Abstract
The primary objective of this study was to determine the role of picornavirus in flu-like episodes (temperature of ≥38.0°C plus one respiratory and one constitutional symptom) among otherwise healthy adults enrolled in a placebo-controlled, double-blind, randomized oseltamivir treatment study. Combined nasal and pharyngeal swabs were collected at baseline for influenza cultures and picornavirus reverse transcription (RT)-PCR. In addition, acute- and convalescent-serum samples were obtained for serological studies of common respiratory pathogens. From a total of 719 subjects enrolled in the clinical trial within 36 h of the onset of symptoms, 475 (66%) had evidence of recent influenza A or B virus infections by means of culture and/or serological testing. Of the 244 remaining patients, 36 (15%) presented a seroconversion for at least one of the common respiratory viruses or atypical pathogens. An RT-PCR assay for the picornavirus 5" noncoding region (NCR) was positive in a subset of 15 (19%) of 78 patients with flu-like illnesses of undetermined etiology. Sequence analysis of the picornavirus 5" NCR amplicons revealed that 14 (93%) of them had greater homology to rhinoviruses, whereas 1 (7%) was related to enteroviruses. Interestingly, median total symptom scores and oral temperatures of picornavirus-positive patients (n = 15) and placebo-treated influenza virus-positive patients (n = 161) were similar over a 3-week period. We conclude that, among the influenza virus-negative preselected cases of this study, rhinoviruses were relatively frequent pathogens associated with important respiratory and systemic symptoms.
The Picornaviridae family, which contains single-stranded RNA viruses, is divided into various genera such as Rhinovirus, Enterovirus, and Hepatovirus. Rhinoviruses are the leading cause of the common cold, a highly prevalent and benign infection of the upper respiratory tract usually devoid of significant systemic symptoms. However, it has been recently suggested that rhinoviruses may also replicate in the tracheobronchial tree and may cause viral pneumonitis in selected settings (10, 14, 23). The full scope of the disease burden associated with rhinovirus infections has not been completely elucidated since virus isolation is difficult and serology testing is limited by the large number (>100) of viral serotypes. The recent development of reverse transcription (RT)-PCR assays based on amplification of the conserved 5" noncoding region (NCR) of picornaviruses has been shown to improve significantly the detection of rhinoviruses in clinical samples (1, 12, 13, 25). In the present study, we used the latter approach to determine the role of picornaviruses in flu-like illnesses of adults enrolled in an influenza virus neuraminidase inhibitor trial based on a stringent clinical case definition.
(This work was presented in part at the Third International Symposium on Respiratory Viral Infections, 1 December 2000, St. Lucia, West Indies [abstr. P4].)
MATERIALS AND METHODS
Study population.
A placebo-controlled, double-blind, randomized study of oseltamivir (Tamiflu; Hoffmann LaRoche) was conducted during 1997 and 1998 for the treatment of influenza virus infections in otherwise healthy adults (18 to 65 years old) (20). Subjects were recruited from Canada, Hong Kong, and Europe (Germany, Switzerland, France, United Kingdom, Norway, Finland, The Netherlands, and Belgium) from 12 December 1997 to 18 April 1998. In that trial, subjects were enrolled based on the presence of a flu-like illness (see below) of <36 h duration at a time when influenza virus was circulating in their community. The clinical case definition used for enrollment was the following: fever ≥38°C with at least one respiratory symptom (cough, sore throat, or nasal congestion) and at least one constitutional symptom (headache, malaise, myalgia, sweat or chills, or fatigue). All patients were allowed to use symptomatic relief medication (acetaminophen) without restriction. Patients recorded their oral temperature and their symptom score, i.e., the presence and severity of all of the above symptoms except malaise on a four-point scale (0, absent; 1, mild; 2, moderate; 3, severe) twice daily for up to 21 days. Nasal and pharyngeal swabs were collected at baseline and combined in the same viral transportation medium. The chilled viral transportation media were shipped within 24 h to a central laboratory where they were immediately frozen at −70°C. Acute (day 1)- and convalescent (day 21)-serum samples were also collected for serological testing.
Virological and serological testing for influenza virus.
