Abstract
Candida species are important nosocomial pathogens associated with high mortality rates. Rapid detection and identification of Candida species can guide a clinician at an early stage to prescribe antifungal drugs or to adjust empirical therapy when resistant species are isolated. Confocal Raman microspectroscopy is highly suitable for the rapid identification of Candida species, since Raman spectra can be directly obtained from microcolonies on a solid culture medium after only 6 h of culturing. In this study, we have used a set of 42 Candida strains comprising five species that are frequently encountered in clinical microbiology to test the feasibility of the technique for the rapid identification of Candida species. The procedure was started either from a culture on Sabouraud medium or from a positive vial of an automated blood culture system. Prior to Raman measurements, strains were subcultured on Sabouraud medium for 6 h to form microcolonies. Using multivariate statistical analyses, a high prediction accuracy (97 to 100%) was obtained with the Raman method. Identification with Raman microspectroscopy may therefore be significantly faster than identification with commercial identification systems that allow various species to be identified and that often require 24 to 48 h before a reliable identification is obtained. We conclude that confocal Raman microspectroscopy offers a rapid, accurate, and easy-to-use alternative for the identification of clinically relevant Candida species.
Yeasts of the genus Candida are increasingly encountered as the cause of nosocomial infections. These opportunistic pathogens are often isolated from critically ill patients on intensive care units (ICUs), e.g., patients receiving broad-spectrum antimicrobial therapy or patients with intravascular devices (28, 42). Candida species are the fourth most commonly encountered nosocomial pathogens in bloodstream infections in the United States, and candidiasis is associated with high mortality rates (5, 14, 17, 31, 43). Of the Candida species encountered in clinical practice, Candida albicans is the most prevalent. C. albicans is often susceptible to the azole group of antifungal agents. However, there is a shift toward the more azole-tolerant species, such as C. glabrata, C. tropicalis, and C. krusei, possibly related to the increasing use of itraconazole and fluconazole, the antifungal drugs of first choice in candidiasis (3, 30, 31, 40, 45). Rapid identification of these species is therefore relevant for the clinician in determining the correct antifungal agent.
Conventional identification of Candida species is based on an extensive series of tests, e.g., carbohydrate fermentation and assimilation, growth at 37 and 42°C, colony and cell morphology, and the ability to form germ tubes (27, 45). Available commercial yeast identification systems are derived from this conventional approach, e.g., the Vitek2 system (bioMerieux, Lyon, France), API 20C (bioMerieux, Basingstoke, United Kingdom), the RapID Yeast Plus system (Innovative Diagnostic Systems, Norcross, Ga.), and the Minitek system (Becton Dickinson Microbiology Systems, Cockeysville, Md.). The performance of commercial identification systems has been extensively evaluated for most clinically relevant Candida species. Once enough biomass was obtained from the initial culture (16 to 24 h for most commonly encountered species), results were obtained after 4 h to several days of incubation, depending on the system. Identification accuracy was reported to be between 59 and 99% and seemed to improve with an increase in the number of tests included in the system (10, 11, 19, 27, 33, 41, 44-46). Most of the rapid (same-day) identification systems are designed to discriminate between two species or to confirm a presumptive identification. Rapid systems enabling identification of various species are at best limited to the more common species seen in the clinical laboratory (45). Therefore, the need for rapid multispecies tests still exists.
Rapid identification of microorganisms in general has been shown to have a major impact on the morbidity, mortality, and duration of hospitalization (2, 7, 16). Doern and coworkers showed that when an empirically started antimicrobial therapy had to be changed based on laboratory results, this change could be made ca. 15 h earlier when rapid techniques were applied (7). For Candida species involved in bloodstream infections on ICUs, it was shown by Ibrahim et al. that initial therapy was inadequate in 95% of the cases because no antifungal agent was included (16). Due to the inadequacy of the initial therapy, a mortality rate of about 60% was observed in the patient group with Candida infections. Hence, early recognition of a Candida infection would help a clinician to select proper treatment. Combined with rapid identification of the causative organism, this treatment could be optimized, if required, at an early stage of the infection.
Vibrational spectroscopic techniques are highly suitable as a basis for the development of rapid identification methods. Fourier transform infrared (FT-IR) spectroscopy and Raman spectroscopy provide information about the molecular composition of a sample. The overall molecular compositions of microbial species and strains are sufficiently different to lead to reproducible differences in FT-IR and Raman spectra, to the extent that the spectra can be used as highly specific spectroscopic fingerprints to enable the identification of microorganisms (6, 8, 9, 12, 13, 15, 22, 25, 26). Recently, a new and rapid method “for recording” Raman spectra of microbial microcolonies directly on solid culture media was reported (23). Reproducible Raman spectra can be obtained from microcolonies 10 to 100 μm in diameter, such as will develop for most commonly encountered microorganisms after about 6 h of culturing (4, 23). A good impression of the potential identification accuracy of Raman spectroscopy-based methods was obtained from a comparison of vibrational spectroscopic methods with genotypic identification methods. Raman spectra were obtained from dried smears on glass slides of overnight cultures of Enterococcus species. A cluster analysis carried out on the Raman database thus established showed that clustering of strains occurred in accordance with genotypic species identification, whereas routine phenotypic methods failed in a number of cases (20).
