Abstract
Hygromycin B is an aminoglycoside antibiotic active against prokaryotic and eukaryotic ribosomes. Ribosomal alterations in bacteria conferring resistance to hygromycin B have not been described, prompting us to use a single rRNA allelic derivative of the gram-positive bacterium Mycobacterium smegmatis for investigation of the molecular mechanisms involved in ribosomal resistance to hygromycin B in eubacteria. Resistance mutations were found to localize exclusively in 16S rRNA. The mutations observed, i.e., 16S rRNA U1406C, C1496U, and U1498C (E. coli numbering), are in close proximity to the hygromycin B binding site located in conserved helix 44 of 16S rRNA. The 16S rRNA positions involved in hygromycin B resistance are highly conserved in all three domains of life, explaining the lack of specificity and general toxicity of hygromycin B.
The ribosome is a target for many important antibacterial drugs (14, 20). Unfortunately, resistance to these drugs has become commonplace (1, 37). At the same time, many available ribosomal drugs are not used in clinical medicine because they are insufficiently specific for bacterial as opposed to eukaryotic ribosomes and hence are too toxic (14). There is thus an increasing need to unravel the details of how these antibiotics interact with ribosomes. With the availability of high-resolution structures of the ribosome, we are beginning to understand the structural basis for antibiotic action on the ribosome (4, 8, 17, 30); crystallographic studies define the drug binding site and may also point to the mechanism of drug action (4, 8). Homology models based on the high-resolution 30S structures allow for a placement of the eukaryotic 18S rRNA relative to prokaryotic 16S rRNA, thereby providing a three-dimensional framework for analysis of selectivity.
The best-studied aminoglycosides are those containing a 2-deoxystreptamine ring (9). By binding to the A (aminoacyl) site of the ribosomal decoding region (helix 44 of 16S rRNA), these drugs interfere with the ribosomal proofreading mechanism, leading to miscoding and/or premature termination (35). One characteristic of A-site binding aminoglycosides is the multitude of nucleotides critical for drug binding, i.e., 16S rRNA C1407, A1408, G1491, A1493, G1494, and U1495 (8, 10, 12) (throughout this paper, Escherichia coli numbering is used for 16S rRNA bases). However, acquired ribosomal drug resistance in bacterial pathogens is confined to alteration of A1408 to G, suggesting that susceptibility of ribosomes to 2-deoxystreptamines is determined largely by the identity of the single nucleotide at 16S rRNA position 1408 (3, 23, 25, 28).
Hygromycin B is a bactericidal aminoglycoside produced by Streptomyces hygroscopicus (22). It has a 5-substituted N-methyl-2-deoxystreptamine aminocyclitol characterized by a dual ether linkage between two of its three sugar moieties, resulting in a fourth, five-member ring. Hygromycin B inhibits protein synthesis in archaebacteria, eubacteria, fungi, and higher eukaryotes (6, 16). In contrast to the typical 2-deoxystreptamines, hygromycin B inhibits protein synthesis by blocking ribosomal translocation without causing significant misreading in vivo (2, 5, 11, 16, 38). Ganoza and Kiel proposed that hygromycin B specifically inhibits the activity of RbbA, a ribosomal ATPase, which, along with elongation factor Tu (EF-Tu) and EF-G, is necessary for decoding and translocation (15). In eukaryotes, hygromycin B interferes with the translocation step by affecting the EF-2-mediated transfer of A-site-bound tRNA to the P site (11). From crystallographic studies, it is clear that hygromycin B binds in a largely sequence-specific manner close to the very top of helix 44 of 16S rRNA, between the A (aminoacyl) and the P (peptidyl) sites. The 16S rRNA nucleotides involved in this interaction are C1403, G1405, G1494, U1495, C1496, and U1498 (4). The localization of hygromycin B binding to this region is consistent with the recently suggested involvement of helix 44 in movements during the translocation step (13).
Because of its universal activity, hygromycin B cannot be used for systemic application in clinical medicine. Regardless of the clinical utility of an antibiotic agent, understanding of the mechanisms which determine its specificity is important for future drug development by rational design. The study of a universal inhibitor of translation is particularly interesting because it allows the elucidation of its lack of specificity.
Little is known concerning the ribosomal components mediating resistance to hygromycin B. In the eukaryote Tetrahymena thermophila, alteration of 16S rRNA nucleotide U1495 to C confers resistance (31). Alterations of ribosomal components conferring resistance to hygromycin B in prokaryotic microorganisms have not yet been described. The difficulty of isolating hygromycin-resistant eubacteria might be because a single mutation may not lead to resistance, so that successive steps of mutational alterations (intra- or intergenic) are necessary.