The presence of influenza viruses was sought by culture and serology. For culture, tertiary cynomolgus monkey kidney cells were inoculated using 300 μl of nasopharyngeal swab eluates collected at baseline. Influenza virus was detected by a positive hemagglutination of turkey erythrocytes after a 7-day incubation period followed by hemagglutination inhibition (HAI) subtyping using reference ferret antisera. Serological testing for influenza A and B virus was performed using an HAI assay. The antigens used at the National Reference Center, Erasmus University, Rotterdam, The Netherlands, were influenza A/Auckland/10/97 (H3N2), A/Nanchang/933/95 (H3N2), A/Johannesburg/82/96 (H1N1), and B/Harbin/07/94. A seroconversion was defined by an increase of fourfold or more in HAI titers between acute- and convalescent-serum samples.
Serological testing for other respiratory pathogens.
An exploratory examination of serum samples collected from the recruited population who did not have evidence of recent influenza virus infection by either culture or serology was carried out at the Erasmus University Hospital by the use of enzyme-linked immunosorbent assays (ELISAs). For the detection of immunoglobulin A (IgA) antibodies against respiratory syncytial virus (RSV), adenovirus, parainfluenza type 1 (PIV-1) PIV-3, and PIV-2, an IgA capture ELISA was used essentially as described previously for influenza A and B virus and RSV (3, 24). ELISA plates were coated with anti-human IgA antibodies and incubated with a 1:100 dilution of paired sera. Capture class-specific immunoglobulins were washed and then incubated with conserved viral antigens. The quantity of bound pathogen-specific IgA was measured following incubation with antigen-specific monoclonal antibodies labelled with horseradish peroxidase. The ratio of absorbance (450 nm) measurements of the convalescent samples to the acute samples was compared with a predefined threshold for seroconversion to each antigen. Detection of IgM-specific antibodies against Mycoplasma pneumoniae (Serion, Wurzburg, Germany) and Chlamydia pneumoniae (Medac, Hamburg, Germany) was performed using commercially available ELISA systems.
RT-PCR assay for picornaviruses.
Presence of picornavirus RNA was sought in a subset of randomly selected patients from European countries who tested negative for influenza virus and other respiratory pathogens. Viral RNA was extracted from 140 μl of nasopharyngeal eluates by the use of the QIAmp Viral RNA Mini kit (Qiagen, Mississauga, Ontario, Canada). The RNA was eluted in 50 μl of Qiagen elution buffer. A one-step RT-PCR assay was developed to amplify a fragment of approximately 400 bp within the 5" NCR of Picornaviridae. Primers were selected from consensus regions of enterovirus and rhinovirus sequences retrieved from GenBank (see Results). The RT-PCR protocol was performed using the C-Therm Polymerase One-Step RT-PCR system (Boehringer Mannheim, Laval, Quebec, Canada), 20 μl of resuspended RNA eluate, 0.4 mM deoxynucleoside triphosphates, 20 U of RNasin (Recombinant Ribonuclease Inhibitor; Promega Corporation, Madison, Wis.), 1.25% dimethyl sulfoxide, 5 mM dithiothreitol, and a 0.3 μM concentration of each of the 5" (5"-CAAGCACTTCTGTTTCCCCGG-3") and 3" (5"-GAAACACGGACACCCAAAGTA-3") primers. The cycling conditions were as follows: 30 min at 60°C and then 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C for 40 cycles, followed by a final extension step of 2 min at 72°C. Amplified products were run on an agarose gel before standard Southern blot hybridization with a 23-bp probe for rhinovirus (5"-GGCAGCCACGCAGGCTGGAACAC-3") labeled at its 3" end with digoxigenin (DIG Oligonucleotide labeling kit; Boehringer Mannheim). Hybridized products were revealed by chemiluminescence using the DIG Luminescent detection kit (Boehringer Mannheim). The sensitivity of the RT-PCR after gel electrophoresis and nonisotopic hybridization was estimated at 500 and 100 copies, respectively, using an RNA standard constructed by cloning an amplified rhinovirus product in the PCR 2.1 plasmid with the TA cloning kit (Invitrogen Corporation, San Diego, Calif.) followed by plasmid transcription with the RNA transcription kit (Stratagene Cloning Systems, La Jolla, Calif.). Because of the important homology between enteroviruses and rhinoviruses within the 5" NCR, positive RT-PCR products were further analyzed by DNA sequencing using a cycle sequencing kit (Big Dye Terminator Cycle Sequencing Ready Reaction; Perkin-Elmer Applied Biosystems, Foster City, Calif.) and the same two PCR primers.
