Abstract
A comparison of quantitative results expressed in hepatitis C virus (HCV) international units per milliliter, obtained from the VERSANT HCV RNA 3.0 (bDNA-3.0) assay, the QUANTIPLEX HCV RNA 2.0 (bDNA-2.0) assay, and the COBAS AMPLICOR HCV MONITOR version 2.0 (HCM-2.0) test was performed. A total of 168 patient specimens submitted to the Mayo Clinic Molecular Microbiology Laboratory for HCV quantification or HCV genotyping were studied. Of the specimens tested, 97, 88, and 79% yielded quantitative results within the dynamic range of the bDNA-3.0, bDNA-2.0, and HCM-2.0 assays, respectively. Overall, there was substantial agreement between the results generated by all three assays. A total of 15 out of 29 (52%) of the specimens determined to contain viral loads of <31,746 IU/ml by the bDNA-3.0 assay were categorized as containing viral loads within the range of 31,746 to 500,000 IU/ml by the bDNA-2.0 assay. Although substantial agreement was noted between the results generated by the bDNA-2.0 and bDNA-3.0 assays, a bias toward higher viral titer by the bDNA-2.0 assay was noted (P = 0.001). Likewise, although substantial agreement was noted between the results generated by the HCM-2.0 and bDNA-3.0 assays, a bias toward higher viral titer by the bDNA-3.0 assay was noted (P ≤ 0.001). The discrepancy between the HCM-2.0 and bDNA-3.0 results was more pronounced when viral loads were >500,000 IU/ml and resulted in statistically significant differences (P ≤ 0.001) in determining whether viral loads were above or below 800,000 IU/ml of HCV RNA, the proposed threshold value for tailoring the duration of combination therapy. The expression of quantitative values in HCV international units per milliliter was a strength of both the bDNA-3.0 and HCM-2.0 assays.
Our current understanding of hepatitis C virus (HCV) pathogenicity has evolved over a relatively short period of time since the virus was described in 1989 (1). This increased knowledge has been accompanied by a rapid change in the diagnostic assays and treatments available. Recent studies have demonstrated that the duration of therapy with alpha interferon (IFN-α) in combination with ribavirin can be optimized based upon the results of HCV genotyping and pretreatment viral load determination (4, 13, 17). These studies were the basis for the establishment of a clinically relevant HCV quantitative threshold of 2,000,000 copies/ml for use in the determination of the length of treatment with IFN-α-ribavirin combination therapy (9). These studies have also emphasized the clinical need for standardized and commercially available methods for accurately and reproducibly quantifying HCV RNA in clinical specimens.
While many of the problems associated with the first-generation branched DNA (bDNA) and reverse transcription-PCR assays have reportedly been resolved (5, 8, 10, 12, 14, 19), the issue of standardization of test reporting has only recently been addressed with the establishment of the World Health Organization (WHO) HCV international unit. The HCV international unit is a standardized unit of measure established by the WHO based on the consensus value obtained from the quantitative analysis of a single HCV RNA sample by numerous participating laboratories throughout the world (18). Recently, Pawlotsky and colleagues have suggested that the threshold value used for tailoring the duration of therapy should be 800,000 HCV IU/ml (16). HCV international unit conversion factors have been established for several of the commercially available assays, including the QUANTIPLEX HCV RNA 2.0 (bDNA-2.0) assay (Bayer Corporation, Tarrytown, N.Y.) (18). The recently introduced VERSANT HCV RNA 3.0 (bDNA-3.0) assay (Bayer Corporation) calculates and reports values in both copies per milliliter and international units per milliliter, whereas the COBAS AMPLICOR HCV MONITOR version 2.0 (HCM-2.0) test (Roche Diagnostics Corporation, Branchburg, N.J.) reports results in international units per milliliter.
Clinicians have been faced with the difficult task of determining appropriate threshold values for the various commercially available quantitative HCV RNA assays (6, 11, 16). With the advent of the standardization of reporting units for the quantification of HCV RNA in clinical specimens, results obtained from assays utilizing vastly different methodologies should now be directly comparable. However, no direct comparison of the bDNA-3.0, bDNA-2.0, and HCM-2.0 assays has yet been published. The primary goal of this study was to directly compare, in HCV international units, the quantitative results obtained from a variety of clinical specimens using these three commercially available quantitative HCV RNA assays and to assess the agreement of quantitative values obtained from each of these assays.
