity change at different temperatures, AGT activity was assayed under the same conditions except for the change in temperatures.
RESULTS
Cloning and analysis of AeAGT cDNA
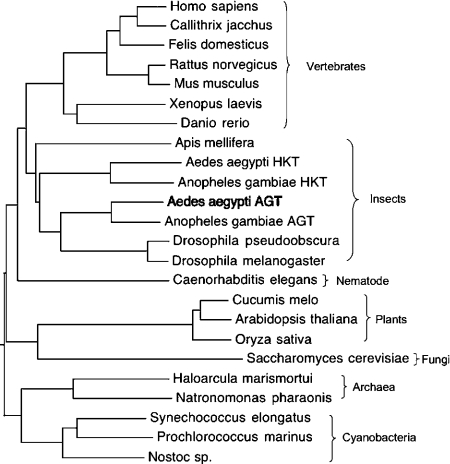
The AeAGT homologous fragment (Ae. aegypti EST sequence, aaest_804) was identified by a BLAST search of the TIGR (The Institute for Genomics Research; Rockville, MD, U.S.A.) Ae. aegypti BAC (bacterial artificial chromosome) ends sequence database with hAGT and DrAGT (Drosophila melanogaster AGT). This AGT homologous fragment matched the C-terminal region of hAGT and DrAGT. A partial 970 bp cDNA fragment was amplified from a first-strand larval/pupal cDNA using specific primers based on the C-terminal sequence of the Ae. aegypti EST sequence and then sequenced. Extension of the 970 bp fragment using a 5′-RACE method led eventually to the isolation of its full-length cDNA that contained 1369 nt with an ORF of 1179 nt coding for 393 amino acids (GenBank® accession no. DQ182329) (Figure 1). BLAST analysis of the deduced sequence revealed a 48% identity with AeHKT [3], 58% identity with DrAGT [3,21] and 45–46% identity with AGTs from humans and mammals. By BLAST searching of the NCBI protein databases (http://www.ncbi.nlm.nih.gov: 80/BLAST/Blast.cgi), it was determined that An. gambiae also has two AGT isoenzymes, one of which recently has been identified as an An. gambiae HKT (AnHKT) with the same function as AeHKT [22]. Cyanobacteria, archaea, yeast, plants, other insects (fruit fly and honeybee), frog, fish, rat, mouse and human have only one AGT. AeAGT and Anopheles AGT have 79% sequence identity, with both of them in the same group as DrAGT in the phylogenetic tree (Figure 2), suggesting that mosquito AGT might be a typical dipteran insect AGT.
Figure 1. The nucleotide and deduced amino acid sequences of AeAGT.
Figure 2. Phylogenetic tree of AGTs.
The alignment was done with CLUSTAL W in biology workbench (http://seqtool.sdsc.edu/CGI/BW.cgi#!) and the dendrogram was edited with TreeEdit (http://evolve.zoo.ox.ac.uk/software.html?id=treeedit).
Dr. melanogaster is a tractable system for studying conserved aspects of eukaryotic gene function and, with the production of other insect genome sequences, it is a useful baseline for evolutionary studies of gene organization [23]. Whole-genome sequence is available for Dr. melanogaster, Ae. aegypti and An. gambiae, the latter of which diverged from Dr. melanogaster approx. 250 million years ago [24], but the assembly is only available for Dr. melanogaster and An. gambiae (URL: http://www.ensembl.org/index.html). Therefore we used AGTs from An. gambiae for a comparative analysis of chromosomal localization and gene organization in an attempt to determine mosquito AGT evolution and function. The chromosomal localization of the two AGTs from An. gambiae is quite different (AnAGT: Chromosome 2R; AnHKT: Chromosome 3L), suggesting that they may function differently in mosquitoes. By analysing the gene organization of AGTs from An. gambiae, fruit fly and humans, we observed that both Anopheles AGT and DrAGT have three exons, which provides partial support for the hypothesis that mosquito AGT and DrAGT might have a common evolutionary origin.