Here we present the results from a study aimed at the development of a rapid and accurate identification method for clinically relevant Candida species. An identification algorithm is described and tested which carries out Candida species identification based on Raman spectra obtained from 6-h microcolonies on a solid culture medium, with or without prior passage through a blood culture system.
MATERIALS AND METHODS
Yeast strains and identification.
A collection of 42 Candida strains was used (Table 1). Strains either were obtained from culture collections (American Type Culture Collection, Manassas, Va.: C. albicans ATCC 90028, C. glabrata ATCC 66032, C. kefyr ATCC 66028, and C. tropicalis ATCC 750; Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands: C. krusei CBS 573) or were clinical isolates identified to the species level by the conventional identification methods mentioned above.
TABLE 1.
Composition of the strain collection used in this study
| Species | Straina | Member of the following data setb:
|
Originc | Identification methodd | Reference or source | |
|---|---|---|---|---|---|---|
| SAB | Blood | |||||
| C. albicans | 2 | x | Blood | CA | 41 | |
| 3 | x | x | Blood | CA | 41 | |
| 11 | x | Blood | CA | 41 | ||
| 12 | x | x | Blood | CA | 41 | |
| 25 | x | Blood | CA | 41 | ||
| 42 | x | x | Tissue | CA | 41 | |
| 6319 | x | x | Oral cavity | GT, CS | 39 | |
| ATCC 28367 | x | x | ||||
| ATCC 38696 | x | x | ||||
| ATCC 90028 | x | x | ||||
| C. glabrata | 32 | x | Blood | CA | 41 | |
| 37 | x | x | Blood | CA | 41 | |
| 46 | x | x | Blood | CA | 41 | |
| 326 I/95 | x | Stomach biopsy | API ID 32C | D. Naumanne | ||
| 33371 I/94 | x | Oral swab | API ID 32C | D. Naumann | ||
| ATCC 66032 | x | |||||
| ATCC 90030 | x | |||||
| C. kefyr | 13 | x | x | Blood | CA | 41 |
| 29 | x | x | Tissue | CA | 41 | |
| 52 | x | x | Feces | CA | 41 | |
| 53 | x | x | Oral rinse | CA | 41 | |
| 146 I/96 | x | BAL | API ID 32C | D. Naumann | ||
| 430 II/96 | x | BAL | API ID 32C | D. Naumann | ||
| ATCC 66028 | x | x | ||||
| C. krusei | 4 | x | Tissue | CA | 41 | |
| 7 | x | x | Blood | CA | 41 | |
| 8 | x | x | Sputum | CA | 41 | |
| 10 | x | x | Tissue | CA | 41 | |
| 14 | x | BAL | CA | 41 | ||
| 15 | x | x | Tissue | CA | 41 | |
| 28 | x | Oral rinse | CA | 41 | ||
| 40 | x | x | Tissue | CA | 41 | |
| 47 | x | x | Tissue | CA | 41 | |
| CBS 573 | x | x | ||||
| C. tropicalis | 19 | x | x | Blood | CA | 41 |
| 45 | x | x | Ascites fluid | CA | 41 | |
| 48 | x | x | Tissue | CA | 41 | |
| 326 II/95 | x | Stomach biopsy | API ID 32C | D. Naumann | ||
| ATCC 750 | x | |||||
| M38 I/96 | x | Oral rinse | API ID 32C | D. Naumann | ||
| M56 I/93 | x | Sputum | API ID 32C | D. Naumann | ||
| M675 I/93 | x | Blood | API ID 32C | D. Naumann | ||
| Total (n) | 42 | 32 | 34 | |||
ATCC, American Type Culture Collection; CBS, Centraal bureau voor Schimmelcultures.
Strains in the SAB data set were only cultured on Sabouraud medium; strains in the blood data set were obtained from spiked blood cultures (see Materials and Methods for details). x, a Raman spectrum of the strain is included.
BAL, bronchoalveolar lavage.
CA, assimilation of carbohydrates; GT, germ tube formation in human serum; CS, chlamydospore formation on cornmeal agar. The API ID 32C was from bioMerieux, Lyon, France.
The strain was kindly provided by the laboratory of D. Naumann, Robert Koch Institute, Berlin, Germany.
Sample preparation.