MATERIALS AND METHODS
Clinical specimens.
One hundred sixty-eight clinical specimens were retrospectively selected for inclusion in this study. Of these, 98 specimens were serum specimens submitted to the Mayo Clinic Molecular Microbiology Laboratory for HCV genotype analysis between March and August 2000. These 98 specimens represented multiple strains of HCV genotypes 1, 2, 3, 4, and 6 selected without prior knowledge of HCV RNA titer. The remaining 70 specimens were retrospectively selected from specimens routinely submitted to the Mayo Clinic Molecular Microbiology Laboratory for HCV quantification between February and April 2000. These 70 specimens consisted of 43 serum specimens yielding quantitative levels of <0.2 Meq/ml and 27 serum specimens yielding quantitative values between 0.2 and 10 Meq/ml by using the bDNA-2.0 assay.
Specimen collection and handling.
Whole-blood specimens (3 to 5 ml) were collected from individual patients using a serum separator tube or a sterile blood collection tube without anticoagulants. After allowing the blood to clot at room temperature for no more than 2 h, serum was collected by centrifugation, placed in sterile tubes, and frozen at −70°C. At the time of this study, all specimens selected for analysis were thawed and aliquoted into 4 individual aliquots which were refrozen at−70°C for eventual use in qualitative HCV RNA detection as well as each of the three quantitative assays. The use of individual specimen aliquots was employed specifically to avoid the introduction of multiple freeze-thaw cycles and potential degradation of HCV RNA during testing by each of the assays included in this study.
Qualitative detection of HCV RNA in serum.
The 168 selected specimens were submitted to HCV qualitative testing by using a modification of the AMPLICOR Hepatitis C Virus test, version 1.0 (Roche Diagnostics Corporation) (7) or the COBAS AMPLICOR Hepatitis C Virus test, version 2.0 (Roche Diagnostics Corporation) in accordance with the manufacturer's directions.
HCV genotype determination.
HCV genotyping was performed by direct sequence analysis of the HCV 5" untranslated region by using previously described methods (7). The set of 98 genotyped specimens included 20 specimens with HCV genotype 1a, 18 specimens with HCV genotype 1b, 8 specimens with HCV genotype 1 (no subtype), 9 specimens with HCV genotype 2a/2c, 13 specimens with HCV genotype 2b, 1 specimen with HCV genotype 2 (no subtype), 16 specimens with HCV genotype 3a, 3 specimens with HCV genotype 4c/4d, 2 specimens with HCV genotype 4e, 1 specimen with HCV genotype 4f, 1 specimen with HCV genotype 4h, 3 specimens with HCV genotype 4 (no subtype), and 3 specimens with HCV genotype 6a.
HCV RNA quantification.
All quantitative testing was performed according to the manufacturer's instructions. Each quantitative assay was performed using a single lot of assay kits. The bDNA-3.0 assay was performed in batches of 96 samples consisting of 5 standards (of which 2 were run in duplicate and 1 was run in triplicate), 3 kit controls, and 84 patient specimens. The bDNA-2.0 assay was performed in batches of 48 samples consisting of 4 standards, 2 kit controls, and 42 patient specimens analyzed in duplicate. Testing by the HCM-2.0 assay was performed in batches of 24 samples consisting of 3 kit controls and 21 patient specimens. All serum aliquots processed for analysis by the HCM-2.0 assays were amplified within the allowable 3-h time limit after sample processing, thus avoiding the freezing and thawing of HCV RNA extracts prior to amplification and detection. Dilution and repeat testing of samples yielding values above the dynamic range of any particular quantitative assay was not performed in light of previous reports suggesting that specimen dilution may compromise the comparison of quantitative results obtained from both diluted and undiluted specimens (10, 14). Any specimen yielding an invalid test result by any assay was retested with a new serum aliquot of the original specimen. The quantitative testing of individual specimen aliquots by each assay was completed within a maximum period of 10 days. This was done in order to reduce potential differences in quantification due to HCV RNA degradation during prolonged storage of serum aliquots at −70°C.
