Abstract
The aacA29b gene, which confers an atypical aminoglycoside resistance pattern to Escherichia coli, was identified on a class 1 integron from a multidrug-resistant isolate of Pseudomonas aeruginosa. On the basis of amino acid sequence homology, it was proposed that the gene encoded a 6′-N-acetyltransferase. The resistance gene was cloned into the pET23a(+) vector, and overexpression conferred high-level resistance to the usual substrates of the aminoglycoside N-acetyltransferase AAC(6′)-I, except netilmicin. The level of resistance conferred by aacA29b correlated perfectly with the level of expression of the gene. The corresponding C-terminal six-His-tagged AAC(6′)-29b protein was purified and found to exist as a dimer in solution. With a spectrophotometric assay, an extremely feeble AAC activity was detected with acetyl coenzyme A (acetyl-CoA) as an acetyl donor. Fluorescence titrations of the protein with aminoglycosides demonstrated the very tight binding of tobramycin, dibekacin, kanamycin A, sisomicin (Kd, ≤1 μM) and a weaker affinity for amikacin (Kd, ≈60 μM). The binding of netilmicin and acetyl-CoA could not be detected by either fluorescence spectroscopy or isothermal titration calorimetry. The inability of AAC(6′)-29b to efficiently bind acetyl-CoA is supported by an alignment analysis of its amino acid sequence compared with those of other AAC(6′)-I family members. AAC(6′)-29b lacks a number of residues involved in acetyl-CoA binding. These results lead to the conclusion that AAC(6′)-29b is able to confer aminoglycoside resistance by sequestering the drug as a result of tight binding.
Aminoglycosides constitute a large family of broad-spectrum antibacterial compounds that have proven efficiency in the treatment of severe infections. Despite the fact that numerous bacterial species have developed resistance to these compounds, their potent bactericidal activity and their ability to interact synergistically with other antimicrobial agents have retained their utility in clinical practice. In particular, their use in combination with an effective β-lactam remains the preferred therapeutic approach to treat the increasing number of nosocomial infections caused by opportunistic pathogens, such as Pseudomonas (6, 13, 30).
Known resistance mechanisms for aminoglycosides can be classified into three types. Modification of the primary target of the drug by mutations in the genes encoding either the 16S rRNA or ribosomal proteins primarily affects the activity of streptomycin and is clinically significant only in Mycobacterium species (11, 19). Aminoglycoside resistance can also result from an insufficient intracellular concentration of the drug. Changes in the permeability of the outer membrane or alteration of specific cytoplasmic transport systems can reduce the uptake of aminoglycosides (8, 12). In addition, resistance caused by the expression of energy-dependent transporters, able to pump aminoglycosides out of the cell, has recently been reported (1, 16, 18). These two mechanisms, independently or in combination, can lead to intrinsic aminoglycoside resistance, as is often observed in Pseudomonas aeruginosa (13, 21, 26). Finally, the enzymatic modification of the antibiotic, resulting in a loss of antibacterial activity due to a diminished affinity for the ribosomal A site target (14), is by far the most important mechanism of aminoglycoside resistance (6, 21, 29). Three activities can cause covalent modification of the aminoglycosides: O phosphorylation, O nucleotidylation, and N acetylation (5, 28). Aminoglycoside N-acetyltransferases (AACs) catalyze the transfer of an acetyl group from acetyl coenzyme A (acetyl-CoA) to an amine function of the aminoglycoside. These enzymes are further subclassified according to the site of the regioselective modification of the aminoglycoside and the pattern of resistance that they confer. The production of AAC(6′)-I, which modifies the 6′ amino group, is associated with resistance to most of the aminoglycosides used in antibacterial chemotherapy; including kanamycin, amikacin, tobramycin, dibekacin, netilmicin, and sisomicin (28). A number of kinetic studies have been performed with AAC enzymes, and they demonstrate that these enzymes exhibit an efficient AAC activity involving a sequential kinetic mechanism (10, 17, 24, 27). Finally, these different mechanisms of resistance can be combined to obtain a very high level of aminoglycoside resistance (21).
The aacA29a and aacA29b genes were identified from a multidrug-resistant clinical isolate of P. aeruginosa, RON-2 (23), that exhibits high-level resistance to various aminoglycosides. Both genes are part of individual gene cassettes located on the In59 class 1 integron carried on the chromosome of the strain (7, 23). In addition to the two aacA29 genes and to the sulI and qacE genes that are usually carried by the class 1 integrons, responsible for sulfonamide and antiseptic resistance, respectively, In59 contains the blaVIM-2 gene encoding a carbapenem hydrolyzing β-lactamase (23). The amino acid sequences deduced from both aacA29 genes were 96% identical to each other, differing by only four amino acids located in the central part of the proteins (23). The AAC(6′)-29 members exhibit significant amino acid sequence similarity to members of the major AAC(6′)-I subfamily (25), including a large number of completely conserved residues. However, both AAC(6′)-29 proteins are shorter than the other members of the subfamily, containing 131 amino acids as opposed to the 144 to 153 amino acid residues present in the other subfamily members. Low-level expression of either the aacA29a or aacA29b gene confers the same atypical aminoglycoside resistance pattern to an E. coli host: decreased susceptibility to amikacin, tobramycin, and kanamycin but an unusual susceptibility to netilmicin. In addition, strains expressing both genes were shown to be resistant to a higher level of aminoglycosides than the strains harboring only one of them (23).