Nonconserved nucleotides in 16S rRNA have been suggested to be important in determining the selectivity of ribosomal drugs, e.g., for an antibacterial agent to affect the prokaryotic ribosome as opposed to the eukaryotic ribosome (3). Using a single rRNA allelic derivative of the gram-positive eubacterium Mycobacterium smegmatis (28), we set out to isolate drug-resistant mutants in order to identify resistance determinants. Our studies revealed that all of the resistance mutations affect nucleotides in 16S rRNA located within the hygromycin B binding site. The universal conservation of the nucleotides involved in mutational resistance explains the universal activity and hence the toxicity of hygromycin B.
MATERIALS AND METHODS
Bacterial strains and media.
E. coli strain XL-1 Blue (Stratagene) was used for cloning and propagation of the plasmids and was grown in Luria-Bertani (LB) medium containing either ampicillin (120 μg ml−1) or gentamicin (5 μg ml−1). The single rRNA allelic strains M. smegmatis mc2155 rrnA− (KO14), M. smegmatis mc2155 rrnB− (KO16), and the parental strain M. smegmatis mc2155 SMR5 (carrying two chromosomal rRNA operons) were cultivated in LB medium containing 0.05% Tween 80 (28). The strains used in this study are listed in Table 1.
TABLE 1.
Hygromycin B resistance mutations, MICs, and growth rates
| Strain | MIC
|
Relative resistancea | Generation time (h) | |
|---|---|---|---|---|
| μg/ml | μM | |||
| Wild type | ||||
| M. smegmatis SMR5 | 16 | 0.03 | 3.4 ± 0.4 | |
| M. smegmatis rrnB− | 16 | 0.03 | 3.2 ± 0.5 | |
| Mutants | ||||
| U1406C | 1,024 | 1.9 | 64 | 5.3 ± 0.8 |
| C1496U | 512 | 0.97 | 32 | 5.2 ± 1.1 |
| U1498C | 128-256 | 0.24-0.48 | 8-16 | 3.8 ± 0.3 |
| Recombinants | ||||
| pMV361ΔKan-Gm-rRNA1406C | 1,024 | 1.9 | 64 | ND |
| pMV361ΔKan-Gm-rRNA1496U | 512 | 0.97 | 32 | ND |
| pMV361ΔKan-Gm-rRNA1498C | 128-256 | 0.24-0.48 | 8-16 | ND |
Relative resistance is the MIC of mutant cells divided by the MIC of wild-type cells.
ND, not done.
For the selection of hygromycin B-resistant mutants, LB medium was supplemented with 1.5% agar and 50 μg of hygromycin B ml−1. M. smegmatis transformed with derivatives of plasmid pMV361ΔKan-Gm (Table 2) was propagated on LB agar plates containing 5 μg of gentamicin ml−1.
TABLE 2.
Plasmids used in this work
DNA techniques and construction of plasmids.
Standard methods were used for restriction endonuclease digestion of DNA and other DNA manipulations (26). The method used for the isolation of genomic DNA is described by Springer et al. (33). Nucleic acid sequencing was done using fluorescence-labeled nucleotides and Taq cycle sequencing (Applied Biosystems). The primers used for sequencing are listed in Table 3.
TABLE 3.
Primers used in this study
| Primer | Sequence | Nucleotide positions |
|---|---|---|
| 16S rRNA (rrs) | ||
| 283 | 5′ GAG AGT TTG ATC CTG GCT CAG GAC 3′ | 11-34 |
| 242 | 5′ CTA CGG GAG GCA GCA GTG GGG 3′ | 342-362 |
| 244 | 5′ CCC CAC TGC TGC CTC CCG TAG 3′ | 362-342 |
| 247 | 5′ TTG TCG CGT TGT TCG TGA AA 3′ | 570-590 |
| 291 | 5′ AAA CTC AAA GGA ATT GAC GGG 3′ | 889-910 |
| 289 | 5′ AAG TCG GAG TCG CTA GTA ATC GC 3′ | 1315-1337 |
| 261 | 5′ AAA GGA GGT GAT CCA GCC GC 3′ | 1527-1507 |
| ITS I | ||
| 294 | 5′ GAT GCT CGC AAC CAC TAT CC 3′ | 2367-2348 a |
Accession number Y08453.