Samples were stored at −80°C in brain heart infusion broth (Becton Dickinson, Franklin Lakes, N.J.) containing 10% glycerol until use. Thirty-two strains were subcultured on Sabouraud-2% glucose (SAB) medium for 6 h at 30°C prior to Raman measurements of microcolonies following overnight passage (30°C) on SAB medium (Merck, Darmstadt, Germany). The data set of spectra obtained from microcolonies prepared in this way is referred to as the SAB data set.
For 34 strains (Table 1), microcolonies were prepared after passage through a blood culture system in order to determine if this process would affect the identification ability of the Raman method. The strains were seeded at 103 CFU/ml in 10 ml of blood from healthy volunteers. The seeded blood samples were used to inoculate mycosis culture vials of the automated BACTEC blood culture system (Becton Dickinson). When the culture vials were flagged as positive by the system (within 24 h for most strains), several drops of the liquid culture medium were plated on SAB medium and cultured for 6 h at 30°C prior to Raman measurements of microcolonies. The data obtained from samples precultured in the BACTEC system are referred to as the blood data set.
Confocal Raman microspectroscopy.
Raman spectroscopic measurements were performed as described previously (23). Briefly, the solid culture medium containing the microcolonies was placed directly under the microscope of a system 1000 Raman microspectrometer (Renishaw plc, Wotton-under-Edge, Gloucestershire, United Kingdom). The microscope was fitted with a 80× near-infrared objective (MIR Plan 80×/0.75; Olympus). Samples were excited by using 100 to 150 mW of 830-nm laser light from a titanium-sapphire laser (model 3900; Spectra Physics, Mountain View, Calif.) pumped by an argon ion laser (series 2000; Spectra Physics). The constant background signal contribution originating from optical elements in the laser light delivery pathway was subtracted from all spectra. The reference spectrum of a tungsten band lamp of known temperature was used to correct for the wavelength-dependent signal detection efficiency of the Raman setup (29, 47). Calibration of the wave number axis was performed by using the known wavelengths of the atomic lines from neon and argon.
For each yeast sample, five microcolonies were selected. Within each microcolony, spectra were obtained from 10 randomly chosen locations by using a signal collection time of 30 s per measurement. For each sample measured, the 50 spectra thus obtained were averaged. The yeast Raman spectra used for this study were obtained over a 3-month period.
Sixty Raman spectra for SAB medium were obtained at random locations in the medium over 30 min of signal collection time. Sixty water spectra were also obtained over 30 min of total signal collection time.
Spectrum treatment.
All spectrum analyses were performed on first derivatives of the measured spectra. This was done in order to minimize the influence of the broad, relatively featureless signal background usually ascribed to fluorescence, on which the Raman spectra are superimposed and which may vary from sample to sample (23).
In an earlier study (23), a method was described for orthogonalizing microbial signal contributions to the background signal contribution of the solid culture medium. This procedure is necessary because the actual signal contribution of the culture medium critically depends on the exact position of the laser focus in the colony and therefore unavoidably varies from one measurement to the next. After the orthogonalizing procedure, Raman spectra obtained from a particular microcolony look the same, irrespective of the intensity of the culture medium signal contribution initially present. Collection of a database of spectra over an extended period of time necessitates the use of culture plates from different batches, which will show slight variations in composition and water concentration. Moreover, inhomogeneities within one culture plate can be encountered at the microscopic scale at which the Raman experiments take place. This means that in order for all spectra of yeast microcolonies to be compared, they must be orthogonalized with respect to all culture medium spectra. In the earlier study, this goal was accomplished by sequentially orthogonalizing a spectrum to all medium spectra (23). Here we have applied a more efficient method. The spectra of all the culture plates and of water were subjected to principal-component (PC) analysis (PCA). The first PCs, accounting for 99% of all signal variance within this data set of spectra, were used to construct a PC subspace. Microcolony spectra were projected onto this PC subspace, and only the spectrum component orthogonal to this PC subspace was retained for further analysis. After this procedure, microcolony spectra are obtained that are both independent of the amount of medium signal contribution originally present and independent of batch-to-batch variations in medium composition (unless these affect the biochemical composition of the cells, e.g., due to effects on growth rate [23]).
All procedures used for spectrum treatment and data analysis were developed by using the Matlab 5.3 software package (The Mathworks Inc., Natick, Mass.) and the multivariate statistical analysis toolbox PLS-toolbox 2.0.0c (Eigenvector Research Inc., Manson, Wash.) unless otherwise stated.
Data analysis. (i) PCA.
Before multivariate statistical analyses, a data reduction was performed by using PCA; this is a well-known method for reducing the dimensionality in a data set (18, 35). The maximum number of n − 1 PCs was calculated (n being the number of spectra in the analysis), typically accounting for 99 to 100% of the variation in the data set.
(ii) HCA.
Hierarchical cluster analysis (HCA) was performed on the n − 1 PC scores obtained for each spectrum by using Ward's clustering algorithm and the squared Euclidean distance measure to generate a dendrogram. For HCA, the SPSS (Chicago, Ill.) statistical software package was used.