Results generated with the bDNA-3.0 assay were converted to HCV international units per milliliter using a conversion factor of 5.2 copies/IU, as currently recommended by the Bayer Corporation. In order to compare the results obtained from the bDNA-2.0 assay directly to the results of the bDNA-3.0 and HCM-2.0 assays, all bDNA-2.0 results were converted from equivalents per milliliter to international units per milliliter by using a previously determined conversion factor of 6.3 eq/IU. The relationship between equivalents per milliliter and international units per milliliter was established based on the results from the original collaborative study establishing the WHO international standard for HCV RNA testing (18) and is currently recommended by the Bayer Corporation.
Statistical analysis.
The agreement between assays, bDNA-3.0 versus bDNA-2.0 and bDNA-3.0 versus HCM-2.0, was assessed by using a weighted kappa statistic (2) on the corresponding matched contingency tables with the four categories of results. For the dichotomous assay assessment, <800,000 IU/ml versus ≥800,000 IU/ml, a simple kappa statistic was used. The interpretation of kappa values references work done by Cohen (2). In addition to the estimated kappa, 95% confidence intervals (95% CIs) were also calculated. The tendency of one assay to report consistently higher or lower than the other, as shown in the matched contingency tables, was assessed by using a Wilcoxon signed-rank test for the four-category assessment and McNemar's test for the two-level assessments. P values of ≤0.05 were considered to be statistically significant.
RESULTS
Qualitative detection of HCV RNA.
One hundred forty-nine of the 168 patient specimens yielded qualitatively detectable HCV RNA at the time of this study. An additional three specimens yielded equivocal test results by the COBAS AMPLICOR Hepatitis C Virus Test, version 2.0. The remaining 16 specimens were negative for the presence of HCV RNA by the COBAS AMPLICOR Hepatitis C Virus Test, version 2.0. No additional qualitative testing could be performed on the three equivocal specimens due to the limited quantities of serum available for this study. All 168 patient specimens were tested by the bDNA-3.0, bDNA-2.0, and HCM-2.0 assays.
Performance characteristics of the bDNA-3.0, bDNA-2.0, and HCM-2.0 assays. (i) bDNA-3.0.
All assay controls and validation requirements were within the acceptable limits for each of the two runs performed, and no retesting of individual specimens was required. A total of 144 (97%) of the qualitatively HCV-RNA-positive specimens yielded quantitative results within the dynamic range of this assay. There were five (3%) specimens yielding a result of <615 IU/ml, and no specimens yielded quantitative values above the upper limit of quantification of the assay (7,692,310 IU/ml). None of the 19 qualitatively HCV-RNA-negative or -equivocal specimens yielded a quantitative result by this assay.
(ii) bDNA-2.0.
Four bDNA-2.0 runs were performed as a part of this study. Two low-titer specimens required retesting due to unacceptably high coefficients of variation (>25%). Both specimens were successfully retested, and both yielded quantitative values below the lower limit of detection of the assay (<0.2 Meq/ml or 31,746 IU/ml). The number of qualitatively HCV-RNA-positive patient specimens containing measurable quantities of HCV RNA was 131 (88%) while 18 (12%) specimens yielded results below the lower detection limit of the assay. None of the specimens were found to contain viral loads above the upper limit of quantification of the assay (120 Meq/ml or 19,047,619 IU/ml). Of the 16 qualitatively HCV-RNA-negative specimens evaluated, 4 yielded quantitative values with an average of 0.245 Meq/ml (38,929 IU/ml). In addition, one of three qualitatively HCV-RNA-equivocal specimens also yielded a quantitative value of 0.453 Meq/ml (71,905 IU/ml).
(iii) HCM-2.0.
Eight separate HCM-2.0 assay runs were performed. All controls and validation requirements were within the acceptable limits of the assay protocol. No additional testing of specimens was performed. Of the 149 qualitatively HCV-RNA-positive specimens, 118 (79%) specimens yielded quantitative results within the dynamic range of the HCM-2.0 assay. Four (3%) specimens contained <600 IU/ml of HCV RNA, and 27 (18%) specimens contained viral loads above the upper limit of quantification (850,000 IU/ml). All 19 qualitatively HCV-RNA-negative or -equivocal specimens yielded results of <600 IU/ml.
Comparison of quantitative results of the bDNA-2.0 and bDNA-3.0 assays.