Plasmid pMV361ΔKan-Gm was constructed by deletion of the aph cassette of pMV361 (34) with NheI-SpeI and insertion of the gentamicin resistance cassette of pMV361-Gm (32) carried on a 3-kb PstI fragment. An approximately 1.1-kbp rRNA gene fragment was generated using primers 291 and 294 with DNA from hygromycin B drug-resistant mutants as the template. Sequencing confirmed that the fragments amplified differed only in the sequence alteration in question. PCR fragments were subcloned into the pGEMTEasy-vector as specified by the manufacturer (Promega). Subsequently, the PCR fragments were isolated by EcoRI digestion and integrated into the single EcoRI site of pMV361ΔKan-Gm. The resulting plasmids were pMV361ΔKan-Gm-rRNA1406C, pMV361ΔKan-Gm-rRNA1496U, and pMV361ΔKan-Gm-rRNA1498C. The single allelic M. smegmatis strain rrnB− (28) was used for the transformation experiments. The strain was made electrocompetent and transformed as described by Sander et al. (29). Primary selection was performed on LB agar plates containing gentamicin (5 μg ml−1). After 5 days of incubation, single colonies were picked and colony purified for further investigations.
Isolation of spontaneous hygromycin B-resistant strains.
Spontaneous hygromycin B-resistant mutants were selected on LB medium containing hygromycin B at 50 μg ml−1. Cultures from single colonies were grown in LB medium supplemented with 0.05% Tween 80 and used for MIC tests in a microtiter plate format. Starting cultures contained 200 μl of bacterial cells at an optical density at 600 nm of 0.025, and hygromycin B was added in twofold dilutions. The MIC was defined as the drug concentration at which the growth of the cultures was completely inhibited after 72 h of incubation at 37°C, corresponding to 24 generations.
RecA-mediated homologous recombination (24) was used to demonstrate a cause-effect relationship of the resistance-associated mutations. Serial dilutions of strains transformed with vectors pMV361ΔKan-Gm-rRNA1406C, pMV361ΔKan-Gm-rRNA1496U and pMV361ΔKan-Gm-rRNA1498C, each carrying the mutation in question on a nonfunctional rRNA fragment, were plated on agar plates containing hygromycin B at 200 to 500 μg ml−1. Introduction of the mutated rrn nucleotide into the single functional rRNA operon of M. smegmatis rrn− by gene conversion (24) results in drug resistance. Drug-resistant recombinants were colony purified and subjected to 16S rDNA sequence determination.
RESULTS
Isolation of mutants.
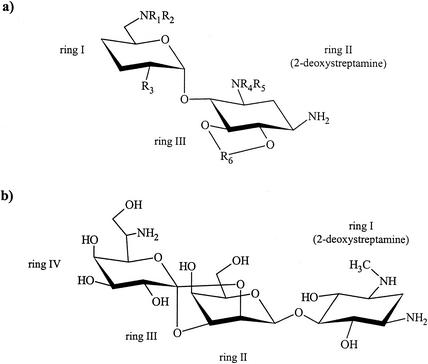
M. smegmatis rrn−, a genetically engineered single rRNA allelic (rrnA or rrnB) derivative of M. smegmatis mc2155 SMR5 (27, 28), was used to select for mutants resistant to hygromycin B (the chemical structure of hygromycin B compared to the classical aminoglycosides is shown in Fig. 1). Drug-resistant mutants were obtained with a frequency of 10−8 to 10−9. No difference in mutational rate was found when plating M. smegmatis rrnA− or rrnB−. Attempts to select for spontaneous hygromycin B-resistant mutants of M. smegmatis mc2155 SMR5 (carrying two functional rRNA operons) were unsuccessful (mutational rate, <10−10).
FIG. 1.
Chemical structures of aminoglycoside antibiotics. (a) The classical 2-deoxystreptamine aminoglycosides are characterized by two variable aminoglycoside rings (rings I and III) that are connected to the 2-deoxystreptamine moiety (ring II). Ring I is always linked to position 4 of the 2-deoxystreptamine, whereas ring III can be connected either to position 5 (4,5-disubstituted ring II; neomycin class) or to position 6 (4,6-disubstituted ring II; gentamicin, kanamycin, amikacin, and tobramycin). (b) Hygromycin B is composed of a 5-substituted 2-deoxystreptamine (ring I). Ring II is connected by a dual ether linkage to ring IV, resulting in an additional fourth, five-member ring (ring III).