(iii) LDA.
For linear discriminant analysis (LDA), only PC scores accounting for more than 1% of the variance in the data set were retained. A two-sided t test was used to individually select PC scores that showed the highest significance in discriminating the different microbial groups presented. The number of PC scores that was used as an input for an LDA model was kept at least two times smaller than the number of spectra in the smallest model group to prevent overfitting in the LDA model (1).
RESULTS AND DISCUSSION
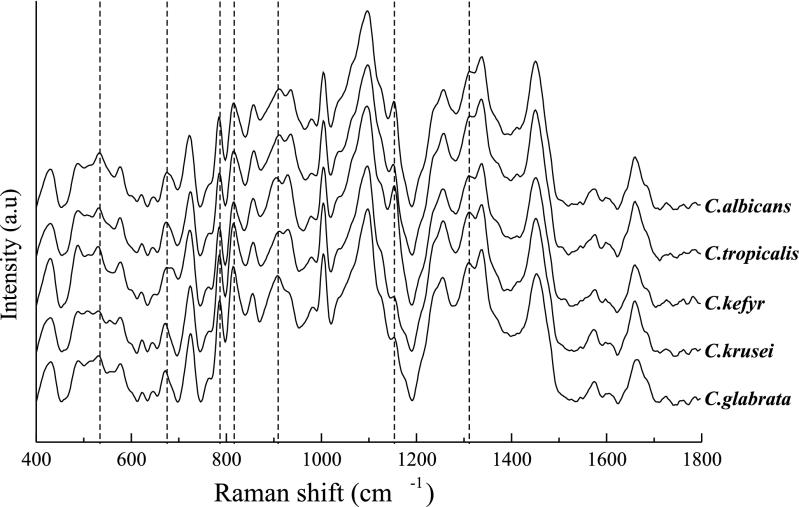
Our aim in this study was to develop a rapid identification scheme for clinically relevant Candida species based on confocal Raman microspectroscopy. From earlier studies, we learned that reproducible Raman spectra can be obtained from microbial microcolonies still growing on a solid culture medium (4, 23). Figure 1 shows typical Raman spectra from the five different Candida species included in this study. The highlighted spectral features show characteristic differences between the different species. The differences, from species to species, in the relative heights of these bands are believed to be due to differences in the biochemical compositions of the cell walls. A precise band assignment is the subject of further investigation.
FIG. 1.
Representative Raman spectra of five Candida species used in this study. Spectra are averages of those obtained from the SAB data set. Broken lines indicate spectral features of which the relative intensities differ between the spectra of different species. a.u, arbitrary units.
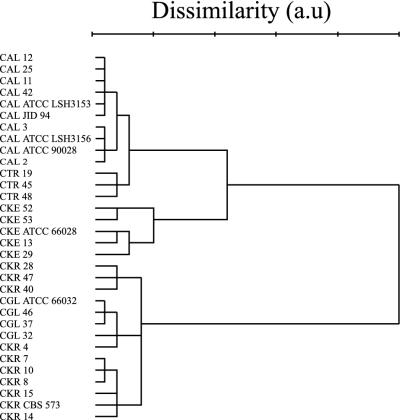
HCA is a nonsupervised method for obtaining information about the dissimilarity between spectra of different species. Figure 2 shows the dendrogram resulting from HCA performed on the Raman spectra in the SAB data set (i.e., spectra obtained from strains cultured on Sabouraud medium only; see Materials and Methods). Separate clusters were formed for the species, with the exception of C. krusei, the spectra of which were distributed between two clusters. One strain, C. krusei 4, was grouped in the C. glabrata cluster by using this objective approach. The spectral differences observed between the averaged spectra of the two C. krusei clusters were only very minor and could not be attributed to specific molecular fractions (Fig. 3). As explained in Materials and Methods, HCA uses squared Euclidean distances between spectra as input parameters. The results that we obtained show that this measure of overall signal variance encountered within a set of spectra obtained from different strains belonging to the same species can be as large as the interspecies signal variance. In order to facilitate species identification, it is necessary therefore to apply supervised analysis methods, which look for the signal variance that is relevant to species discrimination. Here we used the results of the HCA method as a first step in developing a sequential species identification scheme based on LDA.
FIG. 2.
Dendrogram resulting from HCA of Raman spectra of Candida strains in the SAB data set (see Materials and Methods). The squared Euclidean distance measure and Ward's clustering algorithm were used in the analysis. CAL, C. albicans; CGL, C. glabrata; CKE, C. kefyr; CKR, C. krusei; CTR, C. tropicalis; a.u, arbitrary units.
FIG. 3.
Raman spectra arising from the averages of C. krusei cluster 1 (strains 28, 40, and 47) and C. krusei cluster 2 (strains 7, 8, 10 14, 15, and CBS 573) as shown in Fig. 1. The difference spectrum at the bottom shows the spectral differences between cluster 1 and cluster 2. a.u, arbitrary units.