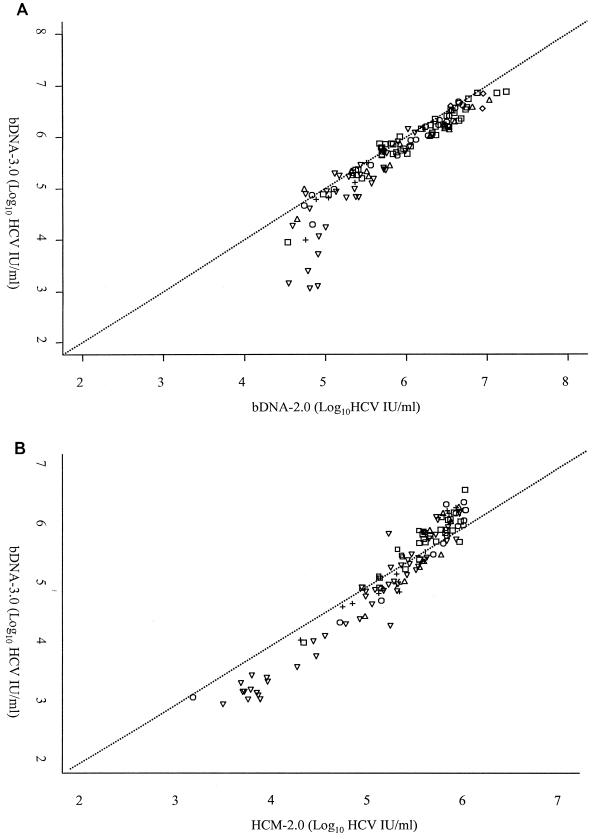
The overall agreement between the bDNA-2.0 and bDNA-3.0 assays for those specimens yielding results within the quantitative range of each of the assays (i.e., a bDNA-2.0 result between 31,746 and 19,047,619 IU/ml and a bDNA-3.0 result between 615 and 7,692,310 IU/ml) is presented graphically in Fig. 1A. The results obtained from 128 specimens including 93 of the 98 specimens of known HCV genotypes, along with 35 specimens containing unknown HCV genotypes, were included in this comparison.
FIG. 1.
Linear comparisons of 128 specimens analyzed by the bDNA-2.0 and bDNA-3.0 assays (A) and 117 specimens analyzed by the HCM-2.0 and bDNA-3.0 assays (B). Comparisons include all specimens yielding values within the quantitative range of each of the assays being compared. HCV genotype is indicated as follows: □, genotype 1; , genotype 2; ▵, genotype 3; +, genotype 4; ◊, genotype 6; and ▿, genotype not determined.
While the overall results obtained from these two assays appeared to be similar in general, they were quite dissimilar within the range extending from 31,746 to 100,000 IU/ml (0.2 to 6.3 Meq/ml) as determined by the bDNA-2.0 assay. Specimens within this lower range exhibit discrepancies of as much as 78,576 IU/ml. While no obvious difference in the quantification of any particular genotype or subtype was observed between the assays included in this comparison, it should be noted that no specimens containing HCV genotype 5 were included in this evaluation and that only three specimens contained HCV genotype 6a.
Table 1 is a matched contingency table comparing the bDNA-2.0 and bDNA-3.0 assays. The table shows substantial agreement between these assays over all four quantitative ranges. The weighted kappa statistic was 0.78 (95% CI, 0.72 to 0.84). Table 1 shows that a total of 9 specimens yielded bDNA-3.0 results which were categorized into higher quantitative ranges than those results obtained by the bDNA-2.0 assay, whereas 30 specimens were categorized into a lower quantitative range by using the bDNA-3.0 assay. These differences in quantification were found to be statistically significant (P = 0.001). For these 39 discrepant results, the HCM-2.0 assay result was in agreement with the bDNA-3.0 assay result in 25 instances.
TABLE 1.
Matched contingency table of the bDNA-2.0 assay versus the bDNA-3.0 assay with quantitative HCV values expressed in HCV international units per millilitera
| bDNA-3.0 result (IU/ml)c | bDNA-2.0 result (IU/ml)b |
Total (%) | |||
|---|---|---|---|---|---|
| ≤31,746 | >31,746-≤500,000 | >500,000-<800,000 | ≥800,000 | ||
| ≤31,746 | 14 | 15 | 0 | 0 | 29 (19.5) |
| >31,746-≤500,000 | 4 | 37 | 8 | 1 | 50 (33.6) |
| >500,000-<800,000 | 0 | 4 | 5 | 6 | 15 (10.1) |
| ≥800,000 | 0 | 0 | 1 | 54 | 55 (36.9) |
| Total (%) | 18 (12.1) | 56 (37.6) | 14 (9.4) | 61 (40.9) | 149 (100.0) |
Weighted kappa, 0.78; 95% CI, 0.72 to 0.84; P, 0.001.