Genetic characterization of the mutants.
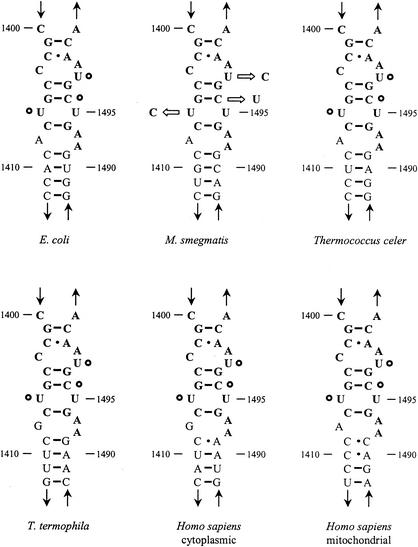
We hypothesized that the resistance mutations would be located close to the ribosomal drug binding site. A total of 35 drug-resistant mutants were investigated. Amplification of the gene encoding the 16S rRNA and subsequent sequence determination revealed that the drug-resistant phenotype coincided with the occurrence of a single point mutation in 16S rRNA in each of the mutants studied. The observed point mutations are all located in the vicinity of the hygromycin B binding site, i.e., positions U1406C (in 24 of the 35 mutants), C1496U (6 of 35), and U1498C (5 of 35) (Fig. 2).
FIG. 2.
Secondary structure and sequence comparison of the hygromycin binding site within the 16S rRNA A-site region of the prokaryotes E. coli and M. smegmatis, the archaeon Thermococcus celer, and the eukaryotic homologues of T. thermophila and Homo sapiens (cytoplasmic and mitochondrial). Universally conserved nucleotides are shown in boldtype (numbered according to E. coli). The hygromycin B resistance mutations at U1406, C1496, and U1498 found in M. smegmatis are indicated by open circles.
Physiological investigations.
Determinations of MICs were performed to investigate the susceptibility of mutants to hygromycin B. The mutant with the most abundant resistance mutation, U1406C, showed a high-level resistant phenotype, as did the mutant with the C1496U mutation; in contrast, U1498C conferred a low level of drug resistance (Table 1). Investigating the growth rate revealed that the generation time of mutants U1406C and C1496U was significantly increased compared to that of the susceptible parental strains (Table 1).
Proof of cause-effect relationship.
Although the frequency of hygromycin B-resistant mutants obtained was well within that known for mutants with single point mutations in M. smegmatis rrn− (24, 29), we wanted to prove directly that the 16S rRNA alterations isolated were responsible for the resistance phenotype. A recombination assay (24) was used to demonstrate a cause-effect relationship for mutations U1406C, C1496U, and U1498C. Plasmids pMV361ΔKan-Gm-rRNA1406C, pMV361ΔKan-Gm-rRNA1496U, and pMV361ΔKan-Gm-rRNA1498C each carry a nonfunctional rrn fragment of 1.1 kbp containing the respective mutation; plasmid pMV361ΔKan-Gm-rRNAwt served as a control. Following transformation of hygromycin B-susceptible M. smegmatis rrn− with pMV361ΔKan-Gm-rRNA1406C, pMV361ΔKan-Gm-rRNA1496U, and pMV361ΔKan-Gm-rRNA1498C, hygromycin B-resistant recombinants were readily observed after plating in the presence of hygromycin B. Subsequent sequence determination of the single functional rRNA operon revealed that the respective sequence alteration was introduced into each of the resistant recombinants; recombinant strains showed the same characteristics as the spontaneous mutant strains (Table 1).
Structural interpretation.