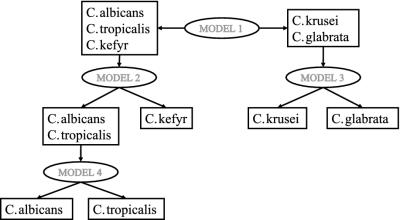
LDA was applied to the PC scores that were most informative for the separation of the different species involved. In order to reduce the complexity of the LDA model used, a method was chosen in which the Candida species were separated at different levels. The use of different levels in a sequential approach to separate microorganisms based on FT-IR was described earlier by Udelhoven et al. (38). For preparing the models in this study, the similarity between the species, as observed in the HCA, was used to distinguish the different levels (Fig. 4). Model 1 separates C. albicans, C. tropicalis, and C. kefyr from C. krusei and C. glabrata. This distinction is based on the largest dissimilarity observed. Model 2 discriminates between C. albicans or C. tropicalis and C. kefyr. Model 3 is designed to discriminate between C. krusei and C. glabrata. Finally, model 4 further separates C. albicans and C. tropicalis. Based on the outcome of model 1, an unknown spectrum is projected on either model 2 or model 3. When model 2 predicts the Raman spectrum as belonging to either C. albicans or C. tropicalis, the spectrum is finally projected on model 4 to distinguish between these species.
FIG. 4.
Schematic representation of the sequential identification procedure based on LDA models 1 to 4. Spectra to be identified are predicted by using model 1; depending on the result, the next projection will be on model 2 or 3, and so forth. Models are produced based on the major groupings found in the dendrogram of Fig. 2.
The prediction accuracy of the identification model was determined by using a “leave-one-out” evaluation (34). The spectra of all but one strain were used to generate LDA models 1 to 4. For strains that were included in both the SAB data set and the blood data set, both spectra were left out. The spectrum or spectra that were left out were used to test the accuracy of the identification model. By repeating this procedure and leaving the spectrum or spectra of each strain out in turn, information was obtained on the reproducibility of the identification procedure, i.e., if there was enough discriminating information in the Raman spectra to identify unknown spectra correctly. When the “leave-one-strain-out” evaluation was performed on the SAB data set, all 32 strains (100%) were correctly identified (Table 2). This result indicates that although HCA showed two separate C. krusei clusters and one misclassification, a supervised method was able to identify characteristic spectral differences between C. krusei and C. glabrata which otherwise remain hidden in non-species-specific signal variance. Furthermore, performing the leave-one-strain-out evaluation with the SAB data set and the blood data set combined again yielded a high prediction accuracy of 97.0% (Table 3). Two strains from the blood data set were misidentified: C. tropicalis 40 was predicted to be C. albicans, and C. krusei M38 I/96 was identified as C. glabrata. Taking the results of the combined data set into account, there were no significant differences in identification accuracy between the SAB data set and the blood data set. This result indicates that pretreatment or culturing of the Candida strains prior to Raman measurements did not significantly influence the accuracy of the LDA model used. Therefore, considering the speed and ease at which various species can be identified, confocal Raman microspectroscopy has the potential to develop into a more powerful and faster technique than the rapid identification systems available today.
TABLE 2.
Results of the leave-one-strain-out evaluation with the SAB data set
| Species | No. of strains with the following Raman identification:
|
|||||
|---|---|---|---|---|---|---|
| C. albicans | C. tropicalis | C. kefyr | C. krusei | C. glabrata | Total | |
| C. albicans | 10 | 10 | ||||
| C. tropicalis | 3 | 3 | ||||
| C. kefyr | 5 | 5 | ||||
| C. krusei | 10 | 10 | ||||
| C. glabrata | 4 | 4 | ||||
| Total | 10 | 3 | 5 | 10 | 4 | 32 |
TABLE 3.
Results of the leave-one-strain-out evaluation with the SAB data set and the blood data set combined
| Species | No. of strains with the following Raman identification:
|
|||||
|---|---|---|---|---|---|---|
| C. albicans | C. tropicalis | C. kefyr | C. krusei | C. glabrata | Total | |
| C. albicans | 17 | 17 | ||||
| C. tropicalis | 1 | 10 | 11 | |||
| C. kefyr | 12 | 12 | ||||
| C. krusei | 16 | 1 | 17 | |||
| C. glabrata | 9 | 9 | ||||
| Total | 18 | 10 | 12 | 16 | 10 | 66 |
Vibrational spectroscopic techniques have been used by several authors to study Candida species (21, 22, 32, 35-38). To our knowledge, however, this is the first time that confocal Raman microspectroscopy has been used for identification purposes. Performing measurements directly with microcolonies on solid culture medium has several advantages in a clinical diagnostic setting. The most obvious advantage is the short time required for obtaining microcolonies with which measurements can be performed. Furthermore, besides the minimal sample processing required, there is no need to use labels or dyes, and there is only a limited need for disposable supplies.
