Abstract
We have molecularly analyzed three genes, sqv-3, sqv-7, and sqv-8, that are required for wild-type vulval invagination in Caenorhabditis elegans. The predicted SQV-8 protein is similar in sequence to two mammalian β(1,3)-glucuronyltransferases, one of which adds glucuronic acid to protein-linked galactose-β(1,4)-N-acetylglucosamine. SQV-3 is similar to a family of glycosyltransferases that includes vertebrate β(1,4)-galactosyltransferases, which create galactose-β(1,4)-N-acetylglucosamine linkages. One model is therefore that SQV-8 uses a SQV-3 product as a substrate. SQV-7 is similar to members of a family of nucleotide-sugar transporters. The sqv genes therefore are likely to encode components of a conserved glycosylation pathway that assembles a C. elegans carbohydrate moiety, the absence of which perturbs vulval invagination.
Most cell-surface and secreted proteins and some lipids are modified by the covalent addition of carbohydrate moieties, which are assembled in the endoplasmic reticulum and Golgi apparatus by the stepwise removal and addition of individual sugar molecules by glycosidases and glycosyltransferases (1, 2). Although entirely eliminating carbohydrate modification from a particular protein can affect its folding, stability, trafficking, or activity (3), mutant mammalian cell lines can survive without apparent decrease in viability even if the carbohydrate modifications on their proteins and lipids are considerably reduced in size and complexity (4). By contrast, the first targeted gene disruptions of glycosyltransferases in mice indicate that multicellular organisms require more elaborate sugar modifications to progress normally through development. For example, loss of the glycosyltransferase GlcNAc-T1, which is required early in the formation of N-linked carbohydrates, has no obvious effect on Chinese hamster ovary cells (5), but mice lacking GlcNAc-T1 die at midgestation (6).
Such observations suggest that the variety and complexity of carbohydrate modifications may be required primarily for cell-cell and cell-matrix interactions. Indeed, in vitro experiments, including the use of inhibitors of glycosylation, competitive oligosaccarides, lectins, carbohydrate-specific antibodies, enzymatic modifiers of carbohydrates, and somatic cell mutants, as well as analyses of the expression patterns and biochemical properties of glycoconjugates, implicate the carbohydrate components of proteoglycans, glycoproteins, and glycolipids in cell-cell and cell-matrix adhesion, recognition, and signaling as well as in extracellular matrix structure and properties (3). However, the in vivo analysis of carbohydrate function in intact multicellular animals has been limited by the small number of mutants with defects in glycosylation. Recent genetic elimination of several glycosyltransferases from mice implicates carbohydrate modifications in normal mouse development (6–8) and confirms the role of fucosylated carbohydrates as ligands for members of the selectin family (9).
We are using the nematode Caenorhabditis elegans to study epithelial invagination, a process that is fundamental to the development of multicellular organisms, in particular to the formation of tubular structures during gastrulation, neurulation, and organogenesis (10, 11). The C. elegans vulva is an epithelial tube that connects the hermaphrodite uterus to the outer epithelium, thereby allowing outward passage of eggs and inward passage of male sperm. We have isolated mutations that perturb C. elegans vulval invagination without affecting vulval cell lineage (12). These mutations define eight genes, sqv-1 to sqv-8, and appear to cause identical vulval defects, specifically, a partial collapse of the invagination and elongation of the central invaginating cells, as well as defects in oocyte and embryonic development. In this paper we describe molecular analyses of sqv-3, sqv-7, and sqv-8 and present evidence that these abnormalities are likely to be caused by specific defects in glycosylation.
MATERIALS AND METHODS
Genetics.
Strains were cultured as described (13) and kept at 20°C. Most mutations and chromosomal rearrangements mentioned in this paper are described in refs. 14 and 15. Exceptions are qC1 dpy-19(e1259) glp-1(q339) (ref. 16; J. Austin and J. Kimble, personal communication) and alleles of sqv-3, sqv-7, and sqv-8 (12).
Molecular Biology.
Standard techniques of molecular biology were used (17). Database searches were performed with the blast program (18) at the National Center for Biotechnology. The human cDNA clone, clone ID no. 132056, was from the Washington University-Merck EST Project (19) and was provided to us by the I.M.A.G.E. consortium (20). All genomic DNA fragments were subcloned into the pBluescript SK− vector (Stratagene). DNA transformation of the strains sqv-8(mn63) unc-4(e120)/mnC1, sqv-3(n2842) unc-69(e587am)/qC1, sqv-7(n2844) unc-4(e120)/mnC1, and sqv-7(n2839) was performed as described (21), and all DNA was coinjected with a plasmid containing the dominant marker rol-6(su1006) (22).
sqv-8 and sqv-3 cDNAs were isolated from mixed-stage libraries (refs. 23 and 24, respectively).