Expression profile of AeAGT during development
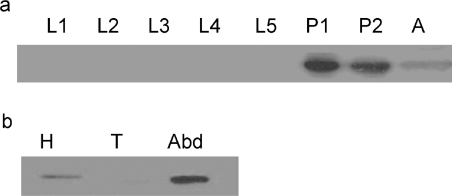
Using Western-blot analysis, AeAGT protein was detected only in the pupal and adult stages of Ae. aegypti (Figure 3a). Further analysis showed that AeAGT was present in both the head and abdomen of adult females (Figure 3b). AeHKT was present in larvae [3], but no larval protein was positively stained using the anti-AeAGT antibodies in Western blot, suggesting that antibodies against AeAGT do not cross-react with AeHKT.
Figure 3. Translational profile of AeAGT.
(a) Developmental-specific expression of the AeAGT protein. An equal amount of total protein (60 μg), extracted from 1- to 5-day-old larvae (L1–L5), newly formed white pupae (P1), 12 h-old black pupae (P2) and 1-day-old female adults (A), were separated by SDS/12% PAGE and were detected on an immunoblot with anti-AeAGT polyclonal antibodies. (b) Tissue-specific expression of the AeAGT protein. Total protein was extracted from the head (H), thorax (T) and abdomen (Abd) of females at 24 h after blood feeding and an equal amount of total protein (40 μg) was resolved by SDS/12% PAGE prior to immunoblotting.
Recombinant AeAGT and hAGT protein production, purification and identification
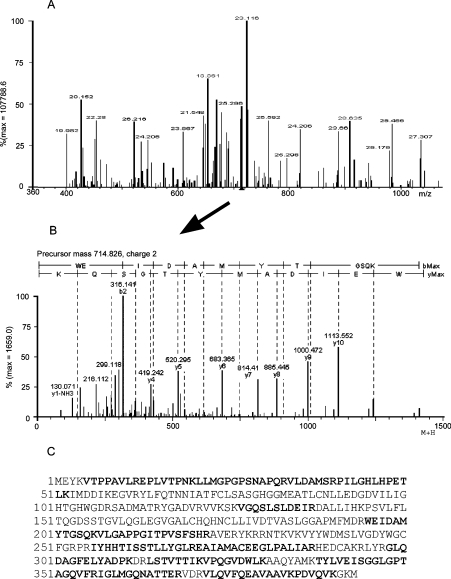
When soluble protein samples from Sf9 cells, collected on the third day after viral inoculation, were analysed by SDS/PAGE, a major protein band with a relative molecular mass of 42 kDa was observed in the AeAGT recombinant virus-infected cells (results not shown). A similar size protein band also was observed in the hAGT recombinant virus-infected cells. Purification of the recombinant AeAGT and hAGT was achieved by DEAE-Sepharose, phenyl-Sepharose, hydroxyapatite and gel-filtration chromatographies. The purified AeAGT was trypsin-digested and then analysed by LC–MS/MS (liquid chromatography tandem MS) with an ion search of the spectral data using ProteinLynx software to verify the AeAGT identity of the recombinant protein (Figure 4). Recombinant hAGT was verified by MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight) (results not shown).
Figure 4. Verification of recombinant AeAGT by peptide MS.
Purified recombinant protein was digested with trypsin, the tryptic peptides were analysed by LC–MS/MS, and collected spectral data were searched against an in-house mosquito protein database. (A) Precursor ions (MS) collected during LC–MS/MS analysis of trypsin-digested protein. Peaks in boldface are those matched to AeAGT sequence. (B) The tandem spectrum (MS/MS) of the doubly charged 714.826 peptide ion from (A) and the derived amino acid sequence. (C) Coverage map shows unambiguous match (boldface) of the spectral data to AeAGT. These data provide evidence that the expressed recombinant protein is AeAGT.