A known problem with Candida infections on ICUs is that therapy is started relatively late, because of the lack of early clinical manifestations and the delay in laboratory detection procedures. Consequently, Candida infections are associated with high mortality rates (24). The results presented here show that high prediction accuracy could be achieved within 6 h after a sample was cultured on SAB medium or when blood cultures became positive. In our routine microbiological laboratory, positive BACTEC cultures due to a Candida infection are analyzed by using the Vitek2 system. After the BACTEC culture vials are flagged as positive by the system, an additional 24 to 48 h is required for identification. Rapid recognition of a species with a possible tolerance toward azole agents (C. glabrata, C. tropicalis, and C. krusei) presents the clinician with an option to monitor very closely the effect of treatment with such an antifungal agent or to choose a different agent at an early stage of the infection. Our current aim is to use Raman identification in a prospective study of blood cultures in our tertiary care hospital. We conclude that confocal Raman microspectroscopy may offer a rapid, accurate, and easy-to-use alternative for the identification of clinically relevant Candida species.
Acknowledgments
We gratefully acknowledge the excellent technical assistance of Tamara van Vreeswijk. Nicole van den Braak, Department of Medical Microbiology and Infectious Diseases, and Carolin Kirschner, Robert Koch Institute, Berlin, Germany, are acknowledged for help in obtaining the Candida strains used.
This research was funded by the European Union Biomed II program (project no. BMH4-97-2054).
REFERENCES
- 1.Bakker Schut, T. C., M. J. Witjes, H. J. Sterenborg, O. C. Speelman, J. L. Roodenburg, E. T. Marple, H. A. Bruining, and G. J. Puppels. 2000. In vivo detection of dysplastic tissue by Raman spectroscopy. Anal. Chem. 72:6010-6018. [DOI] [PubMed] [Google Scholar]
- 2.Barenfanger, J., C. Drake, and G. Kacich. 1999. Clinical and financial benefits of rapid bacterial identification and antimicrobial susceptibility testing. J. Clin. Microbiol. 37:1415-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berrouane, Y. F., L. A. Herwaldt, and M. A. Pfaller. 1999. Trends in antifungal use and epidemiology of nosocomial yeast infections in a university hospital. J. Clin. Microbiol. 37:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choo-Smith, L.-P., K. Maquelin, T. van Vreeswijk, H. A. Bruining, G. J. Puppels, N. A. Thi, C. Kirschner, D. Naumann, D. Ami, A. M. Villa, F. Orsini, S. M. Doglia, H. Lamfarraj, G. D. Sockalingum, M. Manfait, P. Allouch, and H. P. Endtz. 2001. Investigating microbial (micro)colony heterogeneity by vibrational spectroscopy. Appl. Environ. Microbiol. 67:1461-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornwell, E. E., III, H. Belzberg, T. V. Offne, W. R. Dougherty, I. R. Morales, J. Asensio, and D. Demetriades. 1995. The pattern of fungal infections in critically ill surgical patients. Am. Surg. 61:847-850. [PubMed] [Google Scholar]
- 6.Curk, M. C., F. Peladan, and J. C. Hubert. 1994. Fourier transform infrared (FTIR) spectroscopy for identifying Lactobacillus species. FEMS Microbiol. Lett. 123:241-248. [Google Scholar]
- 7.Doern, G. V., R. Vautour, M. Gaudet, and B. Levy. 1994. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J. Clin. Microbiol. 32:1757-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fehrmann, A., M. Franz, A. Hoffmann, L. Rudzik, and E. Wust. 1995. Identification of micro-organisms using mid infrared spectroscopy and quantitative Raman spectroscopy in dairies. J. Mol. Struct. 348:13-16. [Google Scholar]
- 9.Goodacre, R., E. M. Timmins, P. J. Rooney, J. J. Rowland, and D. B. Kell. 1996. Rapid identification of Streptococcus and Enterococcus species using diffuse reflectance-absorbance Fourier transform infrared spectroscopy and artificial neural networks. FEMS Microbiol. Lett. 140:233-239. [DOI] [PubMed] [Google Scholar]