Quantitative values were converted to HCV international units per milliliter by using a conversion factor of 6.3 eq/IU.
Quantitative values were converted to HCV international units per milliliter by using a conversion factor of 5.2 copies/ml.
The agreement between the bDNA-2.0 and bDNA-3.0 assays in predicting viral loads either above or below 800,000 IU/ml yielded results similar to those shown in Table 1. The kappa statistic of agreement was 0.89 (95% CI, 0.81 to 0.96). The test for consistent differences between assays was not found to be statistically significant (P = 0.07). One specimen was classified as <800,000 IU/ml by bDNA-2.0 and ≥800,000 IU/ml by bDNA-3.0, whereas seven specimens were classified as ≥800,000 IU/ml by bDNA-2.0 and <800,000 IU/ml by bDNA-3.0. All remaining 141 quantitative values were in agreement with respect to the proposed threshold value for tailoring the duration of therapy.
Comparison of quantitative results of the HCM-2.0 and bDNA-3.0 assays.
The results for 117 specimens, including 69 specimens of known HCV genotypes and 48 specimens containing unknown HCV genotypes, were included in the graphic comparison of the HCM-2.0 and bDNA-3.0 assays presented in Fig. 1B. As in Fig. 1A, the specimen results included in this figure yielded quantitative results by both assays being compared (i.e., an HCM-2.0 result between 600 and 850,000 IU/ml and a bDNA-3.0 result between 615 and 7,692,310 IU/ml). In contrast to the comparison of the bDNA-2.0 and bDNA-3.0 assays, those values that were <500,000 IU/ml by the HCM-2.0 assay were relatively close to, though consistently lower than, the results obtained with the bDNA-3.0 assay. However, values that were >500,000 IU/ml, as determined by the HCM-2.0 assay, appeared to become increasingly discrepant with rising viral titers. Despite the discrepant viral titers noted between 500,000 and 850,000 IU/ml, the results at the upper end of this comparative range generally appeared to be within 1 log10 of one another.
Again, no obvious differences in the quantification of a particular HCV genotype or subtype were observed between the assays included in this comparison. Only 69 of the 98 specimens containing known HCV genotypes could be included in this comparison due to the fact that a number of specimens yielded quantitative results outside of the dynamic range of at least one of the assays being compared. As previously stated, no specimens containing HCV genotype 5 were included in this evaluation. In addition, all three specimens containing HCV genotype 6a had to be excluded from the comparison of these assays because their viral titers exceeded the upper limit of the HCM-2.0 assay.
Table 2 is a matched contingency table of the HCM-2.0 and bDNA-3.0 assay results obtained from all 149 qualitatively HCV-RNA-positive specimens. Although somewhat lower than in the previous comparison of the bDNA-2.0 and bDNA-3.0 assays, the weighted kappa statistic was 0.73 (95% CI, 0.65 to 0.81), showing substantial agreement between the HCM-2.0 and bDNA-3.0 assays. Despite this overall agreement, the table clearly shows a tendency of the bDNA-3.0 assay to result in higher viral load categorization than the HCM-2.0 assay. This is the case for 32 specimens, while there are only three instances in which the bDNA-3.0 assay yielded a lower viral titer categorization than the HCM-2.0 assay. This bias toward higher viral titer by the bDNA-3.0 assay was statistically significant (P ≤ 0.001).
TABLE 2.
Matched contingency table of HCM-2.0 test versus bDNA-3.0 assay with quantitative values expressed in HCV international units per millilitera
| bDNA 3.0 result (IU/ml)b | HCM-2.0 result (IU/ml) |
Total (%) | |||
|---|---|---|---|---|---|
| ≤615 | >615-≤500,000 | >500,000-<800,000 | ≥800,000 | ||
| ≤615 | 4 | 1 | 0 | 0 | 5 (3.4) |
| >615-≤500,000 | 0 | 72 | 2 | 0 | 74 (49.7) |
| >500,000-<800,000 | 0 | 10 | 5 | 0 | 15 (10.1) |
| ≥800,000 | 0 | 4 | 18 | 33 | 55 (36.9) |
| Total (%) | 4 (2.7) | 87 (58.4) | 25 (16.8) | 33 (22.1) | 149 (100.0) |
Weighted kappa, 0.73; 95% CI, 0.65 to 0.81; P, ≤0.001.