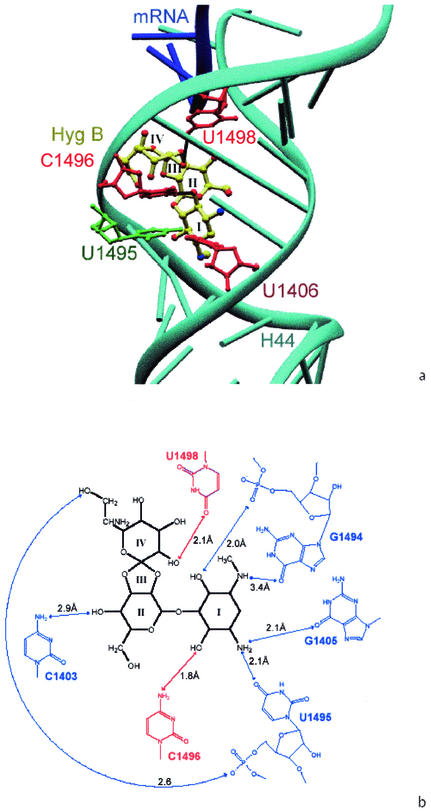
The observed resistance mutations were all located at or near the hygromycin B binding site in helix 44 of the 16S rRNA as observed by X-ray crystallography (4) (Fig. 3). In the crystal structure of hygromycin B bound to the 30S ribosomal subunit from Thermus thermophilus, U1406C did not have any direct interactions with hygromycin B. However, U1406C conferred the highest level of resistance. Presumably, U1406 can influence the binding of hygromycin B through a sheared UU base pair with U1495. U1495 forms part of the pocket in which hygromycin B is bound; it is involved in a base-specific interaction with ring I of hygromycin B, and its backbone phosphate is involved in an interaction with ring IV of the drug. The weak base pairing between U1406 and U1495 is governed by a strong hydrogen bond between N-3 of U1495 and O-4 of U1406. Mutation of U1406 into a cytosine would break this hydrogen bond by placing a N at the 4 position, resulting in either disruption or rearrangement of the base pair, which in turn could change the different local conformation of U1495 and conceivably destroy the hygromycin B binding site.
FIG. 3.
(a) Hygromycin B bound to helix 44 of 16S rRNA (cyan) with rings I, II, III, and IV. The resistance mutations at U1406, C1496, and U1498 found in this study are indicated (in red), and resistance mutation U1495 described by Spangler and Blackburn (31) is also shown (green base). The figure was produced with Ribbons/PovRay (7). (b) Chemical structure diagram of hygromycin B and the proposed interaction with 16S rRNA (blue) (4). Resistance mutations that interact directly with hygromycin B (C1496 and U1498) are shown in red.
Given the uncertainty inherent in the crystal structure determined at approximately 3-Å resolution (4), it cannot yet be resolved whether C1496 is in direct contact with hygromycin B through a hydrogen bond between its N-4 atom and O-31 of ring I of hygromycin B. In case of a direct contact, the C1496U mutation would replace N-4 by O, thus possibly breaking the direct interaction with hygromycin B. In addition, C1496 is involved in a Watson-Crick base pair with G1405, another nucleotide involved in binding of ring I. Mutation of 1496C to U would change the conformation of the base pair and thus probably distort the local geometry and the hygromycin B binding site.
In the crystal structure, U1498 has a clear interaction with hygromycin B. U1498 is not involved in any canonical base pair within 16S rRNA and interacts only via its O-2 with the N-6 of A1499; however, the O-4 atom of U1498 forms a strong and direct hydrogen bond with O-30 of ring IV of hygromycin B (2.1 Å). Mutation of U1498 to a cytosine would break this hydrogen bond by placing an N at the 4 position. Since the base interacts with the remaining 30S subunit only via its O-2 atom, distortion of the local geometry on mutation would therefore not be expected to seriously affect the overall structure of the ribosome. It would, however, seriously and directly affect hygromycin B binding, making this a rather specific resistance mutation. Within all three resistance mutations, U1498C has the smallest effect on gross ribosome function as determined by growth rate (Table 1).
DISCUSSION
Both prokaryotic 70S and eukaryotic 80S ribosomes are inhibited by hygromycin B. Resistance to hygromycin B is associated with drug-modifying enzymes, i.e., the conversion of hygromycin B into the inactive forms 7"-O-phosphorylhygromycin B (6, 22) or 4-O-phosphorylhygromycin B (24a). Besides a possible involvement of nucleotide methylation within 16S rRNA (18), the only ribosomal alteration described to confer resistance to hygromycin B is the 16S rRNA point mutation U1495C in the eukaryote Tetrahymena thermophila (31). Ribosome protection experiments revealed U1495 to be crucial for hygromycin B binding (19). A genetically engineered strain of M. smegmatis rrn− carrying a single functional rRNA operon (28) was exploited to isolate and to map ribosomal alterations causing resistance to hygromycin B in eubacteria.