- 10.Graf, B., T. Adam, E. Zill, and U. B. Gobel. 2000. Evaluation of the VITEK 2 system for rapid identification of yeasts and yeast-like organisms. J. Clin. Microbiol. 38:1782-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heelan, J. S., E. Sotomayor, K. Coon, and J. B. D'Arezzo. 1998. Comparison of the rapid yeast plus panel with the API20C yeast system for identification of clinically significant isolates of Candida species. J. Clin. Microbiol. 36:1443-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helm, D., H. Labischinski, and D. Naumann. 1991. Elaboration of a procedure for identification of bacteria using Fourier-transform IR spectral libraries: a stepwise correlation approach. J. Microbiol. Methods 14:127-142. [Google Scholar]
- 13.Helm, D., H. Labischinski, G. Schallehn, and D. Naumann. 1991. Classification and identification of bacteria by Fourier-transform infrared spectroscopy. J. Gen. Microbiol. 137:69-79. [DOI] [PubMed] [Google Scholar]
- 14.Hoerauf, A., S. Hammer, B. Muller-Myhsok, and H. Rupprecht. 1998. Intra-abdominal Candida infection during acute necrotizing pancreatitis has a high prevalence and is associated with increased mortality. Crit. Care Med. 26:2010-2015. [DOI] [PubMed] [Google Scholar]
- 15.Horbach, I., D. Naumann, and F. J. Fehrenbach. 1988. Simultaneous infections with different serogroups of Legionella pneumophila investigated by routine methods and Fourier transform infrared spectroscopy. J. Clin. Microbiol. 26:1106-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibrahim, E. H., G. Sherman, S. Ward, V. J. Fraser, and M. H. Kollef. 2000. The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest 118:146-155. [DOI] [PubMed] [Google Scholar]
- 17.Jarvis, W. R. 1995. Epidemiology of nosocomial fungal infections, with emphasis on Candida species. Clin. Infect. Dis. 20:1526-1530. [DOI] [PubMed] [Google Scholar]
- 18.Jolliffe, I. T. 1986. Principal component analysis. Springer-Verlag, New York, N.Y.
- 19.Kellogg, J. A., D. A. Bankert, and V. Chaturvedi. 1998. Limitations of the current microbial identification system for identification of clinical yeast isolates. J. Clin. Microbiol. 36:1197-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirschner, C., K. Maquelin, P. Pina, N. A. Ngo Thi, L. P. Choo-Smith, G. D. Sockalingum, C. Sandt, D. Ami, F. Orsini, S. M. Doglia, P. Allouch, M. Mainfait, G. J. Puppels, and D. Naumann. 2001. Classification and identification of enterococci: a comparative phenotypic, genotypic, and vibrational spectroscopic study. J. Clin. Microbiol. 39:1763-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kummerle, M., S. Scherer, and H. Seiler. 1998. Rapid and reliable identification of food-borne yeasts by Fourier-transform infrared spectroscopy. Appl. Environ. Microbiol. 64:2207-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Löchte, T. 1997. Differenzierung und Identifizierung von Mikroorganismen mittels der NIR-FT-Raman-Spektroskopie. Ph.D. thesis. Universität Essen, Essen, Germany.
- 23.Maquelin, K., L. P. Choo-Smith, T. van Vreeswijk, H. P. Endtz, B. Smith, R. Bennett, H. A. Bruining, and G. J. Puppels. 2000. Raman spectroscopic method for identification of clinically relevant microorganisms growing on solid culture medium. Anal. Chem. 72:12-19. [DOI] [PubMed] [Google Scholar]
- 24.Muñoz, P., A. Burillo, and E. Bouza. 2000. Criteria used when initiating antifungal therapy against Candida spp. in the intensive care unit. Int. J. Antimicrob. Agents 15:83-90. [DOI] [PubMed] [Google Scholar]
- 25.Naumann, D., D. Helm, and H. Labischinski. 1991. Microbiological characterizations by FT-IR spectroscopy. Nature 351:81-82. [DOI] [PubMed] [Google Scholar]
- 26.Naumann, D., S. Keller, D. Helm, C. Schultz, and B. Schrader. 1995. FT-IR spectroscopy and FT-Raman spectroscopy are powerful analytical tools for the non-invasive characterization of intact microbial cells. J. Mol. Struct. 347:399-406. [Google Scholar]
- 27.Odds, F. C. 1988. Candida and candidosis, 2nd ed. Baillière Tindall, London, England.