Quantitative values were converted to HCV international units per milliliter by using a conversion factor of 5.2 copies/IU.
When the agreement between the HCM-2.0 and bDNA-3.0 assays in predicting viral loads either above or below 800,000 IU/ml was analyzed, the kappa statistic of agreement was 0.65 (95% CI, 0.53 to 0.78), again defined as substantial agreement. The test for consistent differences between assays also showed the HCM-2.0 assay results to be consistently lower than those obtained using the bDNA-3.0 assay (P ≤ 0.001). While substantial agreement between the two assays still existed with 127 quantitative values in complete agreement, 22 of 55 (40%) specimens determined to contain ≥800,000 IU/ml by bDNA-3.0 were classified as containing <800,000 IU/ml by HCM-2.0.
DISCUSSION
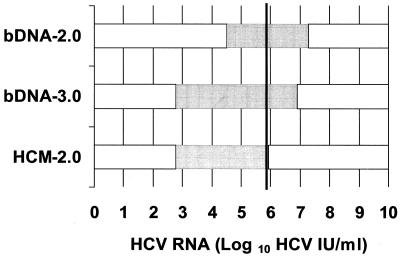
A comparison of the dynamic ranges of each of the assays evaluated in this study is presented in Fig. 2. The ranges are presented relative to one another as well as to the 800,000 IU/ml threshold value for tailoring the duration of therapy proposed by Pawlotsky et al. (16). The bDNA-2.0 assay, while reported to have a clinical sensitivity as high as 96% in an evaluation of chronically infected individuals prior to the initiation of antiviral therapy (5), has a lower limit of sensitivity that is almost 2 log10 higher than that of the other two assays included in this study. The results of our evaluation demonstrated that there were HCV RNAs above the lower limit of detection in only 88% of the specimens tested by the bDNA-2.0 assay (versus 97% of specimens tested by the HCM-2.0 and bDNA-3.0 assays).
FIG. 2.
Comparison of the dynamic ranges of the three quantitative HCV RNA assays expressed in HCV international units per milliliter. The dynamic range of each assay is represented by the shaded portion of the corresponding horizontal bar. The 800,000-HCV-IU/ml threshold for tailoring the duration of therapy proposed by Pawlotsky et al. is indicated by the heavy vertical line.
The bDNA-3.0 assay spans the largest dynamic range of the assays evaluated in this study with a reported level of sensitivity comparable to that of the HCM-2.0 assay as well as a relatively high upper limit of quantification similar to that of the bDNA-2.0 assay. In the current study, 97% of the specimens tested yielded quantitative results with the bDNA-3.0 assay (versus 79% of the specimens tested with the HCM-2.0 assay).
While comparable to the bDNA-3.0 in assay sensitivity, the HCM-2.0 assay has a much lower upper limit of quantification than either the bDNA-2.0 or bDNA-3.0 assays. As a result, 18% of the specimens included in this study were determined to contain viral loads above the upper limit of quantification of the HCM-2.0 assay. By contrast, no specimens yielded viral loads above the upper limit of quantification of the bDNA-2.0 or bDNA-3.0 assays. The upper limit of quantification of the HCM-2.0 assay extends just 50,000 IU/ml beyond the proposed threshold value for tailoring the duration of therapy (800,000 IU/ml). Previous studies have shown that as many as 70% of pretreatment serum samples contain >2,000,000 copies/ml (approximately 800,000 IU/ml) of HCV RNA (13, 17), thus making the HCM-2.0 assay unsuitable for accurate quantification of HCV RNA in the majority of the pretreatment serum samples without including a specimen dilution step. Despite the limitations of the HCM-2.0 assay, the simple determination that the viral load is above the threshold value may be sufficient in the current clinical setting.
