Abstract
PER-1, an extended-spectrum β-lactamase, has been reported only in Europe. We detected PER-1 in 53 of 97 acinetobacters in Korea, mainly in the sputum of intensive care unit patients. Pulsed-field gel electrophoresis analysis suggested that clonal spread had occurred. Only PCR reliably detected PER-1 producers. PER-1 producers may also exist in other Asian countries.
Multidrug-resistant Acinetobacter spp. and Pseudomonas aeruginosa frequently cause serious nosocomial infections. β-Lactamase production is the most important mechanism of β-lactam resistance in gram-negative pathogens. PER-1 is an extended-spectrum β-lactamase which was first found in a P. aeruginosa strain in France (7) and was then subsequently detected in Acinetobacter spp. and P. aeruginosa in Turkey and Italy (5, 12). PER-2 was reported in Salmonella enterica serovar Typhimurium in Argentina (1). However, to our knowledge, the PER-1 enzyme has not been reported in any other countries.
In a Korean hospital, cefepime-resistant acinetobacters increased significantly from 29 to 47% of isolates, but those of P. aeruginosa increased only slightly, from 14 to 17%, between 1991 (2) and 2000 (data not shown), which suggested the presence of PER-1-producing acinetobacters, as cefepime is stable to AmpC but not to PER-1 β-lactamase (5, 11). PER-1 production has been found to be an independent indicator of poor prognosis (13), but its detection by the double-disk synergy (DDS) test is difficult (9).
The aim of this study was to determine the presence of PER-1-producing strains of acinetobacter and P. aeruginosa in Korea. The performance of the DDS test for the detection of PER-1-producing isolates was also evaluated.
Strains of acinetobacter and P. aeruginosa were isolated in 2001 and 2002 from patients in two tertiary-care Korean hospitals and were identified by conventional tests (3, 10) or by using the ATB 32 GN system (bioMerieux, Marcy l'Etoile, France). Antimicrobial susceptibilities were determined by the disk diffusion and the agar dilution methods (6). DDS was tested with cefotaxime, ceftazidime, aztreonam, cefepime, and amoxicillin-clavulanate disks (Beckton Dickinson, Cockeysville, Md.). Clavulanate (GlaxoSmithKline, Greenford, United Kingdom) and tazobactam (Wyeth, Pearl River, N.Y.) were used to add to cefepime disks.
blaPER-1 and blaPER-2 alleles were detected by PCR by using previously reported primers (1, 9), heat-extracted templates, and the mixture containing Taq DNA polymerase and dNTP (Premix, Bioneer, Daejeon, Korea). The amplification conditions used were 30 cycles of denaturation at 94°C for 40 s, annealing at 50°C for 30 s, and extension at 72°C for 50 s. The nucleotide sequences of both strands of the blaPER-1 allele were analyzed with PCR products by the dideoxy-chain termination method with an ABI 3700 DNA sequencer (Perkin-Elmer, Foster City, Calif.).
The isoelectric points of β-lactamases were determined as described previously (4). The transfer of blaPER-1 was tested by plate mating with a rifampin-resistant Acinetobacter baumannii recipient (YMC02/8/P534). Pulsed-field gel electrophoresis (PFGE) of the SmaI-digested genomic DNA was performed and analyzed as described previously (4). The DNA bands separated by PFGE were transferred to a nylon membrane and hybridized by using a digoxigenin-labeled blaPER-1 probe (Roche Diagnostics, Mannheim, Germany).
In a preliminary study, blaPER-1 alleles were detected by PCR in five DDS-positive acinetobacters from a hospital in a southern region of Korea. The nucleotide sequences of blaPER-1 and the pI of the β-lactamase in our isolates were identical to those reported previously (5, 7, 8, 12).
The prevalence of blaPER-1-carrying acinetobacter was determined in another hospital located approximately 300 km north of the first hospital. Of the 97 consecutive isolates of acinetobacter, 61 were resistant to ceftazidime and 53 (54.6%) were positive for the blaPER-1 allele: 51 A. baumannii, 1 Acinetobacter genomospecies 3, and 1 unidentifiable Acinetobacter spp.
blaPER-1 was not detected in 101 consecutive or in 181 ceftazidime-nonsusceptible P. aeruginosa isolates. blaPER-2 was not detected in any of the acinetobacter or P. aeruginosa isolates (data not shown).
blaPER-1 was detected in 46% of acinetobacter and 11% of P. aeruginosa isolates in Turkey (12). It is interesting that blaPER-1-positive acinetobacters were detected in Korea, which is geographically distanced from Europe. The resistance may also exist in other Asian countries.