In the study reported here, we have cloned and overexpressed the aacA29b gene and purified the corresponding six-His-tagged protein. We have studied AAC(6′)-29b-His6 for its ability (i) to confer aminoglycoside resistant to a heterologous host, (ii) to catalyze aminoglycoside acetylation in presence of acetyl-CoA, and (iii) to bind the different substrates of this reaction. These experiments, together with the comparative analysis of the primary sequence of the protein, have allowed us to propose a novel mechanism of aminoglycoside resistance mediated by AAC(6′)-29b.
MATERIALS AND METHODS
Cloning and expression of aacA29b.
The aacA29b gene was amplified from pNOR-2003 plasmid DNA (23) by PCR with the oligodeoxynucleotides O1 (5′-CGGCTTTCATATGTCGATCTTACCTGTGAAAG-3′, complementary to the amino-terminal coding strand of the gene) and O2 (5′-CGGAATTCAATGGTGATGGTGATGGTGGACGGCGCTGCCTGC-3′, complementary to the carboxy-terminal noncoding strand and containing a six-His tagged coding region [boldface letters]) as primers. PCR was performed with Taq DNA polymerase (Stratagene) under standard conditions. The amplification product was ligated into the pCR2.1 vector (TA cloning kit; Invitrogen), and the recombinant plasmid was purified with the Wizard Miniprep DNA kit (Promega). The NdeI-EcoRI-digested insert (restriction sites underlined in the oligodeoxynucleotide sequences) was purified on an agarose gel and ligated into the pET23a(+) expression vector (Novagen) previously digested with the same restriction enzymes. The resulting recombinant plasmid was used as a template to determine the sequence of the insert by the dideoxynucleotide chain termination method (sequencing facility, Albert Einstein College of Medicine, Bronx, N.Y.) and to transform E. coli BL21(DE3) competent cells (Novagen). The transformants were selected on Luria broth (LB) agar containing 100 μg of ampicillin per ml. A single colony was used to inoculate 1 liter of LB medium containing the selective agent and grown at 37°C until the optical density at 600 nm reached 0.5. The temperature was then reduced to 18°C for 1 h before the expression of the aacA29b gene was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The culture was then grown for an additional 4 h at 18°C. Analysis of the overexpressed aacA29b-his6 gene was carried out by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 10 to 15% Phastgels (PhastSystem; Pharmacia).
Cloning of aacA29b in pBAD expression vector.
The aacA29b gene was amplified from pET23a(+)::aacA29b-his6 plasmid DNA by PCR with the oligodeoxynucleotides O2 (defined above) and O3 (5′-GAAGATCTAGGAGGAATAGACCATGTCGATCTTACC TGTGAAAG-3′, complementary to the amino-terminal coding strand of the gene and containing a ribosome binding site [boldface letters]) as primers. PCR was performed with Pfu DNA polymerase (Stratagene) under standard conditions. The amplification product was ligated into the pCR blunt vector (Zero blunt cloning kit; Invitrogen), and the recombinant plasmid was restricted with BglII and EcoRI (restriction sites underlined in the oligodeoxynucleotide sequences). The insert was purified and ligated into the pBAD/Myc-HisA expression vector (Invitrogen) previously digested with the same restriction enzymes. The recombinant plasmid pBAD::aacA29b-his6 was introduced by heat shock into E. coli LMG194 (Invitrogen) competent cells, and the transformants were selected on LB agar containing 100 μg of ampicillin per ml.
Protein purification.
Cells (10 g) were recovered by centrifugation from 6 liters of an LB culture of E. coli BL21(DE3)/pET23a(+)::aacA29b-his6, grown as described above, and were resuspended in 80 ml of 20 mM triethanolamine hydrochloride (TEA; pH 7.8), containing lysozyme (0.2 mg/ml), one Complete protease inhibitor cocktail tablet, 4 mM MgCl2, and 80 μg of DNase I per ml. All subsequent steps were performed at 4°C. After being stirred for 30 min, cells were lysed by ultrasonic disruption for 6 min, and cell debris was removed by centrifugation at 27,000 × g for 45 min. The supernatant was dialysed against 20 mM TEA (pH 7.8) containing 300 mM NaCl for 2 h and centrifuged again for 45 min at 19,000 rpm. The clear crude extract was applied to a column containing 50 ml of Ni-nitrilotriacetic acid (NTA) His.Bind resin (Novagen) equilibrated with the same solution. Proteins were eluted at 1 ml/min with a linear 25 to 250 mM imidazole gradient. The fractions eluting between 100 and 150 mM imidazole and containing a major protein band of approximately 15 kDa by SDS-PAGE—in agreement with the theoretical molecular mass of AAC(6′)-29b-His6—were pooled, concentrated and dialyzed for 4 h against 20 mM TEA (pH 7.8). The 10-ml protein solution obtained was then applied to a 30-ml MonoQ anion-exchange column (Pharmacia), and protein was eluted at 1 ml/min with a 0 to 600 mM NaCl linear gradient. Fractions containing AAC(6′)-29b-His6 eluting between 330 and 500 mM NaCl were pooled, concentrated to 4 ml, and applied to a 320-ml Sephacryl S-200 HR gel filtration column equilibrated with 50 mM HEPES (pH 7.5). The protein was eluted at 0.5 ml/min with 160 ml of the same buffer. The fractions of interest were pooled, concentrated, and stored at −20°C in 50% glycerol.