The hygromycin B resistance mutations observed in this study are all localized to the small-subunit rRNA, i.e., U1406C, C1496U, and U1498C (E. coli numbering). The nucleotides are in the center of 16S rRNA helix 44, in the vicinity of the hygromycin B binding site (4). In contrast to streptomycin (3), an aminoglycoside antibiotic with a different structure, the determinant of specificity was found to be confined to the drug binding site. Attempts to isolate resistant mutants of the parental M. smegmatis SMR5, which carries two rrn operons, were unsuccessful, indicating that rRNA alterations conferring resistance act in a recessive manner. The recessive nature of the resistance determinant prevents the isolation of drug-resistant mutants in microorganisms carrying multiple copies of rRNA operons, which accounts for the lack of described resistant bacterial mutants.
Point mutation U1406C, which mediates the highest level of resistance to hygromycin B, would rearrange the base pair U1406-U1495, resulting in a different local conformation of U1495, a base shown previously to be crucial for hygromycin B binding (31). The C1496U mutation would distort the Watson-Crick base pair C1496-G1405, leading to a change in the local geometry of the hygromycin B binding site. Because of the disruption of a strong hydrogen bond with hygromycin B, mutants carrying U1498C are expected to be unable to bind hygromycin B. Surprisingly, this mutation shows the lowest level of drug resistance. It appears that for hygromycin B binding, the overall conformation of the drug binding pocket may be more important than a single specific chemical bond. This is in contrast to the situation for the classical 2-deoxystreptamine aminoglycosides, where the resistance determinant 16S rRNA A1408 to G disfavors the binding of the aminoglycoside by preventing the formation of the necessary direct hydrogen bonds with atoms O-5′ and N-6′ of ring I (36).
The observed point mutations are in good agreement with crystallographic data (4), supporting the view that hygromycin B does not provoke misreading, as suggested earlier (9). Two adenines, A1492 and A1493, at the top of helix 44 are crucial for decoding. The bases interact directly with the duplex minihelix formed after a correct interaction between the incoming tRNA anticodon and the mRNA codon at the A site (21). Relative to the A site and the bases A1492 and A1493, all three mutations observed in this study are on the other side of helix 44 and thus not in close contact with the decoding center. Under these circumstances, it would not be expected that any of the three mutations would have an effect on the decoding or accuracy of the ribosome. U1498C is the only mutation in close proximity to the mRNA, but it does not seem that U1498C has any strong interactions there. However, indirect (allosteric) effects cannot be excluded. The resistance mutations described here support the view that hygromycin B acts primarily by blocking the proper translocation of the ribosome.
Ribosomal drugs inhibit specific ribosomal functions by discriminating between bacterial and eukaryotic ribosomes to various extents. It was previously suggested that the analysis of resistance mutations in bacteria would allow the prediction of whether cytoplasmic or mitochondrial ribosomes in eukaryotic cells are susceptible to the drug (3). Central to this hypothesis is the concept of informative sequence positions, i.e., the identification of rRNA nucleotides mediating drug resistance in prokaryotic microorganisms which differ between susceptible prokaryotic alleles and the respective eukaryotic homologue. Such nucleotides are likely to be important in determining the specificity of ribosomal drugs, e.g., for an antibacterial agent to affect the prokaryotic as opposed to the eukaryotic ribosome. The identification of a polymorphic nucleotide as a resistance determinant in hygromycin B, a universal ribosomal drug, would effectively falsify this hypothesis.
To investigate whether the broad-spectrum activity of hygromycin B is associated with universal conservation of resistance nucleotides at 16S rRNA positions 1406, 1496, and 1498, in silico sequence comparisons were carried out (Fig. 2). In prokaryotes, as well as in eukaryotes, the nucleotides which confer drug resistance are universally conserved. With the exception of Paenibacillus thiaminolyticus (U1406C), Mycrocystis viridis, Saccharomyces spp. (C1496U), Legionella wadsworthii, Brucella spp., Methylobacterium spp., and Clostridium xylanolyticum (U1498C), all other analyzed organisms (>1,000) carry a susceptible genotype at the respective sequence position (http://www.rna.icmb.utexas.edu/). Two possible explanations for the lack of complete conservation are apparent. The first is the presence of sequencing errors or misalignment of sequences. Alternatively, ribosomes containing a base other than U1406, C1496, or U1498 may show additional sequence covariances that would allow replacement. The observation that ribosomal alterations mediating resistance to hygromycin B exclusively involve universally conserved nucleotides within rRNA is in accordance with the finding that hygromycin B is toxic for almost all organisms.
The findings reported here support the concept that the selection of drug-resistant bacteria, mapping of resistance-conferring mutations, and comparison to the respective eukaryotic homologues offers an important strategy to understand the mechanisms involved in the specificity of ribosomal agents. The possibility of predicting the specificity and toxicity of ribosomal drugs by mapping resistance mutations and in silico sequence comparisons has important implications for future drug development.