RESULTS
Virological and serological results.
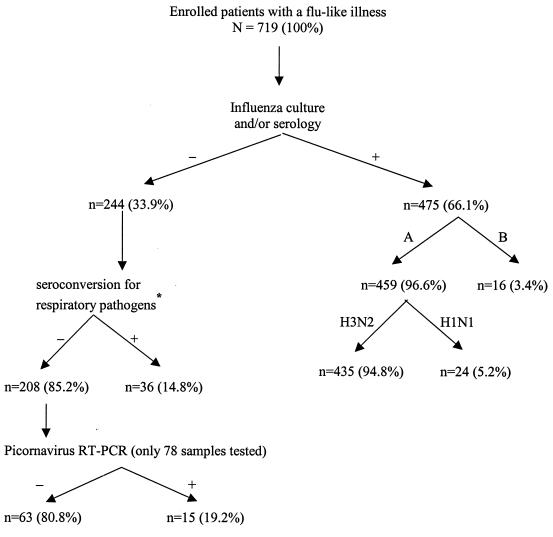
A total of 719 patients fulfilling the clinical case definition for a flu-like syndrome of <36-h duration were enrolled in the clinical trial. An overview of all laboratory results for the study population is outlined in Fig. 1. Two-thirds (475 of 719) of the enrolled patients had an influenza virus infection as confirmed by either cell culture or serology. The overwhelming majority (91.6%) of influenza infections were caused by the A/H3N2 subtype as determined by HAI testing. Of the 244 influenza-negative subjects, 36 (14.8%) presented a seroconversion for at least one of the common respiratory viruses or atypical pathogens. A recent infection was confirmed for the following pathogens: adenovirus (n = 5), PIV-1 to -3 (n = 5), RSV (n = 5), M. pneumoniae (n = 4), C. pneumoniae (n = 10), and mixed infections (n = 7; adenovirus-chlamydia, n = 2; adenovirus-PIV, n = 1; adenovirus-RSV, n = 1; PIV-RSV, n = 2; PIV-chlamydia-RSV, n = 1). Thus, the etiology of the flu-like syndrome was still unknown for 29% (208 of 719) of the patients after influenza cultures and routine serological assays.
FIG. 1.
Overview of laboratory results in a treatment study of influenza-like illness with oseltamivir. ∗, adenovirus (n = 5), PIV (n = 5), RSV (n = 5), M. pneumoniae (n = 4), C. pneumoniae (n = 10), mixed infections (n = 7).
Detection and identification of picornaviruses.
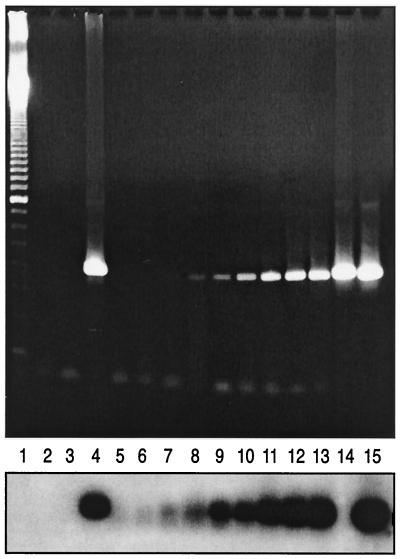
The RT-PCR assay for picornaviruses was performed on a subset of 78 nasopharyngeal swab eluates from randomly selected patients with a flu-like syndrome of unknown etiology. A positive signal was obtained after RT-PCR which was confirmed by nonisotopic hybridization in 15 (19.2%) of the 78 patients (Fig. 2). The sequence of the positive PCR amplicons (approximately 400 bp) was then determined in order to ascribe viruses to a specific genus (Rhinovirus or Enterovirus). For that purpose, the sequence of each clinical sample was aligned with eight rhinovirus (GenBank accession numbers: a10937a, d00239a, x02316a, af108180, af108181, af1108182, af108185, af108187)- and eight enterovirus (GenBank accession numbers: x05690b, x92886b, af083069b, m16560b, x67706b, x84981b, x04468b, j02281b)-matched sequences retrieved from GenBank. A viral sequence was tentatively ascribed to a specific genus when its mean nucleotide homology compared to one group of control sequences was ≥60% but <60% compared to the other group. Using this classification scheme, 14 (93.3%) of the 15 viral strains were identified as rhinoviruses due to a mean nucleotide homology varying from 60.6 to 75.9% compared to the eight rhinovirus control sequences versus a mean homology of 49.8 to 56.6% compared to the eight enterovirus control sequences. However, 1 (6.7%) of the 15 amplified strains gave a faint band after overnight hybridization with the rhinovirus probe and had mean nucleotide homologies of 66.4 and 59.0% compared to control enterovirus and rhinovirus sequences, respectively. Thus, on the basis of sequence analysis, this strain was identified as probable enterovirus.