The sqv-3 cDNA was expressed under the control of the heat-shock promoters by using vectors pPD49.78 and pPD49.83 (25, 26); such expression rescued the vulval defect of transgenic sqv-3(n2842) unc-69(e587am) animals that had been placed at 33°C for 2 hr at any time between the embryonic and late L3 stages.
RESULTS
Molecular Identification of sqv-8.
We identified a 4.6-kb genomic fragment that mapped to the sqv-8 region and fully rescued the vulval and fertility defects of sqv-8 (Fig. 1A; ref. 27; data not shown). This fragment was predicted by the C. elegans Genome Sequencing Consortium (28) to contain one complete coding sequence, ZK1307.5, and the first third of a second coding sequence, ZK1307.9. Inserting a frame shift predicted to eliminate the C-terminal 145 aa of the 356-aa ZK1307.5 protein abolished the ability of the 4.6-kb fragment to rescue sqv-8, suggesting that sqv-8 activity requires an intact ZK1307.5 ORF (Fig. 1A). We used part of the rescuing fragment to isolate 11 sqv-8 cDNA clones: seven had identical end sequences, three others were slightly shorter at their 5′ ends, and the eleventh was likely to be artifactual, because its end sequences were not contained within the ZK1307 cosmid. We determined the sequence of one of the seven longest sqv-8 cDNAs (data not shown) and conclude that this 1.3-kb cDNA is likely to be full length, because its 5′ end contains seven nucleotides of the SL1 trans-spliced leader sequence (29), its 3′ end contains a poly(A) tract, and a probe derived from the sqv-8 cDNA hybridizes to a single band of approximately 1,400 nt on a Northern blot of total RNA (data not shown). Its coding sequence is identical to ZK1307.5.
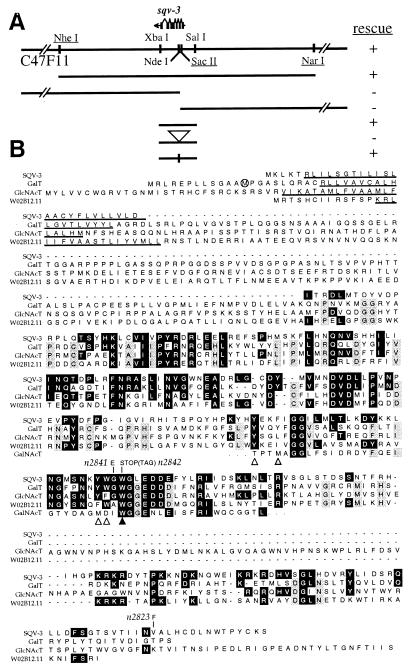
Figure 1.
SQV-8 is similar to a glucuronyltransferase. (A) Cosmid ZK1307 and plasmid clones assayed for sqv-8 rescue activity. + indicates that three or more independently transformed lines con tained rescued sqv-8 animals; − indicates that five or more independently transformed lines contained no rescued sqv-8 animals. Underlined restriction sites are unique in the region of the cosmid that is depicted in detail (i.e., the region from EagI to SacI). The open arrowhead indicates the site at which a frame-shifting insert was introduced, and the vertical line in the bottom clone indicates the site from which this insert was precisely removed. (B) Alignment of the SQV-8, GlcAT-P, and GlcAT-I protein sequences as well as protein sequences predicted from a schistosomal cDNA, and six C. elegans ORFs (W07G9.A, T09E11.1, T15D6.7, C54C8.5, C47F8.4, and E03H4.12) predicted by the Genome Sequencing Consortium. Identities between SQV-8 and any other protein are boxed in black. N-terminal identities are not indicated, because they seem likely to occur randomly in this region. Three or more identical residues that are not shared with SQV-8 are boxed in gray (in some cases two different sets of three identities are in gray within a single column). The underlined amino acids were predicted by the algorithm of (54) (using spans of seven) to have positive hydropathy and therefore may be contained within transmembrane domains. Other stretches of positive hydropathy occur within the regions of amino acid similarity but are not consistently conserved and so are not indicated. The predicted change in amino acid sequence caused by each mutant sqv-8 allele is indicated directly above the amino acid affected, and the DNA sequence of mutant stop codons is noted in parentheses. Cys-317 of GlcAT-P is indicated by a black arrowhead. The region of the alignment from SQV-8 amino acid positions 156–189 is bracketed underneath (see text).
All seven sqv-8 mutant alleles have DNA sequence changes within the ORF defined by the sqv-8 cDNA (Fig. 1B). Three alleles, including the two strongest, n2822 and n2850, as well as mn63, contain nonsense mutations predicted to eliminate, respectively, the final 276, 197, and 129 aa of the 356-aa SQV-8 protein. n2822 additionally contains a missense mutation, as do n2825, n2843, n2847, and n2851.
SQV-8 Is Similar to Two Mammalian β(1,3)-Glucuronyltransferases.