Acknowledgments
This study was supported in part by the Swiss National Science Foundation (NRP Antibiotikaresistenz). D.E.B. was supported by a Human Frontier Science Program postdoctoral fellowship.
We thank T. Prammananan and A. Sturmfels for help in the isolation of drug-resistant mutants, T. Janusic for technical assistance, and P. Sander and V. Ramakrishnan for critical comments on the manuscript. We are indebted to two anonymous reviewers for their careful review.
REFERENCES
- 1.Anderson, R. M. 1999. The pandemic of antibiotic resistance. Nat. Med. 5:147-149. [DOI] [PubMed] [Google Scholar]
- 2.Bakker, E. P. 1992. Aminoglycoside and aminocyclitol antibiotics: hygromycin B is an atypical bactericidal compound that exerts effects on cells of Escherichia coli characteristic for bacteriostatic aminocyclitols. J. Gen. Microbiol. 138:563-569. [DOI] [PubMed] [Google Scholar]
- 3.Böttger E. C., B. Springer, T. Prammananan, Y. Kidan, and P. Sander. 2001. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Rep. 2:318-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodersen, D. E., W. M. Clemons, Jr., A. P. Carter, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143-1154. [DOI] [PubMed] [Google Scholar]
- 5.Cabanas, M. J., D. Vasquez, and J. Modolell. 1978. Dual interference of hygromycin B with ribosomal translocation and with aminoacyl-tRNA recognition. Eur. J. Biochem. 87:21-27. [DOI] [PubMed] [Google Scholar]
- 6.Canino, R., P. Contursi, M. Rossi, and S. Bartolucci. 1998. An autonomously replicating transforming vector for Sulfolobus solfataricus. J. Bacteriol. 180:3237-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carson, M. 1991. Ribbons 2.0. J. Appl. Crystallogr. 24:958-961. [Google Scholar]
- 8.Carter, A. P., W. M. Clemons, Jr., D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S subunit and its interactions with antibiotics. Nature 407:340-348. [DOI] [PubMed] [Google Scholar]
- 9.Davies, J., L. Gorini, and B. D. Davis. 1965. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol. Pharmacol. 1:93-106. [PubMed] [Google Scholar]
- 10.De Stasio, E. A., D. Moazed, H. F. Noller, and A. E. Dahlberg. 1989. Mutations in 16S ribosomal RNA disrupt antibiotic-RNA interactions. EMBO J. 8:1213-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eustice, D. C., and J. M. Wilhelm. 1984. Fidelity of the eukaryotic codon-anticodon interaction: interference by aminoglycoside antibiotics. Biochemistry 23:1462-1467. [DOI] [PubMed] [Google Scholar]
- 12.Fourmy, D., M. I. Recht, S. C. Blanchard, and J. D. Puglisi. 1996. Structure of the A site of E. coli 16S rRNA complexed with an aminoglycoside antibiotic. Science 274:1367-1371. [DOI] [PubMed] [Google Scholar]
- 13.Frank, J., and R. K. Agrawal. 2000. A ratchet-like intersubunit reorganization of the ribosome during translocation. Nature 408:318-322. [DOI] [PubMed] [Google Scholar]
- 14.Gale, E. F., E. Cundliffe, P. E. Reynolds, M. H. Richmond, and J. M. Waring (ed.). 1981. The molecular basis of antibiotic action. John Wiley & Sons, Inc., New York, N.Y.