- 28.Pfaller, M. A., S. A. Messer, A. Houston, M. S. Rangel-Frausto, T. Wiblin, H. M. Blumberg, J. E. Edwards, W. Jarvis, M. A. Martin, H. C. Neu, L. Saiman, J. E. Patterson, J. C. Dibb, C. M. Roldan, M. G. Rinaldi, and R. P. Wenzel. 1998. National epidemiology of mycoses survey: a multicenter study of strain variation and antifungal susceptibility among isolates of Candida species. Diagn. Microbiol. Infect. Dis. 31:289-296. [DOI] [PubMed] [Google Scholar]
- 29.Puppels, G. J., W. Colier, J. H. F. Olminkhof, C. Otto, F. F. M. de Mul, and J. Greve. 1991. Description and performance of a highly sensitive confocal Raman spectrometer. J. Raman Spectrosc. 22:217-225. [Google Scholar]
- 30.Rocco, T. R., S. E. Reinert, and H. H. Simms. 2000. Effects of fluconazole administration in critically ill patients: analysis of bacterial and fungal resistance. Arch. Surg. 135:160-165. [DOI] [PubMed] [Google Scholar]
- 31.Safran, D. B., and E. Dawson. 1997. The effect of empiric and prophylactic treatment with fluconazole on yeast isolates in a surgical trauma intensive care unit. Arch. Surg. 132:1184-1188. [DOI] [PubMed] [Google Scholar]
- 32.Schrader, B., B. Dippel, S. Fendel, S. Keller, T. Löchte, M. Riedl, R. Schulte, and E. Tatsch. 1997. NIR FT Raman spectroscopy--a new tool in medical diagnosis. J. Mol. Struct. 408/409:23-31. [Google Scholar]
- 33.Sheppard, D. C., P. Rene, A. D. Harris, M. A. Miller, M. Laverdiere, E. deSouza, and H. G. Robson. 1999. Simple strategy for direct identification of medically important yeast species from positive blood culture vials. J. Clin. Microbiol. 37:2040-2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stone, M. 1974. Cross-validatory choice and assessment of statistical predictions (with discussion). J. R. Stat. Soc. B 36:111-147. [Google Scholar]
- 35.Timmins, E. M., S. A. Howell, B. K. Alsberg, W. C. Noble, and R. Goodacre. 1998. Rapid differentiation of closely related Candida species and strains by pyrolysis-mass spectrometry and Fourier transform-infrared spectroscopy. J. Clin. Microbiol. 36:367-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Timmins, E. M., D. E. Quain, and R. Goodacre. 1998. Differentiation of brewing yeast strains by pyrolysis mass spectrometry and Fourier transform infrared spectroscopy. Yeast 14:885-893. [DOI] [PubMed] [Google Scholar]
- 37.Tintelnot, K., G. Haase, M. Seibold, F. Bergmann, M. Staemmler, T. Franz, and D. Naumann. 2000. Evaluation of phenotypic markers for selection and identification of Candida dubliniensis. J. Clin. Microbiol. 38:1599-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Udelhoven, T., D. Naumann, and J. Schmitt. 2000. Development of a hierarchical classification system with artificial neural networks and FT-IR spectra for the identification of bacteria. Appl. Spectrosc. 54:1471-1479. [Google Scholar]
- 39.van Belkum, A., W. Melchers, B. E. de Pauw, S. Scherer, W. Quint, and J. F. Meis. 1994. Genotypic characterization of sequential Candida albicans isolates from fluconazole-treated neutropenic patients. J. Infect. Dis. 169:1062-1070. [DOI] [PubMed] [Google Scholar]
- 40.Vazquez, J. A., L. M. Dembry, V. Sanchez, M. A. Vazquez, J. D. Sobel, C. Dmuchowski, and M. J. Zervos. 1998. Nosocomial Candida glabrata colonization: an epidemiologic study. J. Clin. Microbiol. 36:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verweij, P. E., I. M. Breuker, A. J. Rijs, and J. F. Meis. 1999. Comparative study of seven commercial yeast identification systems. J. Clin. Pathol. 52:271-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vincent, J. L., E. Anaissie, H. Bruining, W. Demajo, M. el-Ebiary, J. Haber, Y. Hiramatsu, G. Nitenberg, P. O. Nystrom, D. Pittet, T. Rogers, P. Sandven, G. Sganga, M. D. Schaller, and J. Solomkin. 1998. Epidemiology, diagnosis and treatment of systemic Candida infection in surgical patients under intensive care. Intensive Care Med. 24:206-216. [DOI] [PubMed] [Google Scholar]
- 43.Voss, A., J. L. le Noble, F. M. Verduyn Lunel, N. A. Foudraine, and J. F. Meis. 1997. Candidemia in intensive care unit patients: risk factors for mortality. Infection 25:8-11. [DOI] [PubMed] [Google Scholar]
- 44.Wadlin, J. K., G. Hanko, R. Stewart, J. Pape, and I. Nachamkin. 1999. Comparison of three commercial systems for identification of yeasts commonly isolated in the clinical microbiology laboratory. J. Clin. Microbiol. 37:1967-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warren, N. G., and K. C. Hazen. 1999. Candida, Cryptococcus, and other yeasts of medical importance, p. 1184-1199. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 46.Williams, D. W., and M. A. Lewis. 2000. Isolation and identification of candida from the oral cavity. Oral Dis. 6:3-11. [DOI] [PubMed] [Google Scholar]
- 47.Wolthuis, R., T. C. Bakker Schut, P. J. Caspers, H. P. J. Buschman, T. J. Römer, H. A. Bruining, and G. J. Puppels. 1999. Raman spectroscopic methods for in vitro and in vivo tissue characterisation, p. 431-455. In W. T. Mason (ed.), Fluorescent and luminescent probes for biological activity, 2nd ed. Academic Press Ltd., London, England.