Substrate specificity and kinetic parameters
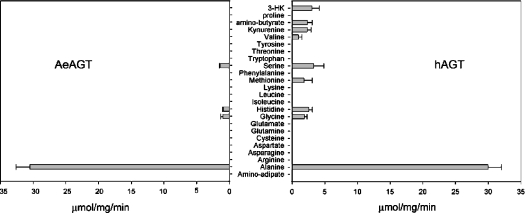
The recombinant protein was tested for AGT activity using alanine and glyoxylate as an amino donor and acceptor respectively, and it displayed relatively high AGT activity. Under the applied assay conditions, the recombinant AeAGT showed a specific activity of 0.5 μmol·s−1·mg of protein−1. When 3-HK or kynurenine was used as the amino donor and glyoxylate as the amino acceptor, transamination of 3-HK or kynurenine was not detected. The possibility that the enzyme catalysed the transamination of the other 19 proteingenic amino acids, aminoadipate and aminobutyrate in the presence of glyoxylate or pyruvate was also tested. The recombinant protein was only able to catalyse the transamination of serine (0.02 μmol·s−1·mg−1), histidine (0.02 μmol·s−1·mg−1) and glycine (0.02 μmol·s−1·mg−1) with pyruvate as the amino group acceptor. The same method was also applied for the recombinant hAGT protein. The hAGT showed a high specific AGT activity and also displayed considerable activities towards kynurenine, 3-HK, aminobutyrate, methionine and valine. The detailed results are shown in Figure 5. Table 1 lists the kinetic parameters of the AeAGT and hAGT and the previously published results for DrAGT and AeHKT [21]. Both AeHKT and hAGT show activity to 3-HK and kynurenine; in contrast, DrAGT and AeAGT do not have any detectable activity towards them using the same method. The substrate specificity results show that AeAGT is similar to DrAGT, and AeHKT is similar to hAGT, but the former has a much greater efficiency in mediating the 3-HK to XA pathway.
Figure 5. Transamination activity of AeAGT and hAGT towards different amino acids.
Glyoxylate was used as the amino group acceptor for all amino acids, except for glycine. When glycine was used as amino group donor, pyruvate was used as an amino group acceptor. Left panel: transamination activity of AeAGT; right panel: transamination activity of hAGT.
Table 1. Kinetic parameters of AeAGT and hAGT towards alanine, kynurenine, 3-HK (DL form) and comparison with previously reported parameters for AeHKT and DrAGT [21].
Amino acids were assayed with glyoxylate as the amino group acceptor. Kynurenine and 3-HK were tested from 0.2 to 10 mM, and alanine was tested from 0.6 to 80 mM. Data are derived from non-linear regression analysis by using the Enzyme Kinetics Module in SigmaPlot 8.2 and the errors represent the S.E. from regression.
| Substrate | Km (mM) | kcat (s−1) | kcat/Km (M−1·s−1) | |
|---|---|---|---|---|
| AeAGT | Alanine | 34±10 | 117±15 | 3450 |
| hAGT | Alanine | 15±2 | 53±3 | 3483 |
| Kynurenine | 5.4±1.7 | 4.3±0.8 | 800 | |
| 3-HK | 9.5±3.9 | 4.4±1 | 450 | |
| AeHKT | Alanine | 18±2 | 20±1 | 1117 |
| Kynurenine | 3.7±0.2 | 12±3.7 | 3250 | |
| 3-HK | 2.6±0.6 | 34.2±5.3 | 13150 | |
| DrAGT | Alanine | 149±44 | 131±20 | 879 |
Effect of pH and temperature on AeAGT and hAGT
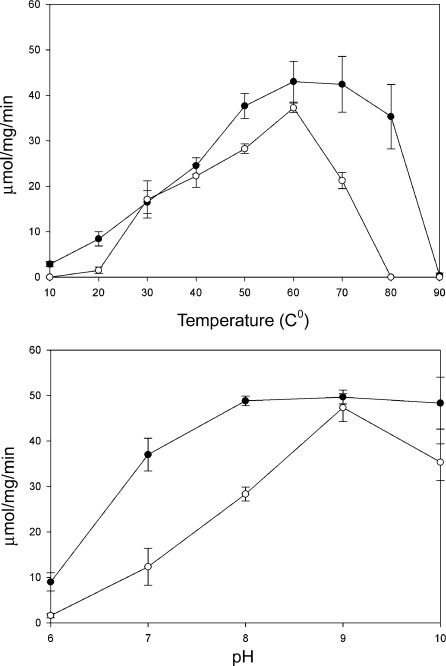
When the phosphate and borate buffer mixture, adjusted to pH 6.0–10.0, was used to prepare the enzyme/alanine/glyoxylate reaction mixtures, both AeAGT and hAGT showed little activity at pH 6.0. However, high activity levels were noted at pH 7.0–10.0, reaching the highest at pH 9.0. Both enzymes showed resistance to heat, but AeAGT, which displayed high activity even at 80 °C, was more resistant to heat than hAGT (Figure 6). Comparison of the temperature and pH-related activity profiles of AeAGT, hAGT, DrAGT and AeHKT again show that AeAGT is similar to DrAGT and AeHKT to hAGT.
Figure 6. Effect of pH and temperature on enzyme activity.