SQV-8 is similar in amino acid sequence to a rat β(1,3)-glucuronyltransferase (GlcAT-P) (30), a human β(1,3)-glucuronyltransferase (GlcAT-I) (31), and several proteins of unknown function, one from the human parasite Schistosoma mansoni (32) and six predicted from C. elegans genomic sequence (Fig. 1B). Glycosyltransferases catalyze the addition of sugars from nucleotide-sugar substrates to proteoglycans, glycoproteins, and/or glycolipids (2). GlcAT-P adds glucuronic acid (GlcA) from UDP-GlcA to galactose (Gal)-β(1,4)-GlcNAc disaccharides on glycoproteins, resulting in the trisaccharide GlcA-β(1,3)-Gal-β(1,4)-GlcNAc (30, 33). A sulfated form of this trisaccharide, SO4–3-GlcA-β(1,3)-Gal-β(1,4)-GlcNAc, is recognized by the mAb HNK-1 (34), which binds a variety of cell-surface proteins involved in cell-cell and cell-matrix adhesion in the vertebrate nervous system (35). GlcAT-I adds GlcA from UDP-GlcA to Gal-β(1,3)-Gal-β(1,4)-xylose-O-Ser, resulting in GlcA-β(1,3)-Gal-β(1,3)-Gal-β(1,4)-xylose-O-Ser, which links the repeating disaccharides of glycosaminoglycans to protein moieties in proteoglycans (31).
Of the 356 aa in SQV-8, 111 (31%) are identical to those of GlcAT-P and 116 (33%) are identical to those of GlcAT-I (Fig. 1B). When N-ethylmaleimide blocks the cysteine at position 317 of GlcAT-P, its enzymatic activity is abolished (30). This cysteine is conserved in SQV-8. Like GlcAT-P, GlcAT-I, and most other glycosyltransferases, SQV-8 has a putative transmembrane domain at its N terminus.
Although six other C. elegans proteins predicted by the Genome Sequencing Consortium (28) are also highly similar to SQV-8, all six predicted proteins are more similar to one another than they are to SQV-8, GlcAT-P, GlcAT-I, and the schistosomal protein and therefore may define a subfamily. None of these six proposed subfamily members have a Cys at the position equivalent to the putative catalytic Cys-317 of GlcAT-P (30). And, for example, in the 34-aa region shown bracketed in Fig. 1B (SQV-8 amino acid positions 156–189), although the sequences of SQV-8, GlcAT-P, and GlcAT-I are identical to one another at 14 positions, 11 of these identities are not contained in W07G9.A, T09E11.1, T15D6.7, C54C8.5, C47F8.4, or E03H4.12. Similarly, although at least four of these latter six amino acid sequences are identical to one another at 28 positions in the bracketed region, 21 of these identities are not contained in SQV-8, GlcAT-P, GlcAT-I, or the schistosomal protein. These six genes do not map near any sqv gene so far identified.
The similarity of SQV-8 to GlcAT-P and GlcAT-I suggests that the loss of SQV-8 might result in the loss of a carbohydrate moiety from one or more glycoconjugates and that the absence of this carbohydrate perturbs C. elegans vulval invagination. The molecular identity of SQV-3 supports this hypothesis.
Molecular Identification of sqv-3.
We identified a 2.4-kb genomic fragment that mapped to the sqv-3 region and fully rescued the sqv-3 vulval defect (although it only partially rescued the fertility defect) (Fig. 2A; data not shown). We used part of the rescuing fragment to isolate sqv-3 cDNA clones and determined that the sqv-3 coding sequence is identical to R10E11.4 predicted from genomic sequence by the C. elegans Genome Sequencing Consortium (28), although the final 30 bp of 3′ untranslated region from this cDNA are absent from the 2.4-kb rescuing fragment (data not shown). Our 1.2-kb sqv-3 cDNA is likely to be full length, because its 5′ end contains nine nucleotides of the SL1 trans-spliced leader sequence, its 3′ end contains a poly(A) tract, and a probe derived from the sqv-3 cDNA hybridized to a single band of 1,300 nt on a Northern blot (data not shown). Each of the three sqv-3 mutant alleles contains a single nucleotide change within the ORF defined by the sqv-3 cDNA. One, n2842, is predicted to result in an in-frame stop codon, and the other two, n2823 and n2841, in changes in amino acid sequence (Fig. 2B).
Figure 2.
SQV-3 is similar to members of a glycosyltransferase family. (A) Cosmid C47F11 and plasmid clones assayed for sqv-3 rescue activity. Symbols are used as in Fig. 1A. (B) Alignment of the SQV-3, GalT, GlcNAcT, and partial human N-acetylgalactosaminyltransferase (GalNAcT) protein sequences and a C. elegans protein sequence, W02B12.11, predicted by the Genome Sequencing Consortium. Identities, transmembrane domains, and predicted changes in mutant alleles are indicated as in Fig. 1B. Open arrowheads indicate Tyr and Trp residues in GalT implicated in binding GlcNAc and/or UDP-Gal, the black arrowhead indicates a Trp residue in GalT required for catalytic activity, and the gray arrowhead indicates a GalT Tyr residue that can be replaced by Phe without affecting catalytic activity (43). The long version of GalT is shown, and the initiator methionine of the short version is circled.