Recent studies have suggested that the upper limit of quantification of the HCM-2.0 assay should be 500,000 IU/ml (10). In addition, if accurate viral loads are to be obtained using this assay, dilutions need to be performed to insure that the viral load is well within the linear range of the assay (10). While we did not confirm our findings through the use of additional dilution studies, the results of our study are consistent with the suggestion of Lee et al. that HCV quantification by the HCM-2.0 assay above a viral load of 500,000 IU/ml may result in high-titer specimens beyond the linear range of the assay being assigned a value within the dynamic range of the assay (10). Our findings demonstrate a lack of agreement between the results obtained from the HCM-2.0 assay and the results of the bDNA-3.0 assay between 500,000 and 850,000 IU/ml.
Although our study was not designed to assess assay specificity, both the bDNA-3.0 and HCM-2.0 assays demonstrated good specificity in our study with none of the 19 HCV-RNA-negative or -equivocal specimens yielding results within the quantitative range of either assay. However, 4 of the 16 qualitatively HCV-RNA-negative specimens yielded a result within the quantitative range of the bDNA-2.0 assay (average value of 0.245 Meq/ml or 38,929 IU/ml) along with one of the three qualitatively HCV-RNA-equivocal specimens (0.453 Meq/ml or 71,905 IU/ml). Additionally, a significant number of HCV-RNA-positive specimens with viral titers between 31,746 and 100,000 IU/ml (0.2 to 6.3 Meq/ml) were shown to be discrepant between the bDNA-2.0 assay and the other two assays studied by as much as 2 log10 (Fig. 1). Our findings suggest that the bDNA-2.0 assay may not yield accurate viral titers below 100,000 IU/ml and are consistent with earlier evaluations of the HCM-2.0 and bDNA-2.0 assays. A recent evaluation of the specificity of the HCM-2.0 assay demonstrated the complete absence of false-positive results in an evaluation of 495 HCV-seronegative blood donors (10). In contrast, Pawlotsky suggested a false-positive rate of as much as 3% in early evaluations of the bDNA-2.0 assay with all falsely positive values falling between 0.2 and 0.5 Meq/ml, which is consistent with the findings of this study (15). While no published data currently exists with regard to the specificity of the bDNA-3.0 assay, refinements in the assay chemistry utilized in the latest version of the assay have reportedly resulted in dramatic improvements in both the sensitivity and specificity of other bDNA assays (3). Our results, though limited in scope, are consistent with these findings.
There have been numerous studies demonstrating the shortcomings of the first generation of commercially available assays for the quantification of HCV. In particular, the inability of the AMPLICOR HCV MONITOR test version 1.0 (Roche Diagnostics Corporation) and the QUANTIPLEX HCV RNA 1.0 assay (Chiron Corporation, Emeryville, Calif.) to quantify HCV RNA from HCV genotypes 1 to 6 equally has been well documented (5, 8, 12, 14). The shortcomings of these early quantitative assays made the task of accurately assessing the influence of HCV viral load and genotype on disease outcome a complex task. While all the questions regarding HCV viral load and genotype have not yet been fully answered, the arrival of second-generation quantitative assays has aided our efforts to better understand the complex relationships between viral and host factors as well as to assess the effectiveness of new therapies. Unfortunately, a lack of standardization among the various quantitative assays has hampered the clinical application of the current recommendations for tailoring the duration of IFN-α-ribavirin combination therapy based on viral load (11). While various conversion equations have recently been suggested by several investigators (6, 11, 16), only the standardization of test reporting along with a thorough evaluation of assay performance characteristics will completely resolve this issue. The introduction of additional assay refinements, including the expression of quantitative values in HCV international units per milliliter, is yet another step forward in the development of not only accurate but also standardized HCV viral load testing.
Acknowledgments
We are very grateful to all of the members of the Mayo Clinic Molecular Microbiology Laboratory, especially Paul Shawn Mitchell, for assistance in this study. We also thank Bayer Corporation and Roche Diagnostics Corporation for providing the quantitative test kits used in this study.