Analytical methods.
The native molecular weight of AAC(6′)-29b-His6 was determined on a 25-ml Superose 12 HR gel filtration column (Pharmacia), eluted at 0.2 ml/min with 50 mM HEPES (pH 7.5), containing 100 mM NaCl and previously calibrated with gel filtration molecular weight standards from Bio-Rad. The protein concentration was estimated by the Bio-Rad protein assay method with bovine serum albumin as a standard. The molecular mass of the protein was determined by electrospray ionization-mass spectrometry (Laboratory for Macromolecular Analysis and Proteomics, Albert Einstein College of Medicine, N.Y.).
Measurement of acetyltransferase activity.
AAC activity was determined spectrophotometrically by measuring the increase in A412 due to the formation of 5-thio-2-nitrobenzoate resulting from the reaction between the sulfhydryl group of the product of the acetyltransfer reaction, CoA-SH, and 5,5′ dithiobis(2-nitrobenzoic acid) (DTNB). The reaction was monitored with a UVIKONXL spectrophotometer (Research Instruments, San Diego, Calif.) either continuously or discontinuously. In both cases, assay mixtures contained 50 mM HEPES buffer (pH 7.5), acetyl-CoA (100 to 300 μM), and aminoglycoside (0 to 100 μM), and the reactions were initiated by the addition of AAC(6′)-29b-His6 (0.3 to 3 μM). In continuous assays, the absorbance of 1 ml of reaction mixture containing DTNB (200 μM) was monitored for 3 to 5 min at room temperature. The discontinuous assays were carried out by measuring the A412 of a 1-ml aliquot of a reaction mixture incubated for different fixed time intervals (0 to 60 min) at 37°C in the presence of DTNB. To ensure that DTNB did not inactivate the enzyme, these essays were repeated in the absence of DTNB, which was added subsequently. In all cases, positive controls were performed with the Salmonella enterica AAC(6′)-Iy enzyme (17).
Fluorescence titration.
Fluorescence measurements were recorded on a FluoroMax-3 spectrofluorometer with excitation and emission slit widths of 3.2 nm. The excitation wavelength was fixed at 295 nm (specific for tryptophan), and the emission spectrum between 310 and 450 nm was recorded. The fluorescence measurements were performed by titrating 3 ml of a 2 μM protein solution in 50 mM HEPES buffer (pH 7.5) containing 50 μM dithiothreitol, 100 μM EDTA, and 0.02% (vol/vol) Tween 80 with aliquots (1.5 to 15 μl) of aminoglycoside or acyl-CoA solutions (2 μM to 50 mM), followed by monitoring the change in fluorescence at 346 nm [λmax of AAC(6′)-29b-His6]. The experiments were performed at 25°C, and the fluorescence of the buffer was subtracted from the observed emission values. The fluorescence intensities of the protein saturated with ligand (F∞) were obtained from the experimental data by plotting (F − Fo)/Fo versus [L] as an exponential curve, where Fo is the fluorescence intensity of the protein alone and F is the fluorescence of the protein in the presence of a given concentration of ligand, [L]. The value of the association constant, Ka, was determined from the plot of log [(Fo − F)/(F − F∞)] versus log [L], assuming the relation that the pKa of the complex equals the absolute value of [L] when log [(Fo − F)/(F − F∞)] = 0 (4). Reported dissociation constants, Kds, are the reciprocal of the association constants. Corrections for the volume change upon ligand addition were ignored, because the volume change at the highest concentration of ligand was <2%.
ITC.
Calorimetric titrations for isothermal titration calorimetry (ITC) were performed with an MCS microcalorimeter from Microcal, Inc. (Northampton, Mass.). Protein solutions were prepared by dialysis against 50 mM HEPES buffer (pH 7.5) containing 0.01% (vol/vol) Tween 80 followed by concentration. The protein concentrations in the titration experiments with acetyl-CoA or aminoglycosides were 200 and 75 μM, respectively. Acetyl-CoA and aminoglycosides were made up to 5 to 7 mM with the final dialysate. Twenty injections of 6 μl of ligand solution each were made via the computer-controlled microsyringe into the 1.5-ml protein solution at 4-min intervals. Measurements were carried out at 27°C with constant stirring at 300 rpm.
Determination of MICs.
The MICs of selected aminoglycosides were determined by the agar dilution technique with an inoculum of 104 CFU per spot. For E. coli BL21(DE3) containing either pET23a(+) or the recombinant plasmid pET23a(+)::aacA29b-his6, MICs were determined on Mueller-Hinton (MH) agar plates. RM agar media (Invitrogen) containing various amounts of l-arabinose were used for E. coli LMG194/pBAD::aacA29b-his6. The plates were read after a 16-h incubation at 37°C.
Materials.