SQV-3 is Similar to Mammalian β(1,4)-Galactosyltransferases and Snail β(1,4)-N-Acetylglucosaminyltransferase (GlcNAcT).
The SQV-3 protein is similar in amino acid sequence to the growing family of human and other vertebrate β(1,4)-galactosyltransferases [the sequence of the original human member of this family (GalT) is depicted in Fig. 2B)] (36–41) and the pond snail Lymnaea stagnalis GlcNAcT (42) (Fig. 2B). The β(1,4)-galactosyltransferases catalyze the addition of Gal from UDP-Gal onto GlcNAc, creating a Gal-β(1,4)-GlcNAc disaccharide; different members of this family have different preferences for the context in which they use GlcNAc as an acceptor. GlcNAcT adds GlcNAc from UDP-GlcNAc onto GlcNAc, creating a GlcNAc-β(1,4)-GlcNAc disaccharide.
Of the 289 aa in SQV-3, 60 (21%) are identical to those of GalT and 61 (21%) are identical to those of GlcNAcT. Like GalT and GlcNAcT, SQV-3 has a putative N-terminal transmembrane domain. Four residues have been implicated in substrate binding and/or catalysis by GalT (43) (Fig. 2). All are conserved in SQV-3, and three of four residues are identical in GlcNAcT. This putative substrate-binding region is also similar to a region in a third glycosyltransferase, a human N-acetylgalactosaminyltransferase (GalNAcT) (44, 45), which has no other obvious sequence identities with GalT, GlcNAcT, or SQV-3 (data not shown). One of the sqv-3 mutant alleles, n2841, results in a nonconservative amino acid change in this region.
The Genome Sequencing Consortium predicts a second C. elegans protein, W02B12.11 (to which cDNA yk258c9 corresponds), that is also highly similar to GalT and GlcNAcT, somewhat more so than is SQV-3. W02B12.11 has 81 identities with GalT and 82 with GlcNAcT, and, unlike that of SQV-3, the N terminus of W02B12.11 is approximately as long as those of GalT and GlcNAcT. W02B12.11 does not map near any sqv gene so far identified.
There is some evidence that, unlike most glycosyltransferases, GalT may be located not only in the Golgi apparatus but also on the cell surface (46) and that cell surface GalT may have a longer N terminus, resulting from an alternative upstream transcriptional start site (47, 48). There is no evidence for analogously longer and shorter versions of SQV-3. We isolated only one class of cDNA, which is predicted to encode a SQV-3 protein with an even shorter N terminus than that of the shorter GalT. This cDNA rescued the sqv-3 mutant vulval defect when expressed under the control of the C. elegans heat-shock promoters (see Materials and Methods), and there is no second ATG upstream of and in-frame with the likely ATG start site in the sqv-3 genomic sequence (data not shown).
That SQV-3 is similar to GalT is of particular interest, because GalT catalyzes the formation of Gal-β(1,4)-GlcNAc, a substrate for the GlcAT-P that is similar to SQV-8.
Molecular Identification of sqv-7.
sqv-7 maps on LG II between dpy-10 on cosmid ZK857 (49) and let-253 on cosmids F31B8 and C24G10 (M. Labouesse, personal communication; map data not shown). The 17.3-kb MluI–PstI subclone from cosmid C52E12, which maps to this interval, rescued both the vulval and fertility defects caused by sqv-7(n2844) and sqv-7(n2839) (Fig. 3A). This MluI–PstI fragment is predicted by the C. elegans Genome Sequencing Consortium to contain two coding sequences: C52E12.2, which corresponds to the unc-104 gene (50), and C52E12.3. The sqv-7 rescue activity seemed unlikely to correspond to C52E12.2 for the following reasons: unc-104(e1265) hermaphrodites have no Sqv vulval defect, dpy-10(e128) unc-104(rh142)/sqv-7(n2839) animals are phenotypically wild type [rh142 is a more severe loss-of-function allele than e1265: although unc-104(e1265) animals are homozygous viable, unc-104(rh142) homozygotes die at a stage long before vulval development would occur], and a construct that lacks the first half of C52E12.2, including its translational initiation codon, retained sqv-7 rescue activity. However, part of the unc-104 genomic region, perhaps the large fourth intron, seems to be required for sqv-7 rescue activity. One possible explanation is that this region may be required for efficient expression of C52E12.3.