REFERENCES
- 1.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 2.Cohen, J. 1960. A coefficient of agreement for nominal scales'. Educ. Psychol. Meas. 20:37-46. [Google Scholar]
- 3.Collins, M. L., B. Irvine, D. Tyner, E. Fine, C. Zayati, C. Chang, T. Horn, D. Ahle, J. Detmer, L. P. Shen, J. Kolberg, S. Bushnell, M. S. Urdea, and D. D. Ho. 1997. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 25:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis, G. L., R. Esteban-Mur, V. Rustgi, J. Hoefs, S. C. Gordon, C. Trepo, M. L. Shiffman, S. Zeuzem, A. Craxi, M. H. Ling, and J. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. International Hepatitis Interventional Therapy Group. N. Engl. J. Med. 339:1493-1499. [DOI] [PubMed] [Google Scholar]
- 5.Detmer, J., R. Lagier, J. Flynn, C. Zayati, J. Kolberg, M. Collins, M. Urdea, and R. Sanchez-Pescador. 1996. Accurate quantification of hepatitis C virus (HCV) RNA from all HCV genotypes by using branched-DNA technology. J. Clin. Microbiol. 34:901-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang, J. W., J. K. Albrecht, S. Jacobs, and J. Y. Lau. 1999. Quantification of serum hepatitis C virus RNA. Hepatology 29:997-998. [DOI] [PubMed] [Google Scholar]
- 7.Germer, J. J., P. N. Rys, J. N. Thorvilson, and D. H. Persing. 1999. Determination of hepatitis C virus genotype by direct sequence analysis of products generated with the Amplicor HCV test. J. Clin. Microbiol. 37:2625-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawkins, A., F. Davidson, and P. Simmonds. 1997. Comparison of plasma virus loads among individuals infected with hepatitis C virus (HCV) genotypes 1, 2, and 3 by QUANTIPLEX HCV RNA assay versions 1 and 2, Roche Monitor assay, and an in-house limiting dilution method. J. Clin. Microbiol. 35:187-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Journal of Hepatology. 1999. EASL International Consensus Conference on Hepatitis C. Paris, 26-28, February 1999, Consensus statement. European Association for the Study of the Liver. J. Hepatol. 30:956-961. [PubMed] [Google Scholar]
- 10.Lee, S. C., A. Antony, N. Lee, J. Leibow, J. Q. Yang, S. Soviero, K. Gutekunst, and M. Rosenstraus. 2000. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J. Clin. Microbiol. 38:4171-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinot-Peignoux, M., N. Boyer, V. Le Breton, G. Le Guludec, C. Castelnau, R. Akremi, and P. Marcellin. 2000. A new step toward standardization of serum hepatitis C virus-RNA quantification in patients with chronic hepatitis C. Hepatology 31:726-729. [DOI] [PubMed] [Google Scholar]
- 12.Martinot-Peignoux, M., V. Le Breton, S. Fritsch, G. Le Guludec, N. Labouret, F. Keller, and P. Marcellin. 2000. Assessment of viral loads in patients with chronic hepatitis C with AMPLICOR HCV MONITOR version 1.0, COBAS HCV MONITOR version 2.0, and QUANTIPLEX HCV RNA version 2.0 assays. J. Clin. Microbiol. 38:2722-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, and J. K. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 14.Mellor, J., A. Hawkins, and P. Simmonds. 1999. Genotype dependence of hepatitis C virus load measurement in commercially available quantitative assays. J. Clin. Microbiol. 37:2525-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pawlotsky, J. M. 1997. Measuring hepatitis C viremia in clinical samples: can we trust the assays? Hepatology 26:1-4. [DOI] [PubMed] [Google Scholar]
- 16.Pawlotsky, J. M., M. Bouvier-Alias, C. Hezode, F. Darthuy, J. Remire, and D. Dhumeaux. 2000. Standardization of hepatitis C virus RNA quantification. Hepatology 32:654-659. [DOI] [PubMed] [Google Scholar]
- 17.Poynard, T., P. Marcellin, S. S. Lee, C. Niederau, G. S. Minuk, G. Ideo, V. Bain, J. Heathcote, S. Zeuzem, C. Trepo, and J. Albrecht. 1998. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. International Hepatitis Interventional Therapy Group (IHIT). Lancet 352:1426-1432. [DOI] [PubMed] [Google Scholar]
- 18.Saldanha, J., N. Lelie, and A. Heath. 1999. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. WHO Collaborative Study Group. Vox Sang. 76:149-158. [DOI] [PubMed] [Google Scholar]
- 19.Yu, M. L., W. L. Chuang, C. Y. Dai, S. C. Chen, Z. Y. Lin, M. Y. Hsieh, L. Y. Wang, and W. Y. Chang. 2000. Clinical evaluation of the automated COBAS AMPLICOR HCV MONITOR test version 2.0 for quantifying serum hepatitis C virus RNA and comparison to the QUANTIPLEX HCV version 2.0 test. J. Clin. Microbiol. 38:2933-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]