Chemicals, buffer, ampicillin, lysozyme, DTNB, acetyl-CoA, and aminoglycosides were purchased from Sigma Aldrich Chemical Co. LB and MH media, DTT, IPTG, and Tween 80 were obtained from Fisher Biotech. BglII, NdeI, and EcoRI were obtained from New England BioLabs, and the Complete protease inhibitor cocktail tablet and DNase I were obtained from Roche Molecular Biochemicals.
RESULTS
Purification and properties of AAC(6′)-29b-His6.
The nucleotide sequence of the aacA29b-his6 gene cloned into pET23a(+) revealed one nucleotide difference from the previously published sequence (23), leading to one amino acid change in the encoded protein (in addition to the six histidine residues added at the C terminus). Amino acid 39 of AAC(6′)-29b-His6 was identified as Leu instead of Phe (Fig. 1). The corresponding recombinant vector was used to overexpress the gene in E. coli BL21(DE3). A three-step purification procedure led to the purification of approximately 120 mg of >95% pure soluble protein from 6 liters of culture. Mass spectrometry analysis of the protein indicated a subunit molecular mass of 15,402 Da compared to the theoretical value of 15,400 Da, and the elution volume of the protein from both Sephacryl S-200 and Superose 12 gel filtration column indicated an apparent molecular mass of approximately 30,000 Da, suggesting that the protein is a dimer in solution.
FIG. 1.
Sequence alignment of AAC(6′)-29b and some members of the major AAC(6′)-I subfamily. The regions predicted to interact with acetyl-CoA based on the structure of AAC(6′)-Iy in complex with CoA are indicated by asterisks. Amino acids cited in the text are in boldface.
Pattern of resistance conferred by AAC(6′)-29b-His6 to E. coli.
In order to verify that the purified protein with the determined amino acid sequence mentioned above and the C-terminal His6 tag was able to confer aminoglycoside resistance, we determined the MICs of various aminoglycosides for E. coli BL21(DE3) harboring either the pET23a(+) vector without insert or the recombinant vector pET23a(+)::aacA29b-his6. The MICs obtained (Table 1) demonstrated that under these experimental conditions, where the structural gene is placed under the control of the strong T7 promoter, the AAC(6′)-29b-His6 protein was able to confer to E. coli BL21(DE3) high-level resistance (up to >256 μg/ml) to tobramycin, dibekacin, kanamycin A, and sisomicin and a lower-level resistance to amikacin, but did not affect the activities of gentamicin, lividomycin A, and netilmicin.
TABLE 1.
MICs of various aminoglycosides for E. coli BL21(DE3) containing either pET23a(+) or the recombinant vector pET23a(+)::aacA29b-His6
| Strain | MIC (μg/ml)a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Net | Gen | LivA | Amk | KmA | Tob | Dbk | Sis | |
| BL21(DE3)/pET23a(+) | 1 | 2 | 8 | 4 | 8 | 2 | 4 | 2 |
| BL21(DE3)/pET23a(+)::aacA29b-His6 | 1 | 2 | 8 | 128 | >256 | >256 | >256 | 256 |
Amk, amikacin; Dbk, dibekacin; Gen, gentamicin; KmA, kanamycin A; LivA, lividomycin A; Net, netilmicin; Sis, sisomicin; Tob, tobramycin.
By cloning the aacA29b-his6 gene under the control of the very tightly regulated PBAD promoter, we demonstrated that the transcription level of the resistant gene was directly correlated with the level of resistance to dibekacin conferred to E. coli LMG194, but did not significantly affect the sensitivity of the strain to netilmicin (Table 2). Indeed, the MIC of dibekacin for E. coli LMG194/pBAD::aacA29b-his6 increased proportionally from 16 to 512 μg/ml with the amount of the added transcriptional inducer l-arabinose over a 10,000-fold range (0.00002 to 0.2%), while the MIC of netilmicin only doubled.
TABLE 2.
MICs of dibekacin and netilmicin for E. coli LMG194 containing the recombinant vector pBAD::aacA29b-His6, depending on the level of transcriptional induction
| Aminoglycoside | MIC (μg/ml) at % of l-arabinose:
|
||||
|---|---|---|---|---|---|
| 0.00002 | 0.0002 | 0.002 | 0.02 | 0.2 | |
| Dibekacin | 16 | 64 | 128 | 256 | 512 |
| Netilmicin | 2 | NDa | ND | ND | 4 |
ND, not determined.
Measurement of AAC activity.