(A, B) The activities of recombinant AeAGT and hAGT at different temperatures (A) and pH values (B). The lines with closed circles and open circles represent the specific activities of AeAGT and hAGT respectively.
DISCUSSION
AGT has been studied in a number of model species. Disruption of its function affects the conversion of glyoxylate into glycine, which may lead to lethal conditions in humans [15,16], and so AGT studies have emphasized its role in the glyoxylate to glycine pathway. Because concrete evidence for the essential role of AGT in maintaining physiological conditions in living organisms has been demonstrated in some species [14–16], it is reasonable to suggest that AGT is indispensable in other animal species as well. Results obtained in the present study that investigated the molecular regulation and biochemical function of AeAGT, in conjunction with results of our previous experiments studying the biochemical characterization of AeHKT [3,21], suggest that AGT is essential in mosquitoes. It seems apparent that the AeHKT protein, translated during larval development, is involved in the detoxification of 3-HK and glyoxylate metabolism, whereas AeAGT, translated in pupae and adults, is primarily responsible for glyoxylate metabolism.
Characterization of AeHKT was initiated following the determination that the transamination of 3-HK to XA was a major pathway in mosquitoes. In mammals, 3-HK either can be hydrolysed by kynureninase to 3-hydroxyanthranilic acid that can be completely oxidized to CO2 and water through a complicated biochemical pathway or it can be used to synthesize NAD(P)+ [25]. Kynurenine to 3-HK is the major pathway in mosquitoes, but because they do not have kynureninase, the hydrolysis pathway for 3-HK is blocked. 3-HK is a natural metabolite, but it is oxidized easily under physiological conditions, stimulating the production of reactive oxygen species [26–28]. To prevent the accumulation of 3-HK, mosquito larvae efficiently convert the chemically reactive 3-HK into the chemically stable XA [2]. Purification of the protein with 3-HK transamination activity resulted in the isolation of a protein sharing high similarity with mammalian AGT [3], which suggests that the primary function of this mosquito AGT, AeHKT, has expanded to include the detoxification of 3-HK.
In a previous study, we compared substrate specificity between AeHKT and DrAGT and our results showed that DrAGT has high AGT activity and is able to catalyse the transamination of a few other amino acids, but it showed no activity towards 3-HK and kynurenine [21]. The BLAST search analysis clearly indicated that Dr. melanogaster has only one AGT. We interpreted the functional difference between AeHKT and DrAGT based on the difference in their food resources. Mosquito larvae are aquatic and their food resources primarily include protein-rich micro-organisms. In mosquito larvae, a considerable portion of tryptophan, obtained from food supplies, is oxidized to 3-HK [2]. Because mosquitoes cannot dispose of 3-HK through hydrolysis and subsequent oxidation like mammals, the transamination of the chemically reactive 3-HK to the relatively chemically stable XA by AeHKT is considered to be the mechanism by which mosquitoes prevent the over accumulation of 3-HK [3]. In contrast, Dr. melanogaster obtains its food from plant resources that are rich in glycolate, the precursor of glyoxylate; therefore we propose that it is more important to prevent the accumulation of glyoxylate in Dr. melanogaster. We believe that such an explanation provides a valid argument regarding the functional evolution of mosquito and DrAGTs.
Interestingly, 3-HK is also the initial precursor for the production of ommochromes that are major eye pigments in mosquitoes. Compound eye development and eye pigmentation occur mainly during the pupal and early adult stages. Coincidently, HKT activity is diminished in pupae and adults with the accumulation of high levels of 3-HK in the compound eyes [29]. The down-regulation of AeHKT is likely to be a tactic for mosquitoes to allow 3-HK to be transported and used for eye pigmentation. Although it seems clear that detoxification of 3-HK is a major function of AeHKT in mosquito larvae, the enzyme has high AGT activity and probably plays a critical role in glyoxylate metabolism as well. Consequently, the interruption of AeHKT expression during the pupal and adult stages would affect the metabolism of glyoxylate in pupae and adults and provides an explanation as to why the highly substrate-specific (i.e. highly active for the glyoxylate to glycine pathway without detectable HKT activity) and stage-specific AeAGT has evolved in mosquitoes.