FIG. 2.
Detection of picornaviruses in nasopharyngeal swab eluates by RT-PCR followed by chemiluminescent hybridization. The upper panel represents the ethidium bromide-stained gel of amplified picornavirus products, whereas the lower panel represents the corresponding hybridization with the rhinovirus probe. Lanes: 1, 100-bp ladder; 2, blank; 3, negative nasopharyngeal sample; 4, positive nasopharyngeal sample; 5 to 13, 50, 100, 250, 500, 1,000, 2,500, 5,000, 10,000, and 25,000 copies of a rhinovirus plasmid, respectively; 14, enterovirus control isolate; 15, rhinovirus control isolate.
Symptom scores and oral temperatures of influenza virus-positive and picornavirus-positive patients.
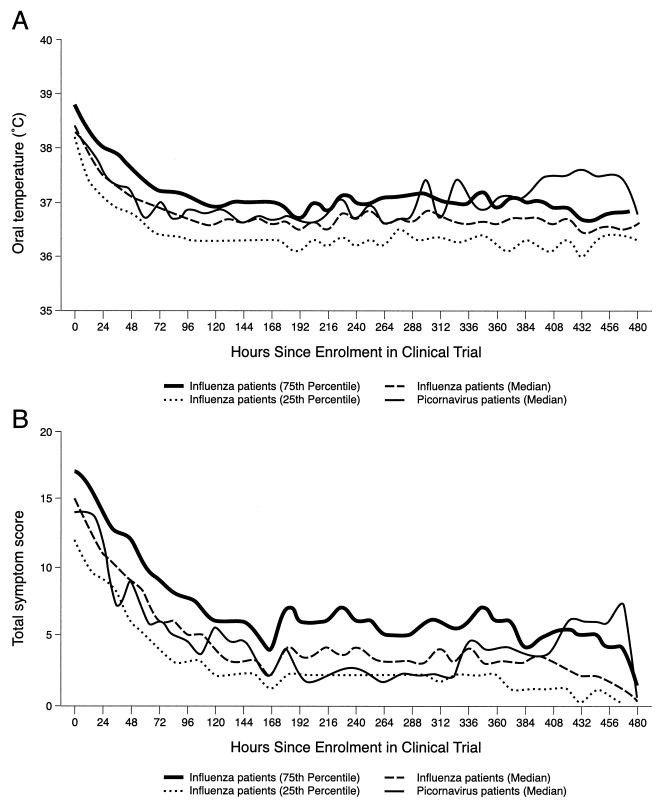
Median oral temperatures and total symptom scores were determined at baseline and for up to 21 days for the 15 picornavirus-positive and the 161 influenza virus-positive patients who received a placebo. As shown in Fig. 3, median temperature and symptom severity curves of the picornavirus-positive patients were generally within the interquartile ranges of the influenza virus-positive patients. However, no formal statistical analysis was performed between the two patient groups due to the small number of picornavirus-positive patients.
FIG. 3.