The AAC activity of AAC(6′)-29b-His6 was studied spectrophotometrically by first continuously measuring the change in the A412. With this assay, carried out at room temperature in the absence or presence of 0.3 μM enzyme obtained from multiple independent purifications, 100 μM acetyl-CoA, and up to 100 μM aminoglycoside, we were unable to detect any significant amikacin-, dibekacin-, kanamycin-, or sisomicin-dependent acetyltransferase activity for AAC(6′)-29b-His6. Under identical conditions, we could demonstrated a robust, and reproducible activity with AAC(6′)-Iy (17) as a positive control (data not shown). To ensure that AAC(6′)-29b-His6 was not inactivated in the presence of DTNB in the assay mixtures, we performed discontinuous assays in the absence of DTNB, incubating the reaction mixture under the same conditions described above, but adding DTNB to aliquots taken after various reaction times (0 to 12 min). No significant CoA-SH production could be detected. Finally, discontinuous assays were carried out by incubating the reaction mixture in the presence of 1.5 to 3 μM AAC(6′)-29b-His6, 50 μM aminoglycoside, 125 to 250 μM acetyl-CoA, and DTNB for 0, 10, 20, 40, or 60 min at 37°C. By using this assay, extremely feeble tobramycin-, kanamycin-, sisomicin-, amikacin- and netilmicin-acetyltransferase activities were detected compared to those of the negative control performed without aminoglycoside or with the 6′-hydroxyl-containing aminoglycoside, lividomycin A (data not shown). The rate of the AAC reaction catalyzed by AAC(6′)-29b-His6 was estimated to be 100 to 1,000 times slower than the corresponding rate of AAC(6′)-Iy (depending on the identity of the aminoglycoside and the concentration of acetyl-CoA).
Titrations of AAC(6′)-29b-His6 with different ligands.
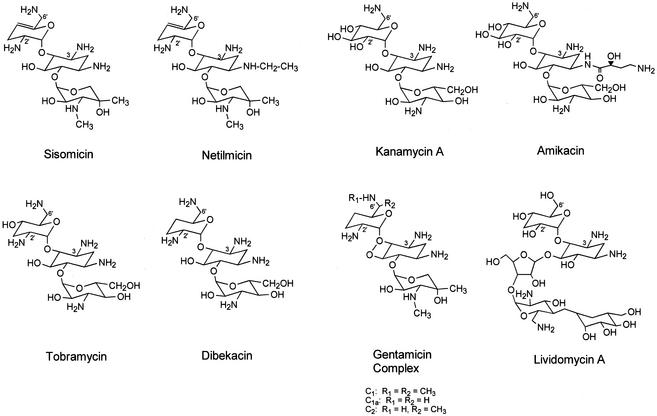
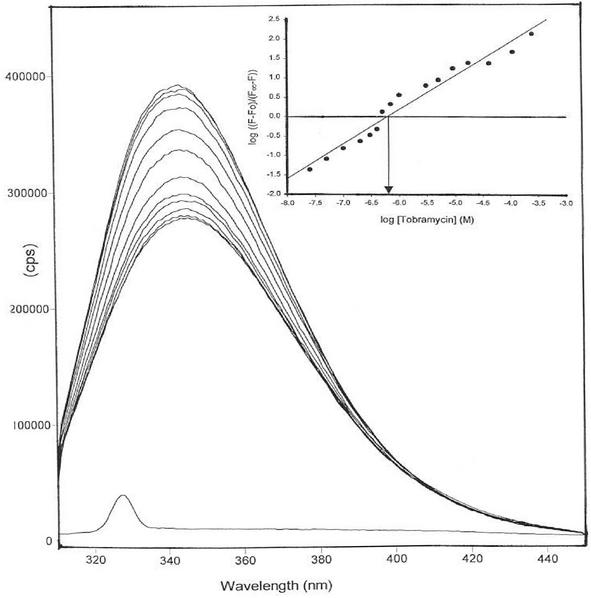
The aminoglycosides tested (Fig. 2) can be separated into three classes, based on the change in the intrinsic protein fluorescence of AAC(6′)-29b-His6 resulting from their addition to solutions of the protein. Titration with amikacin, dibekacin, kanamycin A, or tobramycin resulted in the enhancement of intrinsic protein fluorescence by 30 to 55% (Fig. 3). On the other hand, addition of sisomicin at a concentration of 200 μM led to a decrease of approximately 12% in the intrinsic fluorescence of AAC(6′)-29b-His6 and was associated with a small blue shift in the fluorescence maxima from 346 to 342 nm. Larger amounts of sisomicin caused a larger blue shift to 355 nm, as well as a significant increase in the baseline fluorescence at 440 nm. The third category of aminoglycosides tested included netilmicin, the gentamicin complex, and lividomycin A. No significant change (≤6% after subtraction of the baseline change) was observed in the intrinsic fluorescence of AAC(6′)-29b-His6 upon addition of these compounds (1 to 1,000 μM). In addition, while exothermic binding of tobramycin to AAC(6′)-29b-His6 was revealed by ITC, addition of netilmicin to the protein solution did not result in any enthalpically driven binding (data not shown). The dissociation constants (Kds) calculated for the aminoglycosides for which a change in the intrinsic fluorescence of AAC(6′)-29b-His6 was observed are as follows: amikacin, (59 ± 2.8) × 10−6 M; dibekacin, (0.4 ± 0.02) × 10−6 M; kanamycin A, (0.9 ± 0.03) × 10−6 M; sisomicin, (0.6 ± 0.04) × 10−6 M; and tobramycin, (0.6 ± 0.03) × 10−6 M. Uncertainty is represented by the standard errors of the linear fit (Fig. 3). The slope of the plot of log [(Fo − F)/(F − F∞)] versus log [L] represented unity for all of these ligands (Fig. 3, inset), indicating the formation of a 1:1 complex between the dimeric enzyme and the ligand under the experimental conditions.
FIG. 2.
Structures of aminoglycosides used in this study.
FIG. 3.