All of the blaPER-1-positive acinetobacters were resistant to ceftazidime, cefepime, cefotaxime, and aztreonam, but the resistance rates of the blaPER-1-negative isolates to these drugs and ampicillin-sulbactam were much lower (Table 1). The resistance rates of blaPER-1-positive and -negative isolates were 64 and 30% to amikacin, 75 and 48% to gentamicin, 75 and 52% to tobramycin, 98 and 41% to trimethoprim-sulfamethoxazole, and 83 and 39% to tetracycline, respectively (data not shown). We anticipate difficulties in treating infections due to PER-1-producing acinetobacters, as they are resistant to all cephalosporins, including cefepime, and frequently to aminoglycosides, trimethoprim-sulfamethoxazole, and tetracycline.
TABLE 1.
Susceptibilities of blaPER-1-positive and -negative isolates of acinetobacter
| Antimicrobial agent(s) | blaPER-1 test result (no. of isolates tested) | MIC (μg/ml)
|
Resist- ance (%) | |
|---|---|---|---|---|
| MIC range | MIC90d | |||
| Ampicillin-sulbactam | Positive (53) | 8-64 | 64 | 68 |
| Negative (37) | 0.5-64 | 32 | 27 | |
| Ceftazidime | Positive (53) | >128 | >128 | 100a |
| Negative (37) | 0.5->128 | 128 | 22 | |
| Ceftazidime-clavulanateb | Positive (53) | ≤0.12->128 | >128 | 68c |
| Negative (37) | ≤0.12->128 | >128 | 41c | |
| Cefepime | Positive (53) | 64->128 | >128 | 100 |
| Negative (37) | 0.12-128 | 32 | 30 | |
All of the blaPER-1-positive isolates were also resistant to cefotaxime and aztreonam.
Fixed concentration of 4 μg of clavulanate/ml.
Breakpoint for ceftazidime was applied.
MIC90, MIC at which 90% of the isolates tested are inhibited.
Transfer of a blaPER-1-carrying the 81-MDa plasmid to Escherichia coli was reported when plasmid pUZ8 existed in S. enterica serovar Typhimurium (11). In our study, blaPER-1 was transferred by conjugation to an A. baumannii recipient from 2 of 10 isolates tested, but repeated attempts failed to detect a blaPER-1-carrying plasmid. SmaI-digested genomic DNA bands of approximately 100 to 350 kb hybridized with the blaPER-1 probe, indicating the presence of the gene on the chromosome.
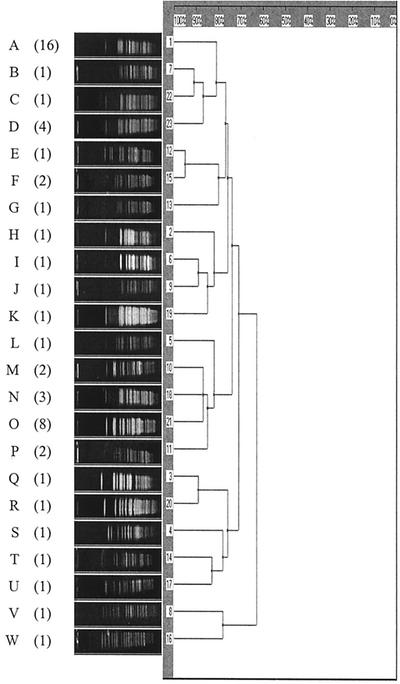
Fifty-three isolates of blaPER-1-positive acinetobacter showed 23 different PFGE patterns, indicating the presence of multiple clones (Fig. 1), but 9 of 16 isolates with pattern A and 7 of 8 isolates with pattern O were from intensive care unit patients, indicating clonal spread. Isolation of 39 (73.6%) strains from sputum samples suggests these were the main source of transmission.
FIG. 1.
Dendrogram of PFGE bands based on the coefficient of Dice. Numbers of multiple isolates with the same band patterns are shown in parentheses. Sixteen isolates had pattern A, and eight had pattern O.
DDS testing using ceftazidime, cefotaxime, or aztreonam disks detected only 17 of the 53 blaPER-1 allele-positive isolates when amoxicillin-clavulanate disks were placed 10 mm distant, edge to edge. The use of 5-mm-distanced cefepime disks detected 52 positive isolates, but the synergistic zones were small and difficult to interpret. Slightly larger synergistic zones were obtained when a cefepime disk and a 20-μg clavulanate disk were used (data not shown). These results indicated unreliability of the DDS tests for the screening of PER-1-producing acinetobacters.
Inhibition zone diameters by disks with cefepime alone and cefepime plus clavulanate or tazobactam were compared, and a difference of ≥5 mm was arbitrarily defined as positive (Table 2). More positive isolates were detected with a cefepime disk containing 20-μg than 10-μg inhibitors, but the zone size differences remained small for both inhibitors. Therefore, PCR is considered the more reliable method for detecting blaPER-1-producing Acinetobacter spp. This is the first report of high prevalence of blaPER-1-positive acinetobacters outside Europe and indicates possible presence of the resistance in other countries.