Median oral temperatures (A) and median total symptom scores (B) over time for 15 picornavirus-positive and 161 placebo-treated influenza virus-positive patients with influenza-like illnesses. Total symptom scores were calculated according to the presence and severity of seven symptoms (cough, sore throat, nasal congestion, headache, myalgia, sweat or chills, and fatigue) on a four-point scale (0, absent; 1, mild; 2, moderate; 3, severe). All patients received no antivirals but were allowed to use symptomatic relief medication (acetaminophen).
DISCUSSION
The most relevant finding of our study is the unanticipated role of picornaviruses in the etiology of flu-like syndromes in influenza virus-negative patients. Of a subset of influenza virus-negative patients randomly chosen, 20% were infected with a picornavirus as determined by an RT-PCR assay for the highly conserved viral 5" NCR (12, 13). With the availability of recent molecular detection methods, the role of rhinoviruses in the pathogenesis of various clinical syndromes has been expanded. Once thought to be only associated with mild self-limiting upper respiratory tract infections, rhinoviruses have been recently linked with more serious lower airway illnesses, including bronchitis and pneumonia (10, 14, 21). These viral agents have also been associated with asthma and cystic fibrosis exacerbations in children (4, 8, 15, 16). Furthermore, both in vitro and in vivo evidence has recently indicated that rhinoviruses can indeed replicate in the tracheobronchial tree (9, 23). In line with a role of rhinoviruses in more severe clinical syndromes, the total symptom scores and median oral temperatures between rhinovirus-positive and influenza virus-positive patients were similar in our preselected cohort of patients (Fig. 3). This suggests that specific antiviral therapy directed against rhinoviruses could be of greater clinical benefit in a subset of patients with more-severe infections. In this regard, some antiviral drugs with activity against rhinoviruses are under evaluation, including capsid binding agents and 3C protease inhibitors (17, 27). Since our subjects were preselected based on the presence of fever and other respiratory and systemic symptoms, our rhinovirus-positive patients likely do not reflect the typical rhinovirus infection. Further studies are needed to identify the subjects at higher risk of complications and the serotypes potentially associated with a more severe outcome as well as the specific symptoms or signs which could be used to differentiate rhinovirus from influenza virus infections.
The present study also provides useful information regarding the use of a clinical case definition for the diagnosis of influenza during flu outbreaks. Indeed, we found that two-thirds of the otherwise healthy adults enrolled in a randomized placebo-controlled study of oseltamivir (20) had a laboratory-confirmed diagnosis of influenza by either culture or serology. This illustrates the excellent positive predictive value of a simple case definition for influenza (fever ≥ 38°C with at least one respiratory and one constitutional symptom) at a time when influenza AH3N2 viruses were known to be circulating in a community during the 1997-to-1998 winter season. Our group (2) and others (19) have reported similar positive predictive values for influenza virus using slightly different case definitions. In these two studies, the combination of fever (above 38°C) and cough achieved the highest positive predictive value for influenza (close to 80%). Thus, during an influenza epidemic, physicians can theoretically rapidly diagnose influenza on clinical grounds in most adults with a typical flu-like syndrome and use this information to institute specific antiviral therapy. However, many factors may potentially limit the usefulness of this approach, such as the lack of an accurate surveillance system with prompt diffusion of the results; the recent use of antipyretics, which may mask the presence of fever; the age of the patients; and the circulating viral strain(s).
No viral and/or atypical pathogens were identified in a substantial proportion of our influenza virus-negative subjects with a flu-like episode (Fig. 1). Indeed, only 15% of the influenza virus-negative subjects presented a seroconversion for one of the common viral and/or atypical pathogens such as adenovirus, PIV, RSV, M. pneumoniae, and C. pneumoniae. Many reasons can explain the high percentage of undetermined infections in our study. First, suboptimal nonmolecular methods were used for the detection of most respiratory pathogens with the exception of picornaviruses. This may have been of particular importance for RSV which has been previously associated with flu-like syndromes in elderly adults (6). In that context, multiplex RT-PCR assays for many respiratory viruses have been recently developed and shown to be more rapid and sensitive than traditional culture or serological assays (2, 5, 7, 11, 22, 26). Second, we did not search for other viruses (for example, coronaviruses [18]) or bacteria that could have been involved in the flu-like episodes of our patients. Finally, we cannot exclude the presence of coinfections in our patients due to the absence of comprehensive molecular testing and the exclusion of influenza virus-positive samples for subsequent studies.