Titration of AAC(6′)-29b-His6 with tobramycin. Curves from the bottom to the top are the fluorescence spectra of buffer baseline AAC(6′)-29b-His6 in the presence of 0, 0.05, 0.1, 0.15, 0.2, 0.3, 0.5, 0.7, 3, 9, 40, or 300 μM tobramycin. (Inset) Representation of the plot used to determine the association constant.
The titration of AAC(6′)-29b-His6 with two acetyl donors, acetyl-CoA and malonyl-CoA (1 to 1,000 μM), did not result in any change in the intrinsic fluorescence of the protein (≤3%). Additionally, no interaction between acetyl-CoA and AAC(6′)-29b-His6 could be detected by ITC (data not shown). Acetyl-CoA injections were associated with a minute heat variations of −0.35 to −0.25 μcal/s, and the heat change was not saturable up to a final concentration of 560 μM acetyl-CoA.
DISCUSSION
Enzymatic modification of the antibiotic is the most prevalent mechanism of bacterial aminoglycoside resistance (6, 21, 29). The AAC(6′) members represent one major subclass of these enzymes, catalyzing N-acetyl transfer from acetyl-CoA to the 6′ amino group of the drug. The aacA29b gene was identified from a multidrug-resistant clinical isolate of P. aeruginosa, in which the gene was located on a class 1 integron with other resistance genes. This clinical strain exhibits high-level resistance to various aminoglycosides (≥256 μg/ml) and an intermediate resistance to netilmicin (8 μg/ml). The AAC(6′)-29b encoded amino acid sequence is highly homologous to the sequences of the enzymes belonging to the major AAC(6′)-I subfamily (25 to 40% identity). However, the AAC(6′)-29b protein, which contains 131 amino acids, is shorter than all other members of the family and confers an atypical aminoglycoside resistance pattern to E. coli.
The MICs determined for E. coli BL21(DE3)/pET23a(+), used as control, and E. coli BL21(DE3)/pET23a(+)::aacA29b-his6 showed that the resistant gene, when overexpressed, conferred to the strain a high level of resistance to most of the usual substrates of AAC(6′)-I enzymes (Table 1). These results demonstrate that the addition of a His6 tag at the C terminus of the protein and the F39L change did not abolish the ability of the protein to confer aminoglycoside resistance. The resistance to amikacin and the susceptibility to gentamicin are characteristic of the AAC(6′)-I-type modification; however, the susceptibility to netilmicin is contrary to the expected pattern of resistance for strains expressing AAC(6′)-I enzymes.
Of the kinetic and mechanistic studies that have been reported for AACs, all reveal a robust acetyltransferase activity (10, 17, 24, 27). The AAC activity of AAC(6′)-29b-His6, determined in this study, was extremely feeble and was similar for netilmicin and aminoglycosides for which a high MIC was determined for E. coli BL21(DE3)/pET23a(+)::aacA29b-his6. Both these qualitative and quantitative findings are inconsistent with the hypothesis that aminoglycoside resistance mediated by the expression of the aacA29b gene is due to N acetylation of the drug.
The ability of different aminoglycosides and acetyl-CoA to bind to AAC(6′)-29b-His6 and their affinity for the protein were then investigated by exploiting the intrinsic fluorescence of AAC(6′)-29b due to three residues (W14, W22, and W103) completely conserved among the AAC(6′)-I family (Fig. 1). The significant change in the intrinsic fluorescence of AAC(6′)-29b-His6 upon addition of either dibekacin, tobramycin, sisomicin, or kanamycin A indicated that the binding of these aminoglycosides modified the environment of at least one of the tryptophan residues. The measured dissociation constants of <10−6 M (see Results) for these compounds demonstrate that they bind very tightly to AAC(6′)-29b-His6, exhibiting at least 10-times-higher affinity than observed for AAC(6′)-Iy (9). In this category of tight-binding ligands, sisomicin was distinguishable from the others, since its binding resulted in a small decrease in intrinsic fluorescence as opposed to a significantly larger increase in fluorescence, suggesting different interactions in the binding site of AAC(6′)-29b. Two structural characteristics distinguish dibekacin from sisomicin, which could be responsible for this different change in the tryptophan environment upon binding: an unsaturation between the carbon 4′ and 5′ present in sisomicin and the substituents on the double prime ring. An increase in the intrinsic fluorescence of the protein was also observed upon the addition of amikacin, but this compound exhibits a lower affinity for AAC(6′)-29b-His6. Amikacin differs from kanamycin A only by a hydroxybutyramide substituent at the N1 position (Fig. 2), suggesting that a substitution in this position adversely affects binding to AAC(6′)-29b, as has been previously reported for different subclasses of aminoglycoside-modifying enzymes (2). The intrinsic fluorescence of AAC(6′)-29b-His6 was not affected by the addition of netilmicin, which differs from sisomicin only by an N1 ethyl substitution (Fig. 2). Using ITC methods, we demonstrated that netilmicin does not bind in an enthalpically driven process to AAC(6′)-29b-His6. Thus, the binding affinities of the various aminoglycosides for the protein (shown above) correlate exactly with the MICs (Table 1). This suggests that resistance mediated by AAC(6′)-29b is due to the sequestration of the drug as a result of very tight binding rather than to inactivation of the aminoglycosides as a result of enzymatic modification. This proposal is supported by the MIC of dibekacin obtained for E. coli LMG194/pBAD::aacA29b-his6 (Table 2), which correlates directly with the amount of AAC(6′)-29b-His6 produced by the bacterium, and does not appear saturable in the range of inducer l-arabinose concentrations studied.