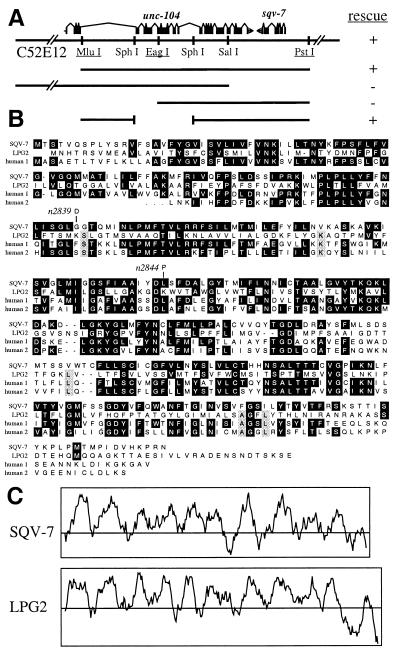
Figure 3.
SQV-7 is similar to LPG2, a protein required for GDP-mannose transport. (A) Cosmid C52E12 and plasmid clones assayed for sqv-7 rescue activity. + indicates that two or more independently transformed lines contained rescued sqv-7 animals. Otherwise symbols are used as in Fig. 1A. (B) Alignment of the SQV-7 and Leishmania LPG2 protein sequences and protein sequences predicted from two human cDNAs. Human 1 is predicted from cDNA accession no. D87449, and human 2 from clone 132056 (see text). The human 2 cDNA is likely to be incomplete at its 5′ end, which is indicated by three dots at the human 2 N terminus. Identities and predicted changes in mutant alleles are indicated as in Fig. 1B. (C) Kyte-Doolittle hydropathy plots of SQV-7 and Leishmania LPG2. In each case, the amino acid sequence is plotted along the horizontal axis, and its corresponding hydropathy (54) is plotted along the vertical axis. Regions above the central horizontal line are of positive hydropathy and, if sufficient length, therefore may be contained within transmembrane domains. The two plots are drawn at the same scale.
To test whether sqv-7 corresponds to C52E12.3, we determined the molecular nature of the two existing sqv-7 mutant alleles. Each contains a single nucleotide change within the predicted C52E12.3 ORF: n2839 and n2844 are missense alleles predicted to result in nonconservative changes in the C52E12.3 amino acid sequence (Fig. 3B). We conclude that sqv-7 corresponds to C52E12.3 or some variant of it that includes at least part of the exons affected by the two mutations described. That the predicted C52E12.3 protein shares extensive amino acid similarity with several previously identified proteins (Fig. 3B; see below) increases the likelihood that this prediction is largely correct. In addition, the cDNA clone yk46f1 (5′ read accession no. D37556, 3′ read accession no. D34487) appears to correspond to part of C52E12.3 (data not shown), although it is incomplete (the first half of the ORF is missing) and contains an inversion when compared with C52E12 cosmid sequence. In the remainder of this paper we will refer to the protein product predicted from the C52E12.3 ORF as SQV-7.
SQV-7 Is Similar to a Putative Nucleotide-Sugar Transporter.
The SQV-7 protein is similar in amino acid sequence to a Leishmania donovani protein, LPG2, which is required for transport of GDP-mannose across membranes (51, 52) (Fig. 3B). Because of its hydrophobicity, subcellular location, and similarity to other proteins implicated in transmembrane transport, LPG2 is likely to be a GDP-mannose transporter (51, 52). Such transporters are required to bring nucleotide sugars from the cytosol, where they are synthesized, into the endoplasmic reticulum and Golgi apparatus, where they are used as sugar-donor substrates by glycosyltransferases (53). Of the 329 aa in SQV-7, 67 (20%) are identical to those of LPG2 (Fig. 3B), and the hydropathy plots (54) of SQV-7 and LPG2 are highly similar (Fig. 3C). From the Washington University-Merck EST Project we identified and determined the sequence of a partial human cDNA (data not shown) that encodes a protein (human 2 in Fig. 3B) that, along with a second human protein (human 1 in Fig. 3B) of unknown function (55), is also closely related in sequence to SQV-7.
DISCUSSION
The sqv Genes Are Likely to Define Components of a Glycosylation Pathway Conserved from Nematodes to Humans.
We have presented evidence that the sqv mutant abnormalities in vulval invagination and early development are likely to result from defects in the assembly of a carbohydrate moiety. SQV-8 is similar to two vertebrate β(1,3)-glucuronyltransferases, and SQV-3 is similar to β(1,4)-galactosyltransferases from vertebrates and GlcNAcT. Because SQV-8 is similar to a protein, GlcAT-P, that uses Gal-β(1,4)-GlcNAc as a substrate, and SQV-3 is similar to a family of proteins that catalyze the formation of this disaccharide, one simple model is that SQV-3 makes Gal-β(1,4)-GlcNAc, which is used as a substrate by SQV-8 (Fig. 4). Such a model in which SQV-8 acts after SQV-3 also might explain why the progeny of sqv-8 mutants tend to arrest at a later stage than do those of sqv-3 mutants (12): perhaps the SQV-3-dependent linkage, but not the further addition of the SQV-8-dependent linkage, is required for the earliest stages of development.
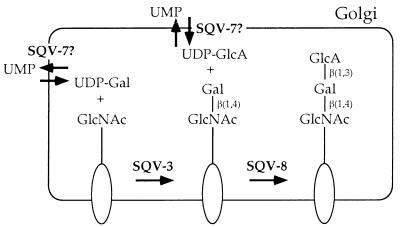
Figure 4.