In summary, the use of a simple case definition during an oseltamivir clinical trial at a time when influenza was known to be circulating in the community was associated with a high positive predictive value for influenza in otherwise healthy adults. Rhinoviruses were common viral pathogens among influenza virus-negative patients with flu-like syndromes, and they were associated with a symptomatology similar to that of influenza viruses in those preselected subjects.
REFERENCES
- 1.Andeweg, A. C., T. M. Bestebroer, M. Huybreghs, T. G. Kimman, and J. C. de Jong. 1999. Improved detection of rhinoviruses in clinical samples by using a newly developed nested reverse transcription-PCR assay. J. Clin. Microbiol. 37:524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boivin, G., I. Hardy, G. Tellier, and J. Maziade. 2000. Predicting influenza infections during epidemics with use of a clinical case definition. Clin. Infect. Dis. 31:1166-1169. [DOI] [PubMed] [Google Scholar]
- 3.Brandenburg, A. H., J. Groen, H. A. van Steensel-Moll, E. C. Claas, P. H. Rothbarth, H. J. Neijens, and A. D. Osterhaus. 1997. Respiratory syncytial virus specific serum antibodies in infants under six months of age: limited serological response upon infection. J. Med. Virol. 52:97-104. [DOI] [PubMed] [Google Scholar]
- 4.Collinson, J., K. G. Nicholson, E. Cancio, J. Ashman, D. C. Ireland, V. Hammersley, J. Kent, and C. O'Callaghan. 1996. Effects of upper respiratory tract infections in patients with cystic fibrosis. Thorax 51:1115-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellis, J. S., D. M. Fleming, and M. C. Zambon. 1997. Multiplex reverse transcription-PCR for surveillance of influenza A and B viruses in England and Wales in 1995 and 1996. J. Clin. Microbiol. 35:2076-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falsey, A. R., J. J. Treanor, R. F. Betts, and E. E. Walsh. 1995. Respiratory syncytial virus and influenza A infections in the hospitalized elderly. J. Infect. Dis. 172:389-394. [DOI] [PubMed] [Google Scholar]
- 7.Fan, J., K. J. Henrickson, and L. L. Savatski. 1998. Rapid simultaneous diagnosis of infections with respiratory syncytial viruses A and B, influenza viruses A and B, and human parainfluenza virus types 1, 2, and 3 by multiplex quantitative reverse transcription-polymerase chain reaction-enzyme hybridization assay (Hexaplex). Clin. Infect. Dis. 26:1397-1402. [DOI] [PubMed] [Google Scholar]
- 8.Folkerts, G., W. W. Busse, F. P. Nijkamp, R. Sorkness, and J. E. Gern. 1998. Virus-induced airway hyperresponsiveness and asthma. Am. J. Respir. Crit. Care Med. 157:1708-1720. [DOI] [PubMed] [Google Scholar]
- 9.Gern, J. E., D. M. Galagan, N. N. Jarjour, E. C. Dick, and W. W. Busse. 1997. Detection of rhinovirus RNA in lower airway cells during experimentally induced infection. Am. J. Respir. Crit. Care Med. 155:1159-1161. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh, S., R. Champlin, R. Couch, J. Englund, I. Raad, S. Malik, M. Luna, and E. Whimbey. 1999. Rhinovirus infections in myelosuppressed adult blood and marrow transplant recipients. Clin. Infect. Dis. 29:528-532. [DOI] [PubMed] [Google Scholar]
- 11.Grondahl, B., W. Puppe, A. Hoppe, I. Kuhne, J. A. Weigl, and H. J. Schmitt. 1999. Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J. Clin. Microbiol. 37:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halonen, P., E. Rocha, J. Hierholzer, B. Holloway, T. Hyypia, P. Hurskainen, and M. Pallansch. 1995. Detection of enteroviruses and rhinoviruses in clinical specimens by PCR and liquid-phase hybridization. J. Clin. Microbiol. 33:648-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyypia, T., T. Puhakka, O. Ruuskanen, M. Makela, A. Arola, and P. Arstila. 1998. Molecular diagnosis of human rhinovirus infections: comparison with virus isolation. J. Clin. Microbiol. 36:2081-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imakita, M., K. Shiraki, C. Yutani, and H. Ishibashi-Ueda. 2000. Pneumonia caused by rhinovirus. Clin. Infect. Dis. 30:611-612. [DOI] [PubMed] [Google Scholar]