The high sequence identity of AeAGT and AeHKT with the AGTs from cyanobacteria, archaea, yeast, plants, other insects (fruit fly and honeybee), frog, fish, rat, mouse and humans suggests that these enzymes have evolved from a common ancestry. The CLUSTAL W alignment and phylogenetic analysis (Figure 2) show that AeAGT seems to be more closely related to DrAGT than AeHKT. Based on substrate specificity, pH and temperature-dependent activity profiles, it seems clear that AeHKT is more closely related to hAGT. The primary function of AeHKT seems to have deviated substantially from the typical dipteran insect AGT [3]. Although AeHKT and AeAGT have significant sequence identity with mammalian AGTs (Figure 7), AeHKT shares similar substrate specificity and other characteristics with hAGT. For example, both hAGT and AeHKT [21] can catalyse the transamination of 3-HK, kynurenine and a number of other amino acids under exactly the same assay conditions, thereby confirming a previous report that showed that native hAGT, purified from human liver, had activity towards 3-HK and kynurenine [30].
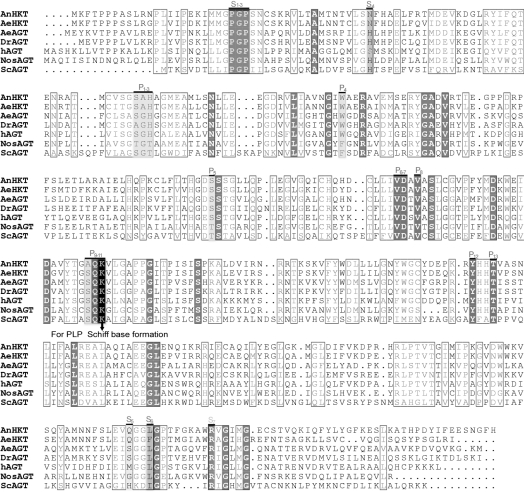
Figure 7. Alignment and active site comparison of AeAGT, AnHKT, DrAGT, hAGT, AeHKT, Nostoc AGT (NosAGT) and yeast AGT (ScAGT) based on known three-dimensional structures and amino acid sequences.
The residues in light or dark grey background, arbitrarily labelled P1 to P13, are PLP-binding sites; similarly, substrate-binding sites are labelled S1 to S7. Identity residues among all seven AGTs are labelled with a dark grey background and boxed, and similar residues are boxed only.
Mammalian AGT is localized either in mitochondria or in peroxisomes, which is dictated by the presence of the N-terminal mitochondrial or C-terminal peroxisomal signal peptide in the protein. The localization of AGT provides useful evidence in predicting the metabolic roles of AGT in mammals and other vertebrates. Using online bioinformatic tools, AeAGT and DrAGT are predicted to be peroxisomal proteins (URL: http://mendel.imp.ac.at/mendeljsp/sat/pts1/PTS1predictor.jsp) and AeHKT is predicted to be a mitochondrial protein (URL: http://ihg.gsf.de/ihg/mitoprot.html). Based on the respective metabolic roles of mammalian peroxisomal and mitochondrial AGTs, it seems reasonable to suggest that AeAGT and DrAGT may be involved in the metabolism of glycolate-derived glyoxylate, whereas AeHKT is involved in converting hydroxyprolone-derived glyoxylate into glycine in addition to its function in the transamination of 3-HK to XA.
Differences in enzyme function, in principle, should be reflected in both the primary and three-dimensional structures. The three-dimensional structures for hAGT [31], yeast AGT [32], Nostoc AGT [33] and AnHKT [34] are available. Based on the primary sequences of AeAGT, AeHKT, AnHKT and DrAGT in comparison with hAGT, yeast AGT, Nostoc AGT and AnHKT sequences and their corresponding three-dimensional structures, all residues involved in PLP binding in those sequences are conserved (Figure 7). Other than the PLP-binding sites, there are seven substrate-binding sites and five of them are conserved (Figure 7). The fourth binding residues are serine in hAGT and asparagine in AnHKT and AeHKT, but the binding site is histidine in the other four AGTs (Figure 7). The fifth binding residue is methionine in hAGT, glutamic residue in AnHKT and AeHKT, but the fifth binding residue becomes serine in AeAGT and DrAGT. The difference in substrate-binding sites may explain the substrate specificities of different AGTs. A three-dimensional structural study is ongoing for DrAGT, AeAGT and AeHKT in our laboratory (Q. Han, H. Robinson, Y. G. Gao and J. Li, unpublished work). We have produced crystals for all three enzymes and diffraction data for AeAGT and AeHKT, as it will be beneficial to have additional evidence to analyse mosquito AGT evolution and to understand the mechanism of catalysis.