A model for sqv gene function. SQV-3, SQV-7, and SQV-8 may catalyze the formation of an oligosaccharide on one or more glycoproteins, glycolipids, or proteoglycans. The oval represents a glycoprotein, glycolipid, or proteoglycan, the longer straight line an oligosaccharide of unspecified structure, and the shorter straight lines sugar-sugar linkages. The biochemical activities proposed for SQV-3, SQV-7, and SQV-8 are based on a subset of those defined for the proteins related to them in amino acid sequence (see text). The terminal oligosaccharide is depicted as an unsulfated version of the HNK-1 epitope but instead may be less closely related to this epitope, may be further modified by the addition of sugars and/or sulfate groups, or may occur as a repeating rather than terminal unit (see text).
Of course, many models in which SQV-8 and SQV-3 have other glycosyltransferase activities are also consistent with the genetic and molecular data. For example, like GlcAT-I, SQV-8 and therefore SQV-3 may be involved in the synthesis of proteoglycans. The sqv-8 and sqv-3 embryonic phenotypes may differ because the existing sqv-8 alleles cause only a partial loss of function or because the function of SQV-8 during early development is redundant with that of one or more other proteins (which therefore can compensate for its loss), whereas the function of SQV-3 is not.
SQV-7 is similar to a putative Leishmania GDP-mannose transporter, LPG2. Human cells have no detectable GDP-mannose transport activity, yet there are at least two human proteins similar to LPG2; thus, it is likely that LPG2, SQV-7, and the two human proteins are members of a family of transporters that have a variety of nucleotide-sugar specificities (52). We therefore propose that SQV-7 may transport a nucleotide-sugar used as a substrate by SQV-3 or SQV-8 (Fig. 4). Because a human UDP-Gal transporter has no sequence similarity to SQV-7 (56), one possibility is that SQV-7 is a UDP-GlcA transporter, although it is also possible that SQV-7 transports UDP-Gal or a different SQV-3 or SQV-8 substrate or that SQV-7 provides a nucleotide-sugar to a third unidentified glycosyltransferase that also can affect vulval invagination.
Other sqv genes might encode additional glycosyltransferases, additional proteins required for the biosynthesis or transport of nucleotide-sugar substrates of SQV-3, SQV-8, or other involved glycosyltransferases, or, possibly, substrates for SQV-3 and SQV-8. The molecular and biochemical analysis of the SQV proteins may allow the systematic identification of many or all components of a conserved glycosylation pathway and may reveal components not yet identified biochemically in other systems.
The sqv Mutant Phenotype Suggests that Glycosylation Can Affect Epithelial Invagination.
In vitro manipulations of invaginating epithelia and observations of glycoconjugate expression previously have implicated glycoconjugates in some examples of epithelial invagination (for example, refs. 57 and 58). Our analysis of the sqv mutants provides in vivo evidence that glycosylation can affect epithelial morphogenesis. The cellular basis of the Sqv defect in vulval invagination is as yet unclear, and we discuss a number of possible models in ref. 12. The future identification of the SQV-dependent carbohydrate, the glycoconjugate(s) modified, and the cellular and subcellular site(s) at which these molecules act should clarify both the cellular basis of the Sqv defect and the precise molecular consequences of losing such a carbohydrate. For example, a SQV-dependent carbohydrate might be required to directly and specifically bind other molecules, as in the case of the selectin ligands (59), or such a carbohydrate might exert its effect largely by virtue of its size and charge, as in the case of glycosaminoglycans (60), or it might modify and thereby mask one or more carbohydrates or glycoconjugates that otherwise can perturb normal development, or such a carbohydrate simply might affect the stability and/or activity of a glycoconjugate required for normal vulval development.