- 15.Johnston, S. L. 1995. Natural and experimental rhinovirus infections of the lower respiratory tract. Am. J. Respir. Crit. Care Med. 152:S46-S52. [DOI] [PubMed] [Google Scholar]
- 16.Johnston, S. L., P. K. Pattemore, G. Sanderson, S. Smith, F. Lampe, L. Josephs, P. Symington, S. O'Toole, S. H. Myint, and D. A. Tyrrell. 1995. Community study of role of viral infections in exacerbations of asthma in 9-11 year old children. BMJ 310:1225-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser, L., C. E. Crump, and F. G. Hayden. 2000. In vitro activity of pleconaril and AG7088 against selected serotypes and clinical isolates of human rhinoviruses. Antivir. Res. 47:215-220. [DOI] [PubMed] [Google Scholar]
- 18.Makela, M. J., T. Puhakka, O. Ruuskanen, M. Leinonen, P. Saikku, M. Kimpimaki, S. Blomqvist, T. Hyypia, and P. Arstila. 1998. Viruses and bacteria in the etiology of the common cold. J. Clin. Microbiol. 36:539-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monto, A. S., S. Gravenstein, M. Elliott, M. Colopy, and J. Schweinle. 2000. Clinical signs and symptoms predicting influenza infection. Arch. Intern. Med. 160:3243-3247. [DOI] [PubMed] [Google Scholar]
- 20.Nicholson, K. G., F. Y. Aoki, A. D. Osterhaus, S. Trottier, O. Carewicz, C. H. Mercier, A. Rode, N. Kinnersley, and P. Ward. 2000. Efficacy and safety of oseltamivir in treatment of acute influenza: a randomised controlled trial. Lancet 355:1845-1850. [DOI] [PubMed] [Google Scholar]
- 21.Nicholson, K. G., J. Kent, V. Hammersley, and E. Cancio. 1996. Risk factors for lower respiratory complications of rhinovirus infections in elderly people living in the community: prospective cohort study. BMJ 313:1119-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osiowy, C. 1998. Direct detection of respiratory syncytial virus, parainfluenza virus, and adenovirus in clinical respiratory specimens by a multiplex reverse transcription-PCR assay. J. Clin. Microbiol. 36:3149-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papadopoulos, N. G., P. J. Bates, P. G. Bardin, A. Papi, S. H. Leir, D. J. Fraenkel, J. Meyer, P. M. Lackie, G. Sanderson, S. T. Holgate, and S. L. Johnston. 2000. Rhinoviruses infect the lower airways. J. Infect. Dis. 181:1875-1884. [DOI] [PubMed] [Google Scholar]
- 24.Rothbarth, P. H., J. Groen, A. M. Bohnen, R. de Groot, and A. D. Osterhaus. 1999. Influenza virus serology—a comparative study. J. Virol. Methods 78:163-169. [DOI] [PubMed] [Google Scholar]
- 25.Steininger, C., S. W. Aberle, and T. Popow-Kraupp. 2001. Early detection of acute rhinovirus infections by a rapid reverse transcription-PCR assay. J. Clin. Microbiol. 39:129-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stockton, J., J. S. Ellis, M. Saville, J. P. Clewley, and M. C. Zambon. 1998. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J. Clin. Microbiol. 36:2990-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zalman, L. S., M. A. Brothers, P. S. Dragovich, R. Zhou, T. J. Prins, S. T. Worland, and A. K. Patick. 2000. Inhibition of human rhinovirus-induced cytokine production by AG7088, a human rhinovirus 3C protease inhibitor. Antimicrob. Agents Chemother. 44:1236-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]