TABLE 2.
Phenotypic differentiation of 51 PER-1-producing isolates by cefepime disk and β-lactamase inhibitor- supplemented cefepime disk
| Inhibitor added to a cefepime disk | No. (%) of isolates with zone diam difference ofa:
|
|
|---|---|---|
| ≥5 mm | <5 mm | |
| Clavulanate, 10 μg | 42 (82.4) | 9 (17.6) |
| Clavulanate, 20 μg | 50 (98.0)b | 1 (2.0) |
| Tazobactam, 10 μg | 39 (76.5) | 12 (23.5) |
| Tazobactam, 20 μg | 51 (100) | 0 (0) |
Difference in zone diameter between a cefepime disk and a cefepime disk plus inhibitor.
Mean inhibition zone diameter difference between cefepime and cefepime plus 20 μg of clavulanate was only 6.7 mm.
Acknowledgments
This study was supported in part by the BK21 Project for Medical Sciences, Yonsei University, in 2002.
We thank Chasoon Lee in the Research Institute of Bacterial Resistance for her excellent technical support.
REFERENCES
- 1.Bauernfeind, A., I. Stemplinger, R. Jungwirth, P. Mangold, S. Amann, E. Akalin, O. Ang, C. Bal, and J. M. Casellas. 1996. Characterization of β-lactamase gene blaPER-2, which encodes an extended-spectrum class A β-lactamase. Antimicrob. Agents Chemother. 40:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chong. Y., K. Lee, and O. H. Kwon. 1993. In-vitro activities of cefepime against Enterobacter cloacae, Serratia marcescens, Pseudomonas aeruginosa and other aerobic gram-negative bacilli. J. Antimicrob. Chemother. 32(Suppl. B):21-29. [DOI] [PubMed] [Google Scholar]
- 3.Kiska, D. L., and P. H. Gilligan. 1999. Pseudomonas, p. 517-525. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 4.Lee, K., J. B. Lim, J. H. Yum, D. Yong, Y. Chong, J. M. Kim, and D. M. Livermore. 2002. blaVIM-2 cassette-containing novel integrons in metallo-β-lactamase-producing Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob. Agents Chemother. 46:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luzzaro, F., E. Mantengoli, M. Perilli, G. Lombardi, V. Orlandi, A. Orsatti, G. Amicosante, G. M. Rossolini, and A. Toniolo. 2001. Dynamics of a nosocomial outbreak of multidrug-resistant Pseudomonas aeruginosa producing the PER-1 extended-spectrum β-lactamase. J. Clin. Microbiol. 39:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing. Ninth information supplement, M100-S9. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 7.Nordmann, P., E. Ronco, T. Naas, C. Duport, Y. Michel-Briand, and R. Labia. 1993. Characterization of a novel extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 37:962-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nordmann, P., and T. Naas. 1994. Sequence analysis of PER-1 extended-spectrum β-lactamase from Pseudomonas aeruginosa and comparison with class A β-lactamases. Antimicrob. Agents Chemother. 38:104-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poirel, L., A. Karim, A. Mercat, I. Le Thomas, H. Vahaboglu, C. Richard, and P. Nordmann. 1999. Extended-spectrum β-lactamase-producing strain of Acinetobacter baumannii isolated from a patient in France. J. Antimicrob. Chemother. 43:157-158. [PubMed] [Google Scholar]
- 10.Schrechenberger, P. C., and A. Graevenitz. 1999. Acinetobacter, Achromobacter, Alcaligenes, Moraxella, Methylobacterium, and other nonfermentative gram-negative rods, p. 539-560. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 11.Vahaboglu, H., L. M. Hall, L. Mulazimoglu, S. Dodanli, I. Yildirim, and D. M. Livermore. 1995. Resistance to extended-spectrum cephalosporins, caused by PER-1 β-lactamase, in Salmonella typhimurium from Istanbul, Turkey. J. Med. Microbiol. 43:294-299. [DOI] [PubMed] [Google Scholar]
- 12.Vahaboglu, H., R. Ozturk, G. Aygun, F. Coskunkan, A. Yaman, A. Kaygusuz, H. Leblebicioglu, I. Balik, K. Aydin, and M. Otkun. 1997. Widespread detection of PER-1-type extended-spectrum β-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob. Agents Chemother. 41:2265-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vahaboglu, H., F. Coskunkan, O. Tansel, R. Ozturk, N. Sahin, I. Koksal, B. Kocazeybek, M. Tatman-Otkun, H. Leblebicioglu, M. A. Ozinel, H. Akalin, S. Kocagoz, and V. Korten. 2001. Clinical importance of extended-spectrum β-lactamase (PER-1-type)-producing Acinetobacter spp. and Pseudomonas aeruginosa strains. J. Med. Microbiol. 50:642-645. [DOI] [PubMed] [Google Scholar]