Acknowledgments
We thank Beth James for help with DNA sequencing and the Caenorhabditis Genetics Center for providing strains containing the unc-104(rh142) and sqv-8(mn63) mutations. This work was supported by Public Health Service Research Grant GM24663. H.R.H. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- GlcA
glucuronic acid
- Gal
galactose
- GlcAT-P
rat β(1,3)-glucuronyltransferase
- GlcAT-I
human β(1,3)-glucuronyltransferase
- GalT
human β(1,4)-galactosyltransferase
- GlcNAcT
snail β(1,4)-N-acetylglucosaminyltransferase
Footnotes
References
- 1.Kornfeld R, Kornfeld S. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 2.Kleene R, Berger E G. Biochim Biophys Acta. 1993;1154:283–325. doi: 10.1016/0304-4157(93)90003-7. [DOI] [PubMed] [Google Scholar]
- 3.Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanley P, Ioffe E. FASEB J. 1995;9:1436–1444. doi: 10.1096/fasebj.9.14.7589985. [DOI] [PubMed] [Google Scholar]
- 5.Stanley P. Annu Rev Genet. 1984;18:525–552. doi: 10.1146/annurev.ge.18.120184.002521. [DOI] [PubMed] [Google Scholar]
- 6.Ioffe E, Stanley P. Proc Natl Acad Sci USA. 1994;91:728–732. doi: 10.1073/pnas.91.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Q, Hasty P, Shur B D. Dev Biol. 1997;181:257–267. doi: 10.1006/dbio.1996.8444. [DOI] [PubMed] [Google Scholar]
- 8.Asano M, Furukawa K, Kido M, Matsumoto S, Umesaki Y, Kochibe N, Iwakura Y. EMBO J. 1997;16:1850–1857. doi: 10.1093/emboj/16.8.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maly P, Thall A, Petryniak B, Rogers C E, Smith P L, Marks R M, Kelly R J, Gersten K M, Cheng G, Saunders T L, et al. Cell. 1996;86:643–653. doi: 10.1016/s0092-8674(00)80137-3. [DOI] [PubMed] [Google Scholar]
- 10.Bard J. Morphogenesis: The Cellular and Molecular Basis of Developmental Anatomy. Cambridge: Cambridge Univ. Press; 1990. [Google Scholar]
- 11.Fristrom D. Tissue Cell. 1988;20:645–690. doi: 10.1016/0040-8166(88)90015-8. [DOI] [PubMed] [Google Scholar]
- 12.Herman T, Hartwieg E, Horvitz H R. Proc Natl Acad Sci USA. 1999;96:968–973. doi: 10.1073/pnas.96.3.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodgkin J, Edgley M, Riddle D L, Albertson D G the Community of C. elegans Researchers, editors. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 491–586. [Google Scholar]
- 15.Hodgkin J. In: C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 881–1047. [Google Scholar]
- 16.Austin J, Kimble J. Cell. 1989;58:565–571. doi: 10.1016/0092-8674(89)90437-6. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Altschul S F, Gish W, Miller W, Myers E W, Lipmann D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Gerhold D, Caskey C T. BioEssays. 1996;18:973–981. doi: 10.1002/bies.950181207. [DOI] [PubMed] [Google Scholar]
- 20.Lennon G G, Auffray D, Polymeropoulos M, Soares M B. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 21.Mello C C, Kramer J M, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kramer J M, French R P, Park E C, Johnson J J. Mol Cell Biol. 1990;10:2081–2089. doi: 10.1128/mcb.10.5.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okkema P G, Fire A. Development (Cambridge, UK) 1994;120:2175–2186. doi: 10.1242/dev.120.8.2175. [DOI] [PubMed] [Google Scholar]
- 24.Barstead R J, Waterston R H. J Biol Chem. 1989;264:10177–10185. [PubMed] [Google Scholar]
- 25.Stringham E G, Dixon D K, Jones D, Candido E P M. Mol Biol Cell. 1992;3:221–233. doi: 10.1091/mbc.3.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mello C, Fire A. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 27.Sigurdson D C, Spanier G J, Herman R K. Genetics. 1984;108:331–345. doi: 10.1093/genetics/108.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The C. elegans Sequencing Consortium. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 29.Krause M, Hirsh D. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terayama K, Oka S, Seiki T, Miki Y, Nakamura A, Kozutsumi Y, Takio K, Kawasaki T. Proc Natl Acad Sci USA. 1997;94:6093–6098. doi: 10.1073/pnas.94.12.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitagawa H, Tone Y, Tamura J, Neumann K W, Ogawa T, Oka S, Kawasaki T, Sugahara K. J Biol Chem. 1998;273:6615–6618. doi: 10.1074/jbc.273.12.6615. [DOI] [PubMed] [Google Scholar]
- 32.Davis R E, Hardwick C, Tavernier P, Hodgson S, Singh H. J Biol Chem. 1995;270:21813–21819. doi: 10.1074/jbc.270.37.21813. [DOI] [PubMed] [Google Scholar]