- 15.Ganoza, M. C., and M. C. Kiel. 2001. A ribosomal ATPase is a target for hygromycin B inhibition on Escherichia coli ribosomes. Antimicrob. Agents Chemother. 45:2813-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzales, A., A. Jimenez, D. Vasquez, J. E. Davies, and D. Schindler. 1978. Studies on the mode of action of hygromycin B, an inhibitor of translocation in eukaryotes. Biochem. Biophys. Acta 521:459-469. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, J. L., J. A. Ippolito, N. Ban, P. Nissen, P. B. Moore, and T. A. Steitz. 2002. The structure of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10:117-128. [DOI] [PubMed] [Google Scholar]
- 18.Kojic, M., N. Milojevic, and B. Vasiljevic. 1999. Gentamicin-resistance determinants confer background-dependent hygromycin B resistance. Microb. Drug Resist 5:177-182. [DOI] [PubMed] [Google Scholar]
- 19.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389-394. [DOI] [PubMed] [Google Scholar]
- 20.Noller, H. F. 1991. Ribosomal RNA and translation. Annu. Rev. Biochem. 60:191-227. [DOI] [PubMed] [Google Scholar]
- 21.Ogle, J. M., D. E. Brodersen, W. M. Clemons Jr., M. J. Tarry, A. P. Carter, and V. Ramakrishnan. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292:897-902. [DOI] [PubMed] [Google Scholar]
- 22.Pardo, J. M., F. Malpartida, M. Rico, and A. Jimenez. 1985. Biochemical basis of resistance to hygromycin B in Streptomyces hygroscopicus—the producing organism. J. Gen. Microbiol. 131:1289-1298. [DOI] [PubMed] [Google Scholar]
- 23.Prammananan, T., P. Sander, B. A. Brown, K. Frischkorn, G. O. Onyi, Y. Zhang, E. C. Böttger, and R. J. Wallace. 1998. A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J. Infect. Dis. 177:1573-1581. [DOI] [PubMed] [Google Scholar]
- 24.Prammananan, T., P. Sander, B. Springer, and E. C. Böttger. 1999. RecA-mediated gene conversion and aminoglycoside resistance in heterozygous rRNAwt/rRNAmut strains. Antimicrob. Agents Chemother. 43:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Rao, R. N., N. E. Allen, J. N. Hobbs, Jr., W. E. Alborn, Jr., H. A. Kirst, and J. W. Paschall. 1983. Genetic and enzymatic basis of hygromycin B resistance in Escherichia coli. Antimicrob. Agents Chemother. 24:689-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Recht, M. I., S. Douthwaite, and J. D. Puglisi. 1999. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 18:3133-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Sander, P., A. Meier, and E. C. Böttger. 1995. rpsL+: a dominant selectable marker for gene replacement in mycobacteria. Mol. Microbiol. 16:991-1000. [DOI] [PubMed] [Google Scholar]
- 28.Sander, P., T. Prammananan, and E. C. Böttger. 1996. Introducing mutations into a chromosomal rRNA gene using a genetically modified eubacterial host with a single rRNA operon. Mol. Microbiol. 22:841-848. [DOI] [PubMed] [Google Scholar]
- 29.Sander, P., T. Prammananan, A. Meier, K. Fischkorn, and E. C. Böttger. 1997. The role of ribosomal RNAs in macrolide resistance. Mol. Microbiol. 26:469-480. [DOI] [PubMed] [Google Scholar]
- 30.Schlünzen, F., R. Zarivach, J. Harms, A. Bashan, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 31.Spangler, E. A., and E. H. Blackburn. 1985. The nucleotide sequence of the 17S ribosomal RNA gene of Tetrahymena thermophila and the identification of point mutations resulting in resistance to antibiotics the paramomycin and hygromycin. J. Biol. Chem. 260:6334-6340. [PubMed] [Google Scholar]
- 32.Springer, B., Y. G. Kidan, T. Prammananan, K. Ellrott, E. C. Böttger, and P. Sander. 2001. Mechanisms of streptomycin resistance: forcing drug resistance conferring mutations into the 16S rRNA. Antimicrob. Agents Chemother. 45:2877-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Springer, B., P. Sander, L. Sedlacek, K. Ellrott, and E. C. Böttger. 2001. Instability and site-specific excision of intergration-proficient mycobacteriophage L5 plasmids: development of stably maintained integrative vectors. Int. J. Med. Microbiol. 290:669-675. [DOI] [PubMed] [Google Scholar]
- 34.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, S. B. Snapper, R. G. Barletta, W. R. Jacobs, and B. R. Bloom. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 35.Umezawa, H., and I. R. Hooper (ed.). 1982. Aminoglycoside antibiotics. Springer-Verlag, New York, N.Y.
- 36.Vicens, Q., and E. Westhof. 2001. Crystal structure of paramomycin docked into the eubacterial ribosomal decoding A site. Structure 9:647-658. [DOI] [PubMed] [Google Scholar]
- 37.Williams, R. J., and D. L. Heymann. 1998. Containment of antibiotic resistance. Science 279:1153-1154. [DOI] [PubMed] [Google Scholar]
- 38.Zierhut, G., W. Piepersberg, and A. Böck. 1979. Comparative analysis of the effect of aminoglycosides on bacterial protein synthesis in vitro. Eur. J. Biochem. 98:577-583. [DOI] [PubMed] [Google Scholar]