The sequence of AAC(6′)-29b was aligned with those of the other AAC(6′)-I family members (Fig. 1) by aligning the last 14 residues of the protein with the carboxy-terminal region of AAC(6′)-I protein containing conserved cationic residues. This alignment revealed an 18-amino-acid internal deletion that includes one of the most conserved residues among the N-acetyltransferase superfamily (22), phenylalanine 131 or 171, numbered according to AAC(6′)-Iy or AAC(6′)-Ib, respectively (Fig. 1). A previous study showed that this residue is important for the ability of AAC(6′)-Ib to modify aminoglycosides possessing a substituent at the N1 position in vivo (3), and the lack of this phenylalanine in AAC(6′)-29b could be responsible for the substantially weaker binding of the N1-substituted aminoglycosides, amikacin and netilmicin.
While significant changes in the fluorescence of AAC(6′)-Iy accompany acyl-CoA binding (9), the addition of neither acetyl-CoA nor malonyl-CoA affected the intrinsic fluorescence of AAC(6′)-29b-His6. ITC of the protein with acetyl-CoA also demonstrated no significant binding of acetyl-CoA to AAC(6′)-29b-His6. This cannot be explained by the presence of bound acetyl-CoA or CoA, since upon heat denaturation of the protein followed by ultrafiltration, no A260 due to CoA was observed in the ultrafiltrates. The recently solved three-dimensional structure of the AAC(6′)-Iy-CoA complex suggests a structural basis for these observations. The deletion in AAC(6′)-29b corresponds to a region in AAC(6′)-Iy that forms an α helix that interacts directly with bound CoA (M. Vetting, unpublished data). In addition, a conserved glycine located in a region that also has interactions with the CoA pyrophosphate (Fig. 1) and previously reported as being important determinant of acetyl-CoA binding to acetyltransferases (15) is substituted for by a negatively charged aspartate in the AAC(6′)-29b sequence, possibly generating unfavorable electrostatic interactions for folding or ligand binding. The inability of AAC(6′)-29b-His6 to bind acetyl-CoA explains the very low acetyltransferase activity and supports a mechanism of resistance involving sequestration of the drug by high-affinity protein binding.
Modification of ribosomal components, decrease of the intracellular concentration of the drug by impermeability or efflux, and enzymatic modification of the drug are the three known aminoglycoside resistance mechanisms found in bacteria. The aminoglycoside phosphotransferase APH (3′)-I, encoded by the conjugative plasmid pIP1518 of E. coli HM69, which confers low-level resistance to tobramycin, represents the only reported example of aminoglycoside resistance due to drug binding (20). In this report, we have shown that AAC(6′)-29b-His6 is able to confer selective, high-level aminoglycoside resistance by binding the drug and not by enzyme-catalyzed acetylation. High-level aminoglycoside resistance can be obtained in P. aeruginosa via this mechanism by a combination of events. This bacterium exhibits an intrinsic, low-level aminoglycoside resistance due to the relative impermeability of its outer membrane and the expression of transporter systems able to efflux aminoglycosides from the cell (1, 26). In addition, a relatively high intracellular concentration of AAC(6′)-29's would be obtained by the presence of two similar structural genes (aacA29a and aacA29b). Finally, we have shown that AAC(6′)-29b-His6 exhibits an extremely high affinity for selected aminoglycosides.
Acknowledgments
This work was supported by NIH grant AI33696. T.-A. Smith was the recipient of an ASM Minority Undergraduate Research Fellowship.
We thank Tarum K. Dam for help with ITC experiments, Matthew Vetting for providing unpublished information, and Subray S. Hegde for helpful discussions.