Both AeHKT and AeAGT, if determined through random cloning or a genome project, would have been listed as AGTs based on their high sequence identity with mammalian AGTs, and their assigned functional identity also would have been assumed because of the level of their sequence identity with mammalian AGTs (see Figure 7). Primary sequence comparison of AeHKT and AeAGT with mammalian AGTs is insufficient to determine if there is any functional difference between the two Ae. aegypti transaminases and the mammalian AGTs, let alone determine the functional difference between the two mosquito AGTs. However, biochemical characteristics and the molecular regulation of these two enzymes during mosquito development, in conjunction with the physiological requirements of mosquito development, clearly demonstrate the functional differences between AeHKT and AeAGT and provide a reasonable explanation as to why two proteins with AGT functions have evolved in mosquitoes. Based on the substrate specificities of the characterized hAGT, AeAGT, AeHKT and DrAGT, AeHKT has a rather broad substrate specificity, followed in decreasing specificity by hAGT, DrAGT and AeAGT; consequently, this result provides additional insight towards a comprehensive understanding of the functional adaptation and evolution of AGTs in living organisms.
Acknowledgments
This work was supported by a grant from the NIH (National Institutes of Health; Bethesda, MD, U.S.A.) (RO1 AI 44399). We are grateful to Dr Brenda Beerntsen (Department of Veterinary Pathobiology, University of Missouri, Columbia, MO, U.S.A.) for critically reading this paper, and to Dr Ruiliang Yao (Department of Pathobiology, University of Illinois, Urbana, IL, U.S.A.) for helping with the in-gel digests of the recombinant enzymes.
References
- 1.Snow R. W., Guerra C. A., Noor A. M., Myint H. Y., Hay S. I. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature (London) 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J., Li G. Transamination of 3-hydroxykynurenine to produce xanthurenic acid: a major branch pathway of tryptophan metabolism in the mosquito, Aedes aegypti, during larval development. Insect Biochem. Mol. Biol. 1997;27:859–867. doi: 10.1016/s0965-1748(97)00068-4. [DOI] [PubMed] [Google Scholar]
- 3.Han Q., Fang J., Li J. 3-Hydroxykynurenine transaminase identity with alanine glyoxylate transaminase. A probable detoxification protein in Aedes aegypti. J. Biol. Chem. 2002;277:15781–15787. doi: 10.1074/jbc.M201202200. [DOI] [PubMed] [Google Scholar]
- 4.Garcia G. E., Wirtz R. A., Barr J. R., Woolfitt A., Rosenberg R. Xanthurenic acid induces gametogenesis in Plasmodium, the malaria parasite. J. Biol. Chem. 1998;273:12003–12005. doi: 10.1074/jbc.273.20.12003. [DOI] [PubMed] [Google Scholar]
- 5.Billker O., Dechamps S., Tewari R., Wenig G., Franke-Fayard B., Brinkmann V. Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite. Cell. 2004;117:503–514. doi: 10.1016/s0092-8674(04)00449-0. [DOI] [PubMed] [Google Scholar]
- 6.Birdsey G. M., Lewin J., Holbrook J. D., Simpson V. R., Cunningham A. A., Danpure C. J. A comparative analysis of the evolutionary relationship between diet and enzyme targeting in bats, marsupials and other mammals. Proc. Biol. Sci. 2005;272:833–840. doi: 10.1098/rspb.2004.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birdsey G. M., Lewin J., Cunningham A. A., Bruford M. W., Danpure C. J. Differential enzyme targeting as an evolutionary adaptation to herbivory in carnivora. Mol. Biol. Evol. 2004;21:632–646. doi: 10.1093/molbev/msh054. [DOI] [PubMed] [Google Scholar]