- 33.Oka S, Terayama K, Kawashima C, Kawasaki T. J Biol Chem. 1992;267:22711–22714. [PubMed] [Google Scholar]
- 34.Voshol H, van Zuylen C W E M, Orberger G, Vliegenthart J F G, Schachner M. J Biol Chem. 1996;271:22957–22960. doi: 10.1074/jbc.271.38.22957. [DOI] [PubMed] [Google Scholar]
- 35.Schachner M, Martini R. Trends Neurosci. 1995;18:183–191. doi: 10.1016/0166-2236(95)93899-9. [DOI] [PubMed] [Google Scholar]
- 36.Shaper N L, Shaper J H, Meuth J L, Fox J L, Chang H, Kirsch I R, Hollis G F. Proc Natl Acad Sci USA. 1986;83:1573–1577. doi: 10.1073/pnas.83.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaper N L, Meurer J A, Joziasse D H, Chou T-D D, Smith E J, Schnaar R L, Shaper J H. J Biol Chem. 1997;272:31389–31399. doi: 10.1074/jbc.272.50.31389. [DOI] [PubMed] [Google Scholar]
- 38.Almeida R, Amado M, David L, Levery S B, Holmes E H, Merkx G, van Kessel A G, Rygaard E, Hassan H, Bennett E, Clausen H. J Biol Chem. 1997;272:31979–31991. doi: 10.1074/jbc.272.51.31979. [DOI] [PubMed] [Google Scholar]
- 39.Sato T, Furukawa K, Bakker H, Van den Eijnden D H, Van Die I. Proc Natl Acad Sci USA. 1998;95:472–477. doi: 10.1073/pnas.95.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo N W, Shaper J H, Pevsner J, Shaper N L. Glycobiology. 1998;8:517–526. doi: 10.1093/glycob/8.5.517. [DOI] [PubMed] [Google Scholar]
- 41.Schwientek T, Almeida R, Levery S B, Holmes E H, Bennett E, Clausen H. J Biol Chem. 1998;273:29331–29340. doi: 10.1074/jbc.273.45.29331. [DOI] [PubMed] [Google Scholar]
- 42.Bakker H, Agterberg M, Van Tetering A, Koeleman C A, Van den Eijnden D H, Van Die I. J Biol Chem. 1994;269:30326–30333. [PubMed] [Google Scholar]
- 43.Aoki D, Appert H E, Johnson D, Wong S S, Fukuda M N. EMBO J. 1990;9:3171–3178. doi: 10.1002/j.1460-2075.1990.tb07515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meurer J A, Naylor J M, Baker C A, Thomsen D R, Homa F L, Elhammer A P. J Biochem (Tokyo) 1995;118:568–574. doi: 10.1093/oxfordjournals.jbchem.a124947. [DOI] [PubMed] [Google Scholar]
- 45.White T, Bennett E P, Takio K, Sorensen T, Bonding N, Clausen H. J Biol Chem. 1995;270:24156–24165. doi: 10.1074/jbc.270.41.24156. [DOI] [PubMed] [Google Scholar]
- 46.Evans S C, Youakim A, Shur B D. BioEssays. 1995;17:261–268. doi: 10.1002/bies.950170313. [DOI] [PubMed] [Google Scholar]
- 47.Shaper N L, Hollis G F, Douglas J G, Kirsch I R, Shaper J H. J Biol Chem. 1988;263:10420–10428. [PubMed] [Google Scholar]
- 48.Lopez L C, Youakim A, Evans S C, Shur B D. J Biol Chem. 1991;266:15984–15991. [PubMed] [Google Scholar]
- 49.Levy A D, Yang J, Kramer J M. Mol Biol Cell. 1993;4:803–817. doi: 10.1091/mbc.4.8.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otsuka A J, Jeyaprakash A, García-Añoveros J, Tang L Z, Fisk G, Hartshorne T, Franco R, Born T. Neuron. 1991;6:113–122. doi: 10.1016/0896-6273(91)90126-k. [DOI] [PubMed] [Google Scholar]
- 51.Descoteaux A, Luo Y, Turco S J, Beverly S M. Science. 1995;269:1869–1872. doi: 10.1126/science.7569927. [DOI] [PubMed] [Google Scholar]
- 52.Ma D, Russell D G, Beverly S M, Turco S J. J Biol Chem. 1997;272:3799–3805. [PubMed] [Google Scholar]
- 53.Abeijon C, Mandon E C, Hirschberg C B. Trends Biochem Sci. 1997;22:203–207. doi: 10.1016/s0968-0004(97)01053-0. [DOI] [PubMed] [Google Scholar]
- 54.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 55.Nagase T, Seki N, Ishikawa K, Ohira M, Kawarabayasi Y, Ohara O, Tanaka A, Kotani H, Miyajima N, Nomura N. DNA Res. 1996;3:321–329. doi: 10.1093/dnares/3.5.321. [DOI] [PubMed] [Google Scholar]
- 56.Miura N, Ishida N, Hoshino M, Yamauchi M, Hara T, Ayusawa D, Kawakita M. J Biochem (Tokyo) 1996;120:236–241. doi: 10.1093/oxfordjournals.jbchem.a021404. [DOI] [PubMed] [Google Scholar]
- 57.Lane M C, Koehl M A R, Wilt F, Keller R. Development (Cambridge, UK) 1993;117:1049–1060. doi: 10.1242/dev.117.3.1049. [DOI] [PubMed] [Google Scholar]
- 58.Ingersoll E P, Ettensohn C A. Dev Biol. 1994;163:351–366. doi: 10.1006/dbio.1994.1154. [DOI] [PubMed] [Google Scholar]
- 59.Rosen S D, Bertozzi C R. Curr Opin Cell Biol. 1994;6:663–673. doi: 10.1016/0955-0674(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 60.Toole B P. In: Cell Biology of Extracellular Matrix. Hay E D, editor. New York: Plenum; 1991. pp. 305–341. [Google Scholar]