REFERENCES
- 1.Aires, J. R., T. Kohler, H. Nikaido, and P. Plesiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bongaerts, G. P. A., and J. S. Vliegenthart. 1988. Effect of aminoglycoside concentration on reaction rates of aminoglycoside-modifying enzymes. Antimicrob. Agents Chemother. 32:740-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chavideh, R., S. Sholly, D. Panaite, and M. E. Tolmasky. 1999. Effects of F171 mutations in the 6′-N-acetyltransferase type Ib [AAC(6′)-Ib] enzyme on susceptibility to aminoglycosides. Antimicrob. Agents Chemother. 43:2811-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chipman, D. M., V. Grisaro, and N. Sharon. 1967. The binding of oligosaccharides containing N-acetylglucosamine and N-acetylmuramic acid to lysozyme. The specificity of binding subsites. J. Biol. Chem. 242:4388-4394. [PubMed] [Google Scholar]
- 5.Davies, J., and G. D. Wright. 1997. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 5:234-240. [DOI] [PubMed] [Google Scholar]
- 6.Dworkin, R. J. 1999. Aminoglycosides for the treatment of gram-negative infections: therapeutic use, resistance and future outlook. Drug Resist. Updates 2:173-179. [DOI] [PubMed] [Google Scholar]
- 7.Fluit, A. C., and F. J. Schmitz. 1999. Class 1 integrons, gene cassettes, mobility, and epidemiology. Eur. J. Clin. Microbiol. Infect. Dis. 18:761-770. [DOI] [PubMed] [Google Scholar]
- 8.Hatch, R. A., and N. L. Schiller. 1998. Alginate lyase promotes diffusion of aminoglycosides through the extracellular polysaccharide of mucoid Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:974-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hegde, S. S., T. K. Dam, C. F. Brewer, and J. S. Blanchard. 2002. Thermodynamics of aminoglycoside and acyl-coenzyme A binding to the Salmonella enterica AAC(6′)-Iy aminoglycoside N-acetyltransferase. Biochemistry 41:7519-7527. [DOI] [PubMed] [Google Scholar]
- 10.Hegde, S. S., F. Javid-Majd, and J. S. Blanchard. 2001. Overexpression and mechanistic analysis of chromosomally encoded aminoglycoside 2′-N-acetyltransferase (AAC(2′)-Ic) from Mycobacterium tuberculosis. J. Biol. Chem. 276:45876-45881. [DOI] [PubMed] [Google Scholar]
- 11.Honoré, N., and S. T. Cole. 1994. Streptomycin resistance in mycobacteria. Antimicrob. Agents Chemother. 38:238-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadurugamuwa, J. L., J. S. Lam, and T. J. Beveridge. 1993. Interaction of gentamicin with the A band and B band lipopolysaccharides of Pseudomonas aeruginosa and its possible lethal effect. Antimicrob. Agents Chemother. 37:715-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livermore, D. M. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34:634-640. [DOI] [PubMed] [Google Scholar]
- 14.Llano-Sotelo, B., E. F. Azucena, L. P. Kotra, S. Mobashery, and C. S. Chow. 2002. Aminoglycosides modified by resistance enzymes display diminished binding to the bacterial ribosomal aminoacyl-tRNA site. Chem. Biol. 9:455-463. [DOI] [PubMed] [Google Scholar]
- 15.Lu, L., K. A. Berkey, and R. A. Casero, Jr. 1996. RGFGIGS is an amino acid sequence required for acetyl coenzyme A binding and activity of human spermidine/spermine N1 acetyltransferase. J. Biol. Chem. 271:18920-18924. [DOI] [PubMed] [Google Scholar]
- 16.Magnet, S., P. Courvalin, and T. Lambert. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45:3375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magnet, S., T. Lambert, P. Courvalin, and J. S. Blanchard. 2001. Kinetic and mutagenic characterization of the chromosomally encoded Salmonella enterica AAC(6′)-Iy aminoglycoside N-acetyltransferase. Biochemistry 40:3700-3709. [DOI] [PubMed] [Google Scholar]
- 18.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meier, A., P. Kirschner, F.-C. Bange, U. Vogel, and E. C. Bottger. 1994. Genetic alterations in streptomycin-resistant Mycobacterium tuberculosis: mapping of mutations conferring resistance. Antimicrob. Agents Chemother. 38:228-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menard, R., C. Molinas, M. Arthur, J. Duval, P. Courvalin, and R. Leclercq. 1993. Overproduction of 3′-aminoglycoside phosphotransferase type I confers resistance to tobramycin in Escherichia coli. Antimicrob. Agents Chemother. 37:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, G. H., F. J. Sabatelli, R. S. Hare, Y. Glupczynski, P. Mackey, D. Shlaes, K. Shimizu, and K. J. Shaw. 1997. The most frequent aminoglycoside resistance mechanisms—changes with time and geographic area: a reflection of aminoglycoside usage patterns? Clin. Infect. Dis. 24(Suppl. 1):S46-S62. [DOI] [PubMed] [Google Scholar]
- 22.Neuwald, A. F., and D. Landsman. 1997. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22:154-155. [DOI] [PubMed] [Google Scholar]
- 23.Poirel, L., T. Lambert, S. Türköglu, E. Ronco, J.-L. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radika, K., and D. B. Northrop. 1984. Substrate specificities and structure-activity relationships for acylation of antibiotics catalyzed by kanamycin acetyltransferase. Biochemistry 23:5118-5122. [DOI] [PubMed] [Google Scholar]
- 25.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams, J. W., and D. B. Northrop. 1978. Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence of antibiotic resistance upon substrate Vmax/Km values. J. Biol. Chem. 253:5908-5914. [PubMed] [Google Scholar]
- 28.Wright, G. D. 1999. Aminoglycoside-modifying enzymes. Curr. Opin. Microbiol. 2:499-503. [DOI] [PubMed] [Google Scholar]
- 29.Wright, G. D., A. M. Berghuis, and S. Mobashery. 1998. Aminoglycoside antibiotics. Structures, functions, and resistance. Adv. Exp. Med. Biol. 456:27-69. [PubMed] [Google Scholar]
- 30.Zembower, T. R., G. A. Noskin, M. J. Postelnick, C. Nguyen, and L. R. Peterson. 1998. The utility of aminoglycosides in an era of emerging drug resistance. Int. J. Antimicrob. Agents 10:95-105. [DOI] [PubMed] [Google Scholar]