- 8.Takayama T., Fujita K., Suzuki K., Sakaguchi M., Fujie M., Nagai E., Watanabe S., Ichiyama A., Ogawa Y. Control of oxalate formation from L-hydroxyproline in liver mitochondria. J. Am. Soc. Nephrol. 2003;14:939–946. doi: 10.1097/01.asn.0000059310.67812.4f. [DOI] [PubMed] [Google Scholar]
- 9.Holbrook J. D., Danpure C. J. Molecular basis for the dual mitochondrial and cytosolic localization of alanine:glyoxylate aminotransferase in amphibian liver cells. J. Biol. Chem. 2002;277:2336–2344. doi: 10.1074/jbc.M107047200. [DOI] [PubMed] [Google Scholar]
- 10.Danpure C. J. Variable peroxisomal and mitochondrial targeting of alanine: glyoxylate aminotransferase in mammalian evolution and disease. BioEssays. 1997;19:317–326. doi: 10.1002/bies.950190409. [DOI] [PubMed] [Google Scholar]
- 11.Watts R. W. Alanine glyoxylate aminotransferase deficiency: biochemical and molecular genetic lessons from the study of a human disease. Adv. Enzyme Regul. 1992;32:309–327. doi: 10.1016/0065-2571(92)90024-t. [DOI] [PubMed] [Google Scholar]
- 12.Rehfeld D. W., Tolbert N. E. Aminotransferases in peroxisomes from spinach leaves. J. Biol. Chem. 1972;247:4803–4811. [PubMed] [Google Scholar]
- 13.Liepman A. H., Olsen L. J. Peroxisomal alanine: glyoxylate aminotransferase (AGT1) is a photorespiratory enzyme with multiple substrates in Arabidopsis thaliana. Plant J. 2001;25:487–498. doi: 10.1046/j.1365-313x.2001.00961.x. [DOI] [PubMed] [Google Scholar]
- 14.Schlosser T., Gatgens C., Weber U., Stahmann K. P. Alanine:glyoxylate aminotransferase of Saccharomyces cerevisiae-encoding gene AGX1 and metabolic significance. Yeast. 2004;21:63–73. doi: 10.1002/yea.1058. [DOI] [PubMed] [Google Scholar]
- 15.Danpure C. J. Primary hyperoxaluria. In: Scriver C. R., Beaudet A. L., Sly W. S., Valle D., Childs B., Kinzler K. W., Vogelstein B., editors. The Molecular and Metabolic Bases of Inherited Disease. New York: McGraw-Hill; 2001. pp. 3323–3367. [Google Scholar]
- 16.Danpure C. J., Jennings P. R., Leiper J. M., Lumb M. J., Oatey P. B. Targeting of alanine:glyoxylate aminotransferase in normal individuals and its mistargeting in patients with primary hyperoxaluria type 1. Ann. N.Y. Acad. Sci. 1996;804:477–490. doi: 10.1111/j.1749-6632.1996.tb18638.x. [DOI] [PubMed] [Google Scholar]
- 17.Rowsell E. V., Carnie J. A., Wahbi S. D., Al-Tai A. H., Rowsell K. V. L-serine dehydratase and L-serine-pyruvate aminotransferase activities in different animal species. Comp. Biochem. Physiol. B. 1979;63:543–555. doi: 10.1016/0305-0491(79)90061-0. [DOI] [PubMed] [Google Scholar]
- 18.Beliveau G. P., Freedland R. A. Metabolism of serine, glycine and threonine in isolated cat hepatocytes Felis domestica. Comp. Biochem. Physiol. B. 1982;71:13–18. doi: 10.1016/0305-0491(82)90168-7. [DOI] [PubMed] [Google Scholar]
- 19.Xue H. H., Sakaguchi T., Fujie M., Ogawa H., Ichiyama A. Flux of the L-serine metabolism in rabbit, human, and dog livers. Substantial contributions of both mitochondrial and peroxisomal serine:pyruvate/alanine:glyoxylate aminotransferase. J. Biol. Chem. 1999;274:16028–16033. doi: 10.1074/jbc.274.23.16028. [DOI] [PubMed] [Google Scholar]
- 20.Han Q., Fang J., Li J. Kynurenine aminotransferase and glutamine transaminase K of Escherichia coli: identity with aspartate aminotransferase. Biochem. J. 2001;360:617–623. doi: 10.1042/0264-6021:3600617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han Q., Li J. Comparative characterization of Aedes 3-hydroxykynurenine transaminase/alanine glyoxylate transaminase and Drosophila serine pyruvate aminotransferase. FEBS Lett. 2002;527:199–204. doi: 10.1016/s0014-5793(02)03229-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossi F., Lombardo F., Paglino A., Cassani C., Miglio G., Arca B., Rizzi M. Identification and biochemical characterization of the Anopheles gambiae 3-hydroxykynurenine transaminase. FEBS J. 2005;272:5653–5662. doi: 10.1111/j.1742-4658.2005.04961.x. [DOI] [PubMed] [Google Scholar]
- 23.Bergman C. M., Pfeiffer B. D., Rincon-Limas D. E., Hoskins R. A., Gnirke A., Mungall C. J., Wang A. M., Kronmiller B., Pacleb J., Park S., et al. Assessing the impact of comparative genomic sequence data on the functional annotation of the Drosophila genome. Genome Biol. 2002;3:RESEARCH0086. doi: 10.1186/gb-2002-3-12-research0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaunt M. W., Miles M. A. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol. Biol. Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- 25.Stone T. W. Neuropharmacology of quinolinic and kynurenic acids. Pharmacol. Rev. 1993;45:309–379. [PubMed] [Google Scholar]
- 26.Okuda S., Nishiyama N., Saito H., Katsuki H. Hydrogen peroxide-mediated neuronal cell death induced by an endogenous neurotoxin, 3-hydroxykynurenine. Proc. Natl. Acad. Sci. U.S.A. 1996;93:12553–12558. doi: 10.1073/pnas.93.22.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okuda S., Nishiyama N., Saito H., Katsuki H. 3-Hydroxykynurenine, an endogenous oxidative stress generator, causes neuronal cell death with apoptotic features and region selectivity. J. Neurochem. 1998;70:299–307. doi: 10.1046/j.1471-4159.1998.70010299.x. [DOI] [PubMed] [Google Scholar]
- 28.Wei H., Leeds P., Chen R. W., Wei W., Leng Y., Bredesen D. E., Chuang D. M. Neuronal apoptosis induced by pharmacological concentrations of 3-hydroxykynurenine: characterization and protection by dantrolene and Bcl-2 overexpression. J. Neurochem. 2000;75:81–90. doi: 10.1046/j.1471-4159.2000.0750081.x. [DOI] [PubMed] [Google Scholar]
- 29.Li J., Beerntsen B. T., James A. A. Oxidation of 3-hydroxykynurenine to produce xanthommatin for eye pigmentation: a major branch pathway of tryptophan catabolism during pupal development in the yellow fever mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 1999;29:329–338. doi: 10.1016/s0965-1748(99)00007-7. [DOI] [PubMed] [Google Scholar]
- 30.Okuno E., Minatogawa Y., Nakamura M., Kamoda N., Nakanishi J., Makino M., Kido R. Crystallization and characterization of human liver kynurenine–glyoxylate aminotransferase. Identity with alanine–glyoxylate aminotransferase and serine–pyruvate aminotransferase. Biochem. J. 1980;189:581–590. doi: 10.1042/bj1890581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X., Roe S. M., Hou Y., Bartlam M., Rao Z., Pearl L. H., Danpure C. J. Crystal structure of alanine:glyoxylate aminotransferase and the relationship between genotype and enzymatic phenotype in primary hyperoxaluria type 1. J. Mol. Biol. 2003;331:643–652. doi: 10.1016/s0022-2836(03)00791-5. [DOI] [PubMed] [Google Scholar]
- 32.Meyer P., Liger D., Leulliot N., Quevillon-Cheruel S., Zhou C. Z., Borel F., Ferrer J. L., Poupon A., Janin J., van Tilbeurgh H. Crystal structure and confirmation of the alanine:glyoxylate aminotransferase activity of the YFL030w yeast protein. Biochimie. 2005;87:1041–1047. doi: 10.1016/j.biochi.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Han G. W., Schwarzenbacher R., Page R., Jaroszewski L., Abdubek P., Ambing E., Biorac T., Canaves J. M., Chiu H. J., Dai X., et al. Crystal structure of an alanine-glyoxylate aminotransferase from Anabaena sp. at 1.70 Å resolution reveals a noncovalently linked PLP cofactor. Proteins. 2005;58:971–975. doi: 10.1002/prot.20360. [DOI] [PubMed] [Google Scholar]
- 34.Rossi F., Garavaglia S., Giovenzana G. B., Arca B., Li J., Rizzi M. Crystal structure of the Anopheles gambiae 3-hydroxykynurenine transaminase. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5711–5716. doi: 10.1073/pnas.0510233103. [DOI] [PMC free article] [PubMed] [